Abstract
It is widely accepted that different forms of stress activate a common target, p53, yet different outcomes are triggered in a stress-specific manner. For example, activation of p53 by genotoxic agents, such as camptothecin (CPT), triggers apoptosis, while non-genotoxic activation of p53 by Nutlin-3 (Nut3) leads to cell-cycle arrest without significant apoptosis. Such stimulus-specific responses are attributed to differential transcriptional activation of various promoters by p53. In this study, we demonstrate that CPT, but not Nut3, induces miR-203, which downregulates anti-apoptotic bcl-w and promotes cell death in a p53-dependent manner. We find that acetylation of K120 in the DNA-binding domain of p53 augments its association with the Drosha microprocessor and promotes nuclear primary miRNA processing. Knockdown of human orthologue of Males absent On the First (hMOF), the acetyltransferase that targets K120 in p53, abolishes induction of miR-203 and cell death mediated by CPT. Thus, this study reveals that p53 acetylation at K120 plays a critical role in the regulation of the Drosha microprocessor and that post-transcriptional regulation of gene expression by p53 via miRNAs plays a role in determining stress-specific cellular outcomes.
Keywords: apoptosis, Drosha, miRNA, p53
Introduction
Cellular stress responses occur due to sudden environmental changes. Such changes can damage macromolecules, including DNA, mRNA, proteins, and lipids, which need to be replenished (Kultz, 2005). Depending on the stress encountered, cells either re-establish cellular homeostasis or adopt an altered state in the new environment. Stress responses are mediated by multiple mechanisms, including induction of cell death, growth arrest, and activation of specific gene expression programmes through various molecular mechanisms (Kultz, 2005). The tumour suppressor p53 (also known as TP53 in humans and Trp53 in mice) is a confirmed sensor of different types of cellular stress, such as DNA damage, oncogenic activities, erosion of telomeres, and hypoxia. It can modulate gene expression to coordinate various cellular responses to control cell survival, DNA repair, senescence, cell-cycle regulation, or elimination of damaged cells by promoting apoptotic cell death or autophagy. In the unstressed steady state, cellular expression of p53 is kept low by negative regulators, such as human double minute 2 (HDM2 in humans and MDM2 in mice) and HDMX2 (MDMX in mice) (Toledo and Wahl, 2006). A small molecule that inhibits the interaction between HDM2 and p53, Nutlin-3 (Nut3), is able to de-repress and activate p53 similarly to genotoxic stimuli, such as topoisomerase inhibitors (doxorubicin (DXR), camptothecin (CPT), and etoposide (ETP)) and the transcription inhibitor actinomycin D (ActD). Upon DNA damage, p53 is stabilized and activated. Mutations of p53 directly or indirectly modulate its expression and/or activity and are found in nearly all human cancers, illustrating the central role of p53 as a gatekeeper of the human genome. Mechanistically, p53 acts as a transcription factor, binding DNA in a sequence-specific manner. It is composed of several domains critical for its transcription activity, such as an amino (N)-terminal transactivation domain (TAD), a DNA-binding domain (DBD), and a carboxyl (C)-terminal regulatory domain (CRD). Cellular responses by p53 are mediated by transcriptional regulation of protein-coding genes as well as small non-coding microRNA (miRNA) genes.
miRNAs are ∼22-nt regulatory RNAs that negatively modulate the stability and/or translational potential of target mRNAs (Siomi and Siomi, 2010). As miRNA-dependent gene regulation requires neither transcription nor protein synthesis, miRNAs are well positioned to play a significant gene regulatory role in the rapid restoration of homeostasis upon stress responses that have compromised the transcription or translation machineries (Mendell and Olson, 2012). Following DNA damage, p53 induces the primary transcripts of specific miRNAs (pri-miRNAs), such as miR-34a and miR-107. In turn, these miRNAs collectively repress a number of genes, and in this way contribute to promote growth arrest and apoptosis (Hermeking, 2007). In addition, p53 can control the processing of a limited population of pri-miRNAs by associating with RNA helicase p68 (also known as DDX5), a component of the Drosha microprocessor complex. Drosha and its cofactors are essential for the processing of pri-miRNAs to generate precursor miRNAs (pre-miRNAs) in the nucleus during miRNA biogenesis (Suzuki et al, 2009). Consequently, we can speculate that deregulation of p53 expression or function due to a mutation of p53 or p53 regulatory proteins can lead to altered expression of miRNAs in addition to changes in transcriptional targets of p53. However, the contribution of p53-modulated miRNAs during DNA damage response is unclear.
Given the essential role of p53 in a wide variety of cellular processes, it is understandable that p53 is constrained by a complex regulatory network that includes positive and negative effector proteins. Many conserved amino-acid residues in p53 undergo different types of post-translational modifications, including phosphorylation, ubiquitination, acetylation, methylation, sumoylation, and neddylation (Dai and Gu, 2010). The impact of these post-translational modifications on the miRNA processing function of p53 has not been addressed previously. Accumulating evidence suggests that the acetylation of lysine (K) residues significantly affects p53 activity during stress responses by modulating the stability and the transcriptional activity of p53 (Dai and Gu, 2010). Currently, 10 acetylated lysine residues (K120, K164, K305, K320, K370, K372, K373, K381, K382, and K386) have been identified in p53 (Dai and Gu, 2010). The acetylation of six of these lysines (K370, K372, K373, K381, K382, and K386), all clustered in the CRD, is mediated by the acetyltransferase p300/CREB-binding protein (p300/CBP) (Dai and Gu, 2010). These six acetylation sites are not only critical to the specificity of the DNA binding and transcriptional activation ability of p53, but also contribute to the stabilization and subsequent elevation of total p53 levels, as acetylation of these lysine residues inhibits ubiquitination by HDM2/HDMX and subsequent degradation of p53 (Dai and Gu, 2010). However, mutations in lysines in the CRD have not been found in human cancers (according to the UMD_TP53 Mutation database at http://p53.free.fr/). Recently, two additional acetylation sites (K120 and K164) were identified in the DBD (Sykes et al, 2006; Tang et al, 2006). Although rare, K120R and K164R mutations can be found in tumours and cancer cell lines (from the UMD_TP53 Mutation database). Acetylation of K120 is mediated by the human orthologue of Males absent On the First (hMOF) and Tat-Interactive Protein of 60 kDa (Tip60), and K164 is acetylated by CBP/p300 similarly to the C-terminal acetylation sites (Dai and Gu, 2010). An acetylation-defective mutation at K120 (K120R) abrogates p53-mediated apoptosis, but not growth arrest or senescence, suggesting that K120 acetylation can determine the type of cellular outcome mediated by p53 (Sykes et al, 2006; Tang et al, 2006; Li et al, 2012). However, the molecular mechanism by which acetyl-K120 p53 triggers a specific outcome is not fully understood.
In this study, we demonstrate that DNA damaging agents activate both the transcriptional activity and the miRNA processing activity of p53, ultimately resulting in cell death. On the other hand, Nut3 inactivates HDM2, an E3 ubiquitin ligase, which results in p53 degradation and induces p53-dependent transcription, but shows little effect on the miRNA processing activity of p53. Likewise, Nut3 does not cause cell death, suggesting that miRNA processing by p53 is required for the induction of apoptosis. Acetylation of K120, induced by DNA damaging agents, plays a critical role in the stimulation of miRNA processing by p53 through an enhanced association with p68 and Drosha and cropping of pri-miRNA. Our study suggests that CPT-mediated cell death is due to the induction of miR-203, which downregulates pro-survival Bcl-w (BCL2L2) and mediates the apoptotic response. Thus, the acetylation of the K120 in p53 is critical for the post-transcriptional regulation of miRNAs and for determining the specificity of the p53-dependent cellular response.
Results
The DNA damaging agent CPT mediates apoptosis via downregulation of Bcl-w
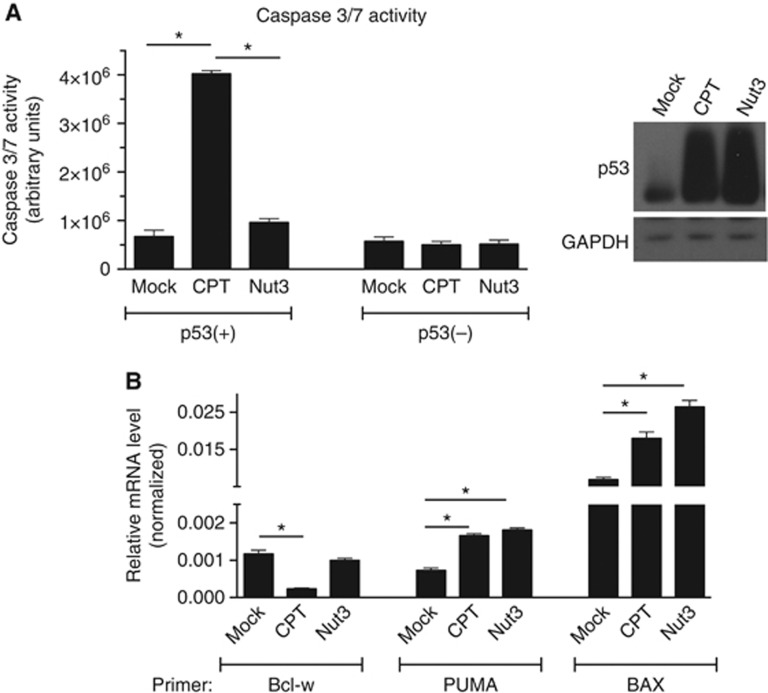
It has been shown previously that the DNA damaging agents CPT and doxorubicin DXR induce p53 accumulation and cause apoptosis. Nut3 stabilizes p53 and induces the accumulation of p53, but fails to promote apoptosis and mediate cell-cycle arrest. In agreement with a previous report, in human colon carcinoma HCT116 cells carrying a wild-type p53 gene (p53(+) cells), elevation of caspase-3/7 activity was observed upon CPT treatment but not upon Nut3 treatment (Figure 1A). CPT triggers apoptotic cell death in a p53-dependent manner, as HCT116 cells with a homozygous deletion of p53 (p53(−) cells) did not have elevated caspase-3/7 activity upon CPT treatment.
Figure 1.
CPT mediates apoptotic cell death. (A) Caspase-3/7 activities were measured using a luminescent assay by adding a profluorescent caspase-3/7 consensus substrate to cell lysates from p53(+) cells treated with DMSO (mock), CPT, or Nut3 for 16 h. *P<0.01; error bars represent standard deviation (s.d.). n=3. Total cell lysates from the same samples were subjected to immunoblot analysis with anti-p53 or anti-GAPDH (loading control) antibodies. (B) mRNA levels of pro-apoptotic genes (PUMA and BAX), which are transcriptional targets of p53, and anti-apoptotic Bcl-w normalized to GAPDH were monitored by qRT-PCR analysis in p53(+) cells treated with DMSO (mock), CPT, or Nut3 for 16 h.
Source data for this figure is available on the online supplementary information page.
To elucidate the molecular mechanism of CPT-induced, p53-dependent cell death, we examined the expression of pro-apoptotic proteins Puma and Bax and the pro-survival protein Bcl-w (a member of the Bcl-2 family) upon CPT or Nut3 treatment. As previously reported (Toledo and Wahl, 2006), both Puma and Bax, which are transcriptional targets of p53, were induced by both CPT and Nut3 treatment to similar levels (Figure 1B). Unlike Puma and Bax, Bcl-w mRNA was reduced to ∼20% of the control level by CPT but not by Nut3 (Figure 1B). When Bcl-w was experimentally downregulated, Nut3 was able to mediate cell death similarly to CPT (Supplementary Figure S1). These results suggest that the downregulation of Bcl-w in combination with the induction of Puma and Bax might play a critical role in the induction of cell death by CPT.
Induction of miR-203 mediates Bcl-w downregulation and cell death upon CPT treatment
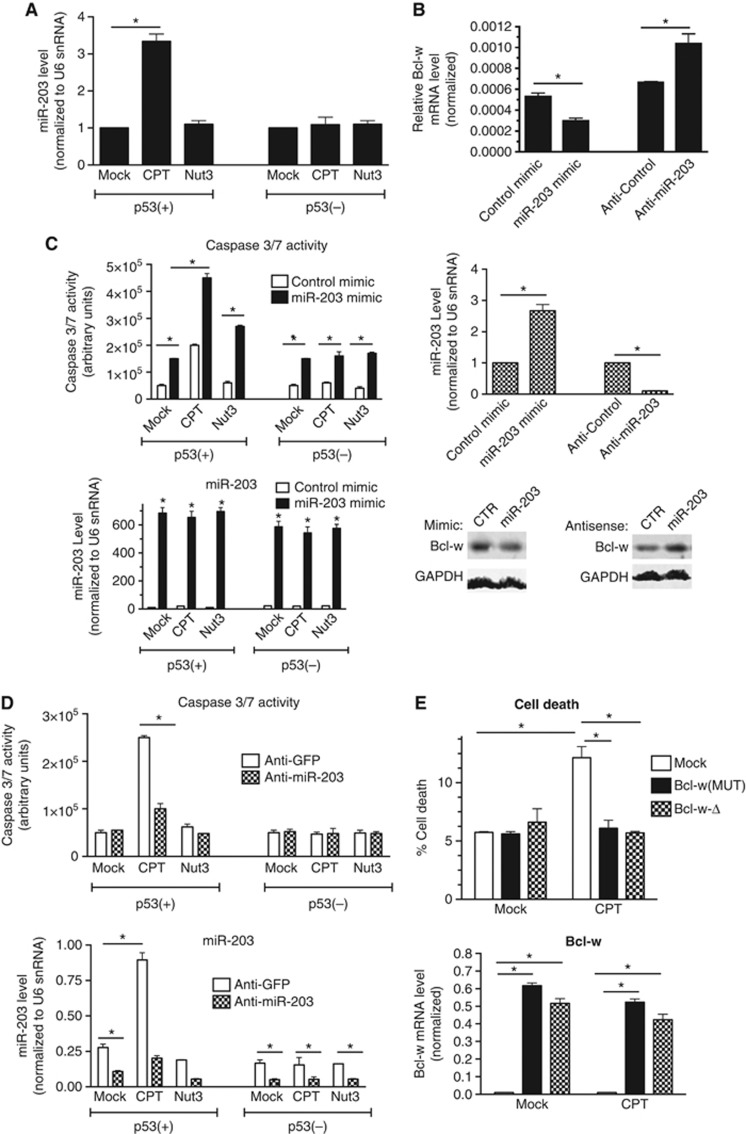
It has been reported that miR-203 targets an evolutionarily conserved miR-203 recognition element (MRE) located at the 3′ untranslated region (UTR) of Bcl-w mRNA and downregulates Bcl-w expression in bladder cancer cells (Bo et al, 2011). Thus, we hypothesized that the induction of miR-203 by CPT might lead to the downregulation of Bcl-w in p53(+) cells (Bo et al, 2011). We found that miR-203 in p53(+) cells is upregulated ∼3-fold over steady-state levels after CPT treatment (Figure 2A). The miR-203 level in p53(−) cells was unchanged upon CPT treatment (Figure 2A), indicating that induction of miR-203 by CPT is p53 dependent. Interestingly, treatment of p53(+) cells with Nut3 did not induce miR-203 (Figure 2A), supporting our hypothesis that regulation of the miR-203-Bcl-w axis is CPT specific and p53 dependent. To test whether miR-203 targets Bcl-w in p53(+) cells, we transfected cells with either a chemically modified RNA with the mature miR-203 sequence (miR-203 mimic), which elevates endogeneous miR-203 three-fold (Figure 2B, bottom panel), or antisense oligonucleotides against miR-203 (anti-miR-203), which downregulate miR-203 to <10% of endogeneous level (Figure 2B, middle panel), followed by analysis of Bcl-w mRNA and protein. Four previously validated targets of miR-203 (Akt2, Saini et al, 2011; Abl1, Bueno et al, 2008; Craig et al, 2011; Src, Saini et al, 2011; and SOCS3, Ru et al, 2011) were repressed by miR-203 mimic and induced by anti-miR-203, confirming the expected gain- and loss-of-function effects of these reagents in p53(+) cells (Supplementary Figure S2). Transfection of miR-203 mimic reduced endogeneous Bcl-w mRNA to ∼50%, and expression of anti-miR-203 derepressed Bcl-w mRNA by ∼60% (Figure 2B, top panel). Similar results were obtained by analysing the Bcl-w protein level (Figure 2B, bottom panel). We also confirmed that the luciferase activity of a reporter construct containing the MRE identified in the 3′UTR of Bcl-w mRNA (Bo et al, 2011) at the 3′ end of the luciferase reporter gene (WT) was reduced by miR-203 mimic (Supplementary Figure S3, WT). Conversely, a reporter construct with four substitutions in the MRE (MUT) that disrupt complementarity with the miR-203 seed sequence was resistant to overexpression of miR-203 (Supplementary Figure S3, MUT), indicating that miR-203 targets the MRE in the 3′UTR of Bcl-w mRNA in p53(+) cells.
Figure 2.
Downregulation of Bcl-w by miR-203 leads to apoptosis. (A) p53(+) or (−) cells were treated with DMSO (mock), CPT, or Nut3 for 16 h. Total RNA was extracted after drug treatment and analysed by qRT–PCR to examine miR-203 levels normalized to U6 snRNA levels (right panel). *P<0.05; error bars represent standard deviation (s.d.). n=3. miR-203 levels were quantified by qRT–PCR from total RNA extracted following drug treatment (bottom panel). (B) p53(+) cells were transfected with control mimic, miR-203 mimic, control antisense (anti-GFP), or anti-miR-203, followed by qRT–PCR analysis to examine Bcl-w mRNA levels normalized to GAPDH (top panel) and miR-203 normalized to U6 snRNA (middle panel). *P<0.05; error bars represent standard deviation (s.d.). n=3. Immunoblot analysis of Bcl-w protein in the presence of miR-203 mimic or anti-miR-203 was also examined in the same cells (bottom panel). (C) p53(+) cells were transfected with control mimic or miR-203 mimic, followed by treatment with DMSO (mock), CPT, or Nut3 for 24 h, followed by examining caspase-3/7 activities (top panel). miR-203 level normalized to U6 snRNA was examined by qRT–PCR analysis (bottom panel). *P<0.05; error bars represent standard deviation (s.d.). n=3. (D) p53(+) cells were transfected with control antisense (anti-GFP) or anti-miR-203 followed by treatment with DMSO (mock), CPT, or Nut3 for 24 h, followed by examining caspase-3/7 activities (top panel). Results are the average of three independent experiments performed in triplicate. miR-203 levels were measured by qRT–PCR to confirm effective silencing of miR-203 by anti-miR-203 (bottom panel). *P<0.01; error bars represent standard deviation (s.d.). n=3. (E) p53(+) cells were transfected with empty vector (mock), Bcl-w cDNA expression construct bearing mutations in the miR-203 MRE (Bcl-w(MUT)) or deleted in 3′UTR (Bcl-w-Δ) constructs and treated with DMSO (mock) or CPT for 16 h. Cell death was assessed by trypan blue uptake in a minimum of 200 cells (left panel). Transfection efficiency was assessed by qRT-PCR of Bcl-W mRNA (normalized to GAPDH) from remaining cells after trypan blue staining. *P<0.01; error bars represent standard deviation (s.d.). n=3.
Source data for this figure is available on the online supplementary information page.
To confirm the induction of miR-203 leading to cell death in a p53-dependent manner, miR-203 mimic was transfected into p53(+) or p53(−) cells, followed by CPT or Nut3 treatment. Exogeneous expression of miR-203 triggered cell death in both p53(+) and p53(−) cells as measured by caspase-3/7 activity (Figure 2C). Transfection of miR-203 mimic in p53(+) cells was sufficient to weakly induce the activation of caspase-3/7, and miR-203 mimic with CPT treatment synergistically increased the caspase activity (Figure 2C). Consistent with the results shown in Figure 1A, Nut3 treatment alone did not promote cell death; however, miR-203 mimic with Nut3 treatment in p53(+) led to cell death (Figure 2C), suggesting that miR-203-mediated repression of Bcl-w in addition to transcriptional induction of pro-apoptotic Puma and Bax by CPT or Nut3 treatment synergistically promotes cell death. Conversely, we examined whether anti-miR-203 transfection, which abrogates endogeneous miR-203 activity, also abrogates apoptotic cell death induced by CPT. To this end, endogeneous miR-203 was blocked by transfection of anti-miR-203, followed by CPT treatment. Transfection of anti-miR-203, which decreased the amount of miR-203 induced upon CPT treatment by 70% (Figure 2D, bottom panel), prevented CPT-mediated cell death in p53(+) cells as measured by caspase-3/7 activity (Figure 2D, top panel). Neither CPT nor Nut3 induced cell death, and anti-miR-203 showed little effect in p53(−) cells (Figure 2D, top panel). Finally, to provide evidence that miR-203-mediated downregulation of Bcl-w is responsible for CPT-mediated cell death in p53(+) cells, Bcl-w expression constructs either lacking the 3′UTR (Bcl-w-Δ) or containing a 3′UTR bearing nucleotide substitutions within the MRE (Bcl-w(MUT)) (see Supplementary Figure S3), were transfected into p53(+) cells, followed by stimulation with CPT (Figure 2E). Unlike cells transfected with empty vector (mock), which suffered from induced cell death, Bcl-w-Δ or Bcl-w(MUT)-transfected cells were resistant to cell death upon CPT treatment (Figure 2E). These results indicate that the CPT-mediated activation of p53 induces miR-203, which downregulates Bcl-w and promotes cell death.
Genotoxic stimuli specifically elevate the activity of p53 to promote pri-miRNA processing
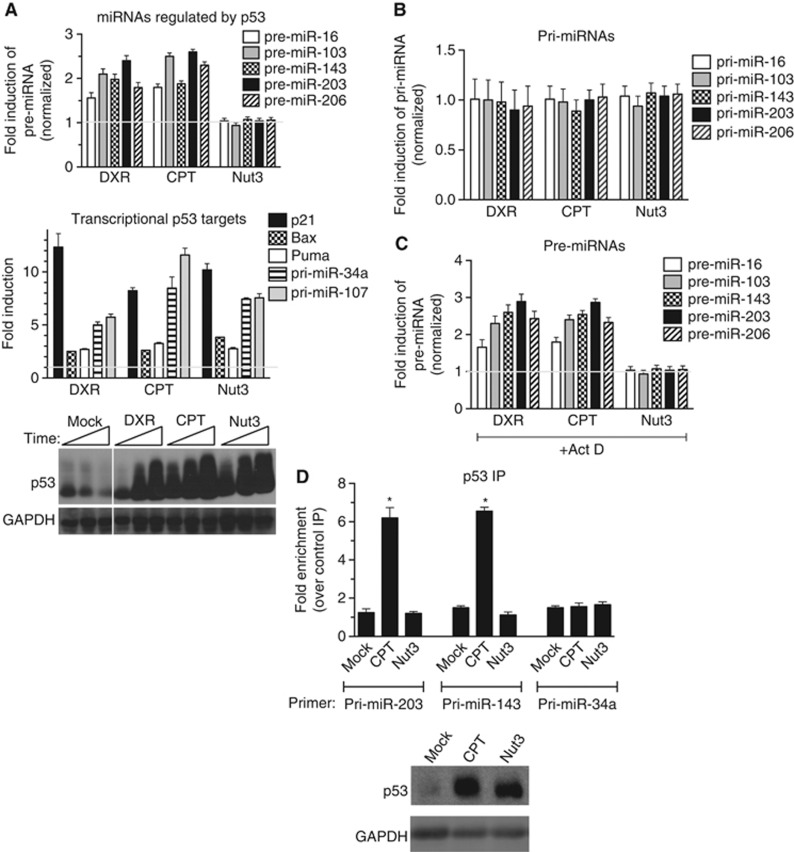
A number of post-translational modifications modulate the activity of p53 and are implicated in contributing to specific cellular outcomes (Kruse and Gu, 2009). It was recently identified that upon DXR treatment, p53 post-transcriptionally induces a subset of miRNAs, including miR-203, miR-16, miR-103, and miR-206, by facilitating the first cropping step from pri-miRNA to pre-miRNA catalysed by Drosha in the nucleus (Suzuki et al, 2009). Therefore, we hypothesized that CPT but not Nut3 mediates the induction of miR-203 and promotes cell death. p53(+) cells were treated with DXR, CPT, or Nut3 for 6 h and then harvested for pre-miRNA analysis. As previously reported, DXR induced the expression of a set of pre-miRNAs, including miR-203 (Figure 3A, top panel). Similarly, CPT induced expression of these pre-miRNAs, whereas Nut3 treatment did not, despite the fact that similar levels of p53 protein were detected in all conditions (Figure 3A, bottom panel). Induction of these miRNAs upon CPT or DXR treatment is p53 dependent, as these agents failed to induce miRNAs in p53(−) cells (Supplementary Figure S4). Consistent with the amount of p53 protein, the induction of transcriptional targets of p53 (p21, Puma, Bax, pri-miR-341, and pri-miR-107) was comparable in DXR-, CPT-, and Nut3-treated cells (Figure 3A, middle panel), illustrating that DXR/CPT-mediated activation of p53 leads to the activation of both the transcription and the miRNA processing activity of p53, whereas Nut3-mediated activation of p53 selectively activates its transcriptional activity. Induction of pre-miRNAs by CPT and DXR is a post-transcriptional effect of p53, as pri-miRNA levels of these miRNAs were not altered by CPT or DXR treatment (Figure 3B). Induction of pre-miRNAs was observed when cells were treated with CPT or DXR in the presence of the RNA polymerase II inhibitor Actinomycin D (Act D) (Figure 3C). As a control of specificity, miRNAs regulated by the signal transducers of the TGFβ signalling pathway, Smad proteins (miR-21, miR-199a, and miR-421) (Davis et al, 2008, 2010), have been examined after DXR treatment in p53(+) cells and were not affected by DXR (Supplementary Figure S5), indicating the specificity of miRNAs that are regulated by p53 upon DXR or CPT stimulation.
Figure 3.
Nut3-mediated activation of p53 does not induce miRNA processing. (A) qRT–PCR analysis of pre-miRNAs regulated by p53 (miR-16, -103, -143, -203, and -206) with 6 h of DXR, CPT, or Nut3 treatment in p53(+) cells is shown. As controls, pre-miRNAs regulated by Smad (miR-21, -199a, and -421) and levels of transcriptional targets of p53 (p21, Puma, Bax, pri-miR-34a, and pri-miR-107) were examined. Results are shown as the fold induction upon treatment with DXR, CPT, or Nut3 as compared to DMSO treatment. Immunoblot analysis of total p53 protein level after DMSO (mock), DXR, CPT, or Nut3 treatment for 6 h is shown from p53(+) cell lysates. GAPDH is shown as a loading control (bottom panel). (B) qRT–PCR analysis of pri-miRNA regulated by p53 (miR-16, -103, -143, -203, and -206) with 6 h of DXR, CPT, or Nut3 treatment in p53(+) cells. (C) qRT–PCR analysis of pre-miRNA induced by p53 (miR-16, -103, -143, -203, and -206) with 6 h of co-treatment with actinomycin D (ActD) and DXR, CPT, or Nut3 in p53(+) cells. (D) p53(+) cells were treated with DMSO (mock), CPT, or Nut3 for 8 h, followed by RNA-IP analysis. Results are shown as the fold enrichment of pri-miRNA of processing targets of p53 (miR-203 and miR-143) in the IP samples with anti-p53 antibody over control IP samples with non-specific IgG. The transcriptional target of p53, pri-miR-34a, was measured as a control. Total cell lysates from the same samples were subjected to immunoblot analysis with anti-p53 or anti-GAPDH (loading control) antibodies.
Source data for this figure is available on the online supplementary information page.
Next, we performed immunoprecipitation (IP) of p53 followed by detection of associated RNAs in p53(+) cells treated with CPT or Nut3. The results indicated that treatment with CPT, but not with Nut3, promotes the association of p53 with pri-miRNAs of p53-regulated targets (miR-203 and miR-143), whereas no interaction was detected with control pri-miRNA (miR-34a) (Figure 3D).
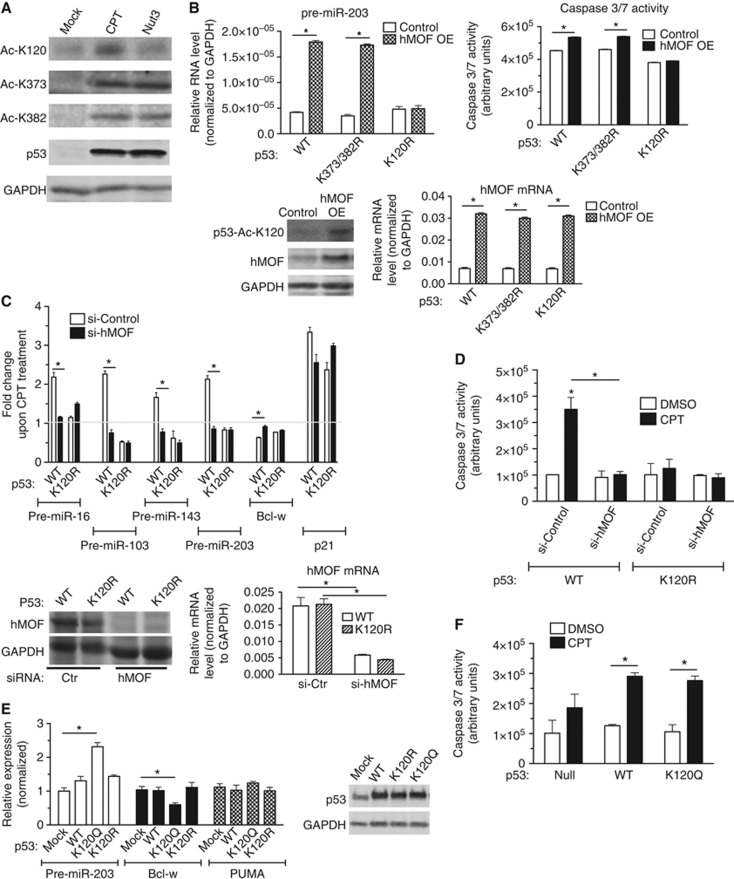
Acetylation of K120 is critical for the miRNA processing activity of p53
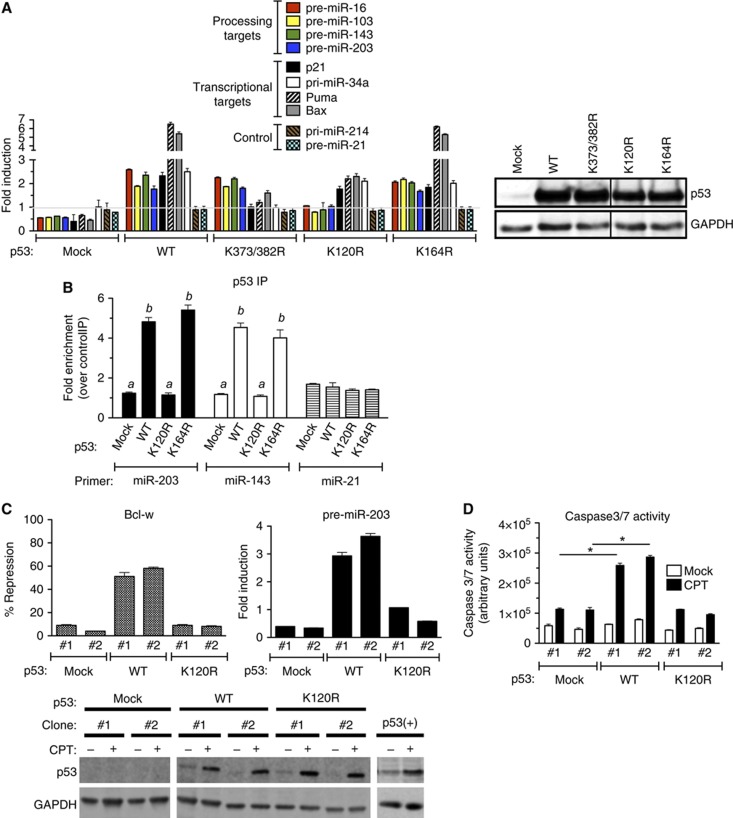
Among the various post-translational modifications that have been identified in p53, the acetylation of the K120 residue in the DBD has been found to play a critical role in mediating apoptosis, as a single amino-acid substitution at K120 to Arg (K120R) abrogates p53-dependent apoptosis with no major effect on mediating cell-cycle arrest (Sykes et al, 2006; Tang et al, 2006; Li et al, 2012). Thus, we examined whether this modification plays a role in facilitating the miRNA processing activity of p53. Wild-type p53 (WT) or two acetylation-deficient K mutants in the DBD (K120R and K164R) were transiently expressed in p53(−) cells, followed by the quantitative analysis of p53-regulated targets (pre-miRNA-16, -103, -143, and -203), transcriptional targets (p21, Puma, Bax, and pri-miR-34a) or control miRNA (pri-miRNA-21 and -214) (Figure 4A, left panel). K to R mutation in the C-terminal region (CTD) of p53 (K373/382R double mutant) was used as a control (Figure 4A). K120R mutant expressed at a level similar to WT and K372/382R p53 (Figure 4A, right panel), greatly reduced CPT-mediated miRNA processing (Figure 4A, Processing targets) while still being able to mediate the induction of transcriptional targets, such as p21 and pri-miR-34a (Figure 4A). Similar result was obtained in p53-deficient human lung carcinoma H1299 cells exogeneously expressing p53(K120R) (Supplementary Figure S6). As previously reported (Sykes et al, 2006; Tang et al, 2006), induction of Puma and Bax was reduced in K120R cells (Figure 4A). Control miRNAs were not affected by the introduction of WT or K120R p53 (Figure 4A). A mutation of K164, which is also a known site of acetylation (Tang et al, 2006), had miRNA processing ability similar to WT (Figure 4A, K164R). RNA-IP analysis demonstrated that WT and K164R were recruited to pri-miRNAs of the p53 processing targets (miR-203 and miR-143) upon CPT stimulation, but K120R was unable to associate with these pri-miRNAs (Figure 4B). This result is in agreement with the lack of miRNA processing in K120R mutant-expressing cells (Figure 4A). Therefore, K120 in the p53 DBD is critical for association with the miRNA processing target and induction of miRNA processing.
Figure 4.
Modification at the K120 residue is critical for the miRNA processing activity of p53. (A) p53(−) cells were transfected with empty expression vector (mock), expression vector carrying wild-type p53 (WT), K120R mutant, K164R mutant, or K373/382 double mutant (K373/382R), followed by DMSO or CPT treatment for 16 h. Total RNA was subjected to qRT–PCR analysis of p53-processed targets (miR-16, 103, -143, and -203), p53 transcriptional target (p21, Puma, Bax, and pri-miR-34a), or control (pri-miR-214 and pre-miR-21). Results are shown as the fold change of pre-miRNA upon CPT treatment as compared to DMSO treatment (left panel). Total lysates of the same cells or p53(+) cells following DMSO treatment were subjected to immunoblot analysis of p53 and GAPDH (loading control; right panel). (B) RNA-IP was performed on p53(−) cells transfected with the expression vector carrying p53 WT, K120R, or K164R. pri-miRNA levels of processing targets of p53 (miR-203 and miR-143) or control (miR-21) associated with p53 WT or mutants were analysed by qRT-PCR. Results are shown as the fold induction of p53-associating pri-miRNAs upon CPT treatment for 8 h over DMSO treatment. Immunoblot analysis of p53 and GAPDH is shown in (B). (C) Two independent clones of p53(−) cells stably expressing an empty vector (mock), WT, or K120R were treated with DMSO (mock) or CPT for 16 h and subjected to qRT-PCR analysis of pre-miR-203 and Bcl-w mRNA normalized to GAPDH. Result is shown as % repression and fold induction of Bcl-w and pre-miR-203 by comparing the levels in CPT-treated cells with DMSO-treated cells. The levels of p53 and GAPDH (loading control) protein following DMSO (mock) or CPT treatment were examined by immunoblotting using the same stable clones. (D) p53(−) cells stably expressing empty vector (mock), WT, or K120R were treated with DMSO (mock) or CPT for 16 h and subjected to the caspase-3/7 activity assay. *P<0.01; error bars represent standard deviation (s.d.). n=3.
Source data for this figure is available on the online supplementary information page.
Next, using two independent stable clones of p53(−) cells expressing WT or K120R p53 at levels similar to endogeneous p53 in p53(+) cells (Figure 4C, bottom panel), we examined the CPT-mediated regulation of Bcl-w. In WT clones, a 2- to 3-fold induction of miR-203 led to ∼50% repression of Bcl-w upon CPT treatment (Figure 4C, top panel). Unlike in WT clones, downregulation of Bcl-w was blunted in K120R clones to a similar degree as clones transfected with empty vector (mock) (Figure 4C, top panel), suggesting that K120 acetylation plays a critical role in CPT-induced downregulation of Bcl-w. In agreement with the lack of downregulation of Bcl-w, activation of caspase-3/7 by CPT was reduced in K120R clones to a level similar to mock-transfected cells (Figure 4D), confirming that the K120R mutant of p53 is unable to mediate apoptotic cell death due to its inability to promote miR-203-mediated downregulation of Bcl-w. Thus, our study uncovers novel mechanistic insight into the critical role of the acetyl-K120 form of p53 upon DNA damage stress in the regulation of miRNA biogenesis and the induction of apoptosis.
The acetyltransferase hMOF is required for induction of the miRNA processing activity of p53
To provide evidence that the regulation of miRNA processing by CPT relies on acetylation of K120 instead of other types of K modifications, we tested whether treatment with CPT (and not Nut3) induces acetylation of K120 by immunoblot analysis using antibodies against total p53 or acetyl-K120, -K373, or -K382 p53 (Figure 5A). Levels of total p53, Ac-K373, and Ac-K382 were similar following both CPT and Nut3 treatment; only CPT elevated acetylation of K120 (Figure 5A). Next, we tested whether forced expression of an acetyltransferase responsible for K120 acetylation could augment the levels of p53-processed miRNAs in p53(+) cells. Unlike K164 and the C-terminal K residues (K370, 372, 373, 381, 382, and 386), which are acetylated by CBP/p300, K120 is acetylated by a distinct acetyltransferase hMOF or the closely related protein Tip60 (Brooks and Gu, 2011). We transfected WT, K120R, or K373R/K382R p53 into p53(−) cells with or without an hMOF expression plasmid. Overexpression of hMOF (4.5-fold over endogeneous) elevated K120 acetylation (Figure 5B, bottom panels) as well as the miRNA processing activity of WT and K373R/K382R with the exception of K120R (Figure 5B, top left panel). Marginal increase in caspase-3/7 activity was observed in WT cells but not in K120R cells upon overexpression of hMOF (Figure 5B, top right panel). When >80% of endogeneous hMOF protein was downregulated by siRNA (si-hMOF) in p53(−) cells stably expressing WT p53 (Figure 5C, bottom panels), CPT-dependent induction of pre-miR-203, downregulation of Bcl-w (Figure 5C, top panel), and apoptosis (Figure 5D, WT) were abrogated. si-hMOF exhibited a small effect on the transcriptional activity of p53 (Figure 5C; Supplementary Figure S7, p21). No significant effect was observed by si-hMOF in K120R cells (Figure 5C and D). These results indicate that acetylation of K120 by hMOF is critical for the p53-dependent induction of miRNA processing and cell death upon CPT treatment.
Figure 5.
Acetylation at K120 by hMOF facilitates miRNA processing. (A) Total cell lysates from p53(+) cells treated with DMSO (mock), CPT, or Nut3 for 6 h were subjected to immunoblot analysis with anti-Ac-K120-p53, anti-Ac-K373-p53, anti-Ac-K382-p53, total p53 antibody, and anti-GAPDH (control) antibodies. (B) p53(−) cells were transfected with the expression vector carrying wild type (WT), K373/382R, or K120R mutant p53 with co-transfection of control plasmid or hMOF expression plasmid, followed by CPT treatment for 16 h. qRT–PCR analysis of pre-miRNAs of the p53-processed miRNAs (miR-203) or hMOF mRNA normalized to GAPDH are shown. *P<0.05; error bars represent standard deviation (s.d.). Total cell lysates of WT-transfected cells with control plasmid or hMOF expression plasmid were subjected to immunoblot analysis using anti-p53-Ac-K120, anti-hMOF, and anti-GAPDH antibodies (loading control). (C) p53(−) cells stably transfected with the expression vector carrying WT or K120R mutant p53 were transfected with siRNA against hMOF (si-hMOF) or control siRNA (si-Control), followed by DMSO or CPT treatment for 16 h. qRT–PCR analysis of pre-miR-16, -103, -143, -203, Bcl-w, p21 mRNA (control), or hMOF mRNA normalized to GAPDH are shown. Results are shown as the fold induction of pre-miRNAs or p21 upon CPT treatment as compared to DMSO treatment. *P<0.01; error bars represent standard deviation (s.d.). n=3. (D) p53(−) cells stably transfected with the expression vector carrying WT or K120R mutant p53 were transfected with siRNA against hMOF (si-hMOF) or control siRNA (si-Control). After DMSO or CPT treatment for 16 h, cells were subjected to caspase-3/7 assay. *P<0.05; error bars represent standard deviation (s.d.). (E) p53(−) cells were transfected with the empty expression vector (mock), the expression vector carrying wild-type (WT) p53, the K120 acetylation mimic (K120Q), or the K120 acetylation-deficient mutant (K120R), followed by qRT–PCR analysis of pre-miR-203, Bcl-w, and Puma normalized to GAPDH. Immunoblot analysis of p53 and GAPDH (control) is also shown. *P<0.05; error bars represent standard deviation (s.d.). n=3. (F) p53(−) cells stably expressing empty vector (mock), WT, or K120Q were treated with DMSO (mock) or CPT for 16 h and subjected to the caspase-3/7 activity assay. *P<0.01; error bars represent standard deviation (s.d.). n=3.
Source data for this figure is available on the online supplementary information page.
We also generated an acetyl-K120 mimic by replacing K120 with glutamine (Q) (K120Q), as K-to-Q substitutions in histone H4 (Hecht et al, 1995) and p53 (Luo et al, 2004) are known to be functionally comparable to acetylated K residues. In contrast to WT p53, which required CPT stimulation to promote pri-miR-203 processing, K120Q expressed at a similar level to WT (Figure 5E, right panel), was able to mediate pri-miR-203 processing in the absence of CPT stimulation (Figure 5E, left panel), suggesting that acetylation of K120 is sufficient for the induction of miRNA processing by p53. Consistently with the elevated level of pre-miR-203 in the absence of CPT treatment, Bcl-w expression was reduced in K120Q cells in comparison with WT or K120R mutant cells (Figure 5E, left panel). Unlike pri-miR-203 processing, Puma mRNA was not significantly altered in K120Q cells (Figure 5E, left panel). Despite the reduction in Bcl-w in K120Q cells, the basal activity of caspase-3/7 was similar to WT p53 cells (Figure 5F), suggesting that K120Q-mediated downregulation of Bcl-w is not sufficient to promote cell death. These results indicate that acetylation of K120 is sufficient for the miRNA processing activity of p53, but not for the transcriptional activation of p53. It also indicates that the induction of apoptosis by DNA damage agents requires both miRNA processing-mediated gene regulation, i.e., downregulation of Bcl-w, and the transcriptional induction of Puma or Bax.
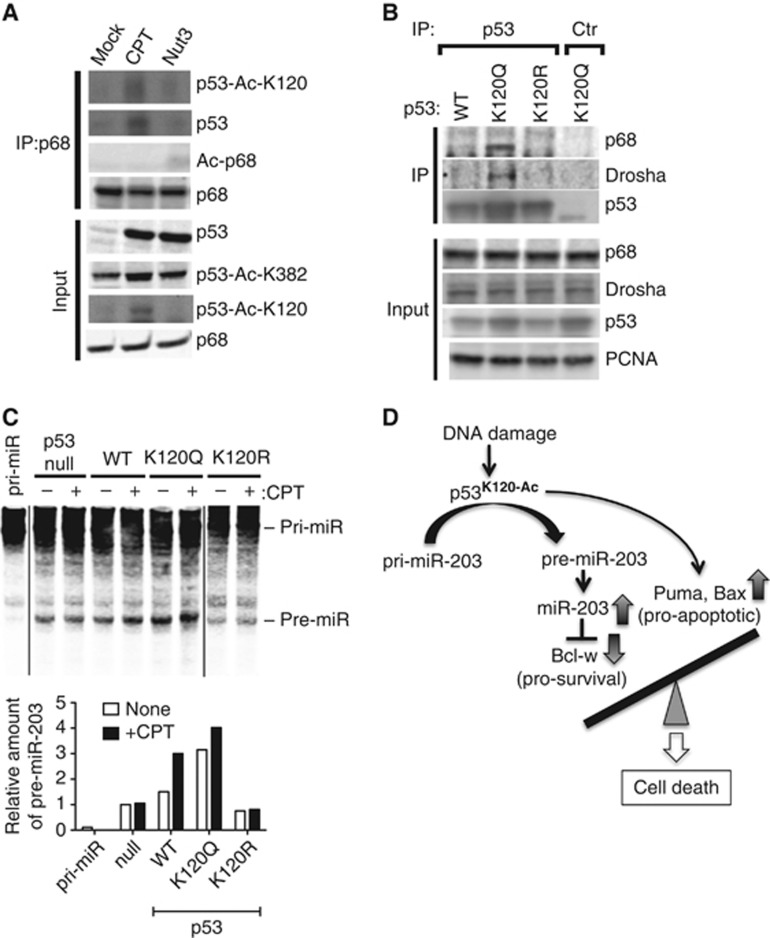
Acetylation of K120 facilitates stable interaction with p68 and Drosha
p53 associates with the Drosha complex through RNA helicase p68 (DDX5), a subunit of the Drosha microprocessor complex (Suzuki et al, 2009). We tested whether K120 acetylation of p53 stabilizes the interaction with p68, in turn promoting p53-dependent pri-miRNA processing. p68 was immunoprecipitated from nuclear extracts of cells stimulated with CPT or Nut3, followed by immunoblot analysis using antibodies against total p53 or K120-acetylated p53 (p53-Ac-K120) (Figure 6A). Although levels of total p53 and K382-acetylated p53 were similar following both CPT and Nut3 treatment, only CPT increased K120 acetylation (Figure 6A, input) and the interaction between p53 and p68 (Figure 6A, IP: p68). We also confirmed that the p53 associated with p68 upon CPT treatment was K120 acetylated (Figure 6A, p53-Ac-K120). Lysine acetylation of p68 was not detected following CPT or Nut3 treatment (Figure 6A, Ac-p68). These results suggest that the differential effect of CPT and Nut3 on p53-mediated miRNA processing is due to acetyl-K120, which facilitates stable association between p53 and the Drosha complex. The acetyl-K120 mimic K120Q was constitutively associated with p68 and Drosha in the absence of CPT treatment (Figure 6B), while WT p53 associated with p68 upon DNA damage stress (Figure 6A), suggesting that K120 acetylation is sufficient to promote interaction of p53 with the Drosha complex. K120R mutant of p53 was unable to interact with p68 both in the presence and absence of CPT stimulation (Figure 6B; Supplementary Figure S8). Finally, we performed in vitro pri-miRNA processing assays by adding biotin-labelled pri-miR-203 with total cell lysates from p53-null, p53(WT, K120Q or K120R) stable clones with or without CPT treatment (Figure 6C). In accordance with the result of RT–PCR analysis of pre-miR-203 (Figure 4A), two-fold induction of a band corresponding to pre-miR-203 was detected in the reaction with p53(WT) cell lysates upon CPT treatment (Figure 6C, WT). Cell lysates of K120Q cells in the absence of CPT treatment exhibited a high level of pre-miR-203 band similarly to the level of CPT-treated p53(WT) cells, which was not significantly augmented by CPT (Figure 6C, K120Q). The levels of pre-miR-203 band in K120R cells were low and unchanged upon CPT treatment (Figure 6C, K120R). These results provide direct evidence of functional significance of K120 acetylation of p53 in the regulation of the Drosha microprocessor activity upon DNA damage stimuli. In summary, this study uncovers a critical role of K120 acetylation in the DBD of p53 in regulating the miRNA biogenesis pathway and in determining a stress-specific cellular outcome (Figure 6D).
Figure 6.
Acetylation at K120 of p53 promotes stable interaction with the Drosha complex. (A) Nuclear extracts from p53(+) cells treated with DMSO (mock), CPT, or Nut3 for 6 h were subjected to IP with anti-p68 antibody, followed by immunoblotting with anti-p53-Ac-K120, anti-p53, anti-Ac-p68, or anti-p68 antibody. Input samples were subjected to immunoblotting with anti-p53, anti-p53-Ac-K120, anti-p53-Ac-K382, or anti-p68 antibody. (B) p53(−) cells were transfected with the expression vector carrying the wild-type p53 (WT) or the K120 acetylation mimic (K120Q) mutant. Nuclear extracts were subjected to IP with anti-p53 antibody or the anti-GAPDH antibody (Ctr) as a negative control, followed by immunoblotting with anti-p68, anti-Drosha, or anti-p53 antibodies. Nuclear extracts (input) were subjected to immunoblotting with anti-p53, anti-p68, anti-Drosha, and anti-proliferating cell nuclear antigen (PCNA) antibody as the loading control for nuclear extracts. (C) In vitro pri-miR-203 processing assays were performed by applying biotin-labelled pri-miR-203 with total cell lysates from p53(null), p53(WT), or p53 mutant (K120Q and K120R) cells following DMSO (mock) or CPT treatment (top panel). Relative amount of pre-miR-203 quantitated by LI-COR is shown (bottom panel). (D) Schematic diagram of the mechanism of DNA damage stress-induced cell death mediated by the acetyl-K120 form of p53. Upon DNA damage stimuli, such as DXR and CPT, K120 of p53 is acetylated, which preferentially promotes miRNA processing activity of p53. One of the p53 miRNA processing targets, miR-203, downregulates anti-apoptotic Bcl-w and induces apoptotic cell death.
Source data for this figure is available on the online supplementary information page.
Discussion
The mechanism of stimulus-specific regulation of p53, in particular the different consequences mediated by genotoxic stimuli and the HDM2 inhibitor, has been investigated previously (Donner et al, 2007). One proposed mechanism for the varying responses is the differential accessibility of the genomic binding sites of p53, which in turn controls different subsets of downstream targets in distinct cellular environments. However, genome-wide analysis of p53 occupancy suggests that the regulation of specific subsets of p53 targets cannot be attributed to the association of p53 to specific target genes, as the pattern of p53 occupancy does not change dramatically between different stimuli, despite different biological outcomes (Wei et al, 2006). Our study shows that the p53-dependent modulation of miRNA biogenesis is a defining mechanism for the specificity of DNA damaging agent-dependent cell death as a consequence of K120 acetylation.
It has been shown that p53 modulates miRNA biogenesis through associating with p68 to enable processing of pri-miRNA to pre-miRNA by Drosha, similarly to the function of TGFβ signal transducers, the Smad proteins. In response to a DNA damage stimulus, the level of a small subset of miRNAs is post-transcriptionally upregulated (Suzuki et al, 2009). In addition to this miRNA processing regulation, p53 also transcriptionally regulates the level of some miRNAs, such as miR-34a, miR-107, miR-125b, miR-149*, miR-146, miR-192, miR-215, and miR-1246. Conversely, p53 itself is regulated by a miRNA, miR-125b (Le et al, 2009), which decreases upon DNA damage (Junttila and Evan, 2009). Therefore, during DNA damage, a decrease in the miRNA-dependent repression of p53 is coupled with p53 activation, which results in the upregulation of protein-coding genes and miRNAs, as well as the downregulation of miRNA targets to promote cell-cycle arrest and apoptosis. This post-transcriptional mechanism of miRNA regulation might be critical and relevant to a rapid modulation of the expression of a large number of genes under various stress stimuli that can compromise transcription activities.
Interestingly, p53 mutants frequently identified in cancers, such as C135Y, R175H, and R273H, fail to interact with the Drosha complex and to facilitate miRNA processing (Suzuki et al, 2009), suggesting that the tumorigenesis caused by these p53 mutants might be, in part, due to deregulation of miRNA biogenesis. We propose that the K120R mutant found in tumours may also contribute to tumorigenesis and resistance to chemotherapy through deregulation of miRNA biosynthesis.
Although multiple lysine residues in p53 are acetylated, the observation that only K120 and K164 mutations can be found in tumours and tumour-derived cell lines supports the physiological significance of the modification of these two amino acids. It has been reported that acetyl-K120 is essential for the induction of apoptosis but dispensable for cell-cycle arrest or senescence (Sykes et al, 2006; Tang et al, 2006; Li et al, 2012). On the basis of the observation that the K120R mutant can activate p21 but not the pro-apoptotic gene Puma, it has been suggested that K120 acetylation conveys the transactivation of specific gene promoters, thus prompting specific cellular outcomes (Sykes et al, 2006; Tang et al, 2006; Li et al, 2012). Consistently, the crystal structure of the K120-acetylated form of the p53 DBD indicates that acetyl-K120 does not alter the overall structure of the DBD but instead increases DBD binding to a specific DNA sequence (Arbely et al, 2011). In addition to affecting DNA binding, our study reveals that K120 acetylation of p53 mediates the stable association of p53 with the Drosha complex, and thus stimulates miRNA processing. Other transcription factors, such as the Smad proteins and oestrogen receptor (ER) α, also interact with the Drosha complex via p68 and modulate the processing activity of Drosha (Davis-Dusenbery and Hata, 2010). The N-terminus Mad-homology 1 (MH1) domain of Smads, already known as a sequence-specific DBD, directly interacts with a double-stranded region of several pri-miRNAs in a sequence-dependent manner (Davis et al, 2010). Therefore, it is intriguing to speculate that K120 acetylation may contribute to p53 binding affinity and specificity not only to DNA, but also to a double-stranded region of pri-miRNAs.
Downregulation of miR-203 has been reported in wide a range of malignancies, including cancers of the bladder, breast and prostate, gastric and colorectal cancers, hepatocarcinomas, melanomas and lymphomas, suggesting that miR-203 functions as a tumour suppressor (Bueno et al, 2008; Lena et al, 2008; Furuta et al, 2010; Craig et al, 2011; Li et al, 2011; Luzna et al, 2011; Saini et al, 2011; Chen et al, 2012; Xu et al, 2012). miR-203 levels are also modulated by Epstein-Bar virus and human papilloma virus type 16 (HPV16) infection (Yu et al, 2012). Interestingly, the HPV16 oncoproteins E6 and E7 modulate miR-203 expression in human foreskin keratinocyte (HFK) in a p53-dependent manner; in fact, E6/E7 cannot induce miR-203 in HFK in which p53 has been reduced by short-hairpin RNAs (McKenna et al, 2010). This finding is consistent with our observation that p53-mediated processing is critical for the induction of miR-203.
Downregulation of anti-apoptotic Bcl-w by miR-203 is necessary for cell death upon CPT treatment, as an expression construct of Bcl-w lacking the MRE blocked CPT-mediated apoptosis. Gene targeting of Bcl-w in mouse indicates that Bcl-w has an essential function in spermatogenesis and survival of damaged epithelial cells in the gut, but it has a dispensable role in the physiological cell death process of other tissues (Pritchard et al, 2000; Yan et al, 2000). In addition to regulating Bcl-w expression, p53 is known to transcriptionally activate the pro-apoptotic genes Bax and Puma (Dai and Gu, 2010). Previous studies demonstrate that the p53-dependent induction of Bax and Puma upon stress is greatly decreased in cells expressing the K120R mutant, while activation of HDM2 and p21 is unaltered compared to WT p53-expressing cells, suggesting that K120 acetylation of p53 promotes preferential binding and transcriptional activation of the promoters of these pro-apoptotic genes (Sykes et al, 2006; Tang et al, 2006). Thus, we speculate that induction of K120 acetylation by CPT promotes cell death through the combined effects of (i) downregulation of pro-survival Bcl-w by miR-203 and (ii) induction of pro-apoptotic Bax/Puma proteins.
Materials and methods
Cell culture
HCT116 p53(+/+) and HCT116 p53-null cell lines were provided by Lieberman’s laboratory and cultured as previously described (Bunz et al, 1998). Cos7 cells were purchased from American Type Culture Collection (ATCC). All cell cultures were maintained in DMEM supplemented with 10% fetal bovine serum (FBS; Hyclone). Genotoxic drugs were purchased from Sigma-Aldrich and used at the following concentrations: 4 μM (S)-(+)-Camptothecin (CPT); 1 μl/ml dimethyl sulfoxide (DMSO); 10 μM Nutlin-3 (Nut3); 0.1 μg/ml Doxorubicin (DXR); 400 nM Trichostatin-A (TSA); 20 μM Etoposide (ETP); 0.2 μg/ml Actinomycin D (ActD).
Antibodies and siRNAs
Anti-p53 (DO-1) (Cat. No. OP43, EMD Millipore; Clone FL-393, Santa Cruz); anti-GAPDH (Clone MAB374, EMD Millipore); anti-p68 (Clone 05-850, Upstate); anti-p53-Ac-K373 (Clone 04-1137, EMD Millipore); anti-acetylated-lysine (Clone 9441, Cell Signaling); anti-p53-Ac-K382 (Clone 04-1146, EMD Millipore); anti-p53-Ac-K120 (a gift from Gu laboratory; Tang et al, 2006); anti-c-kit (used as a negative control for RNA-IP; Clone H-300, Santa Cruz); anti-cleaved Caspase-3 (Clone 5A1E, Cell Signaling); and anti-Bcl-w (Clone 31H4, Cell Signaling) antibodies were used. Non-targeting control siRNA was purchased from Qiagen (#1027280). siRNAs against hMOF#1: 5′-AAAGACCATAAGATTTATT-3′, hMOF#2: 5′-GTGTCCAGTCTCGAGTGA-3′.
Immunoblotting
Cells were lysed in TNE buffer (1% Nonidet P-40, 10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl) or low stringency RIPA buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS). Total cell lysates or proteins immunoprecipitated with antibodies were separated by SDS–PAGE, transferred onto PVDF or nitrocellulose membranes (EMD Millipore), immunoblotted with antibodies, and visualized using an enhanced chemiluminescence detection system (Amersham Bioscience or Pierce).
miRNA mimic
miR-203 mimic was purchased from Invitrogen. Negative control siRNA was purchased from Qiagen (1022076). Both mimic and negative controls were used according to the manufacturer’s directions.
Antisense miRNA
2′-O-methyl modified RNA oligonucleotides complementary to miRNA (anti-miR) or GFP (control) sequence were purchased from IDT. Anti-miRNAs were transfected to host cells at a concentration of 106 nM using RNAiMax (Invitrogen) according to the manufacturer’s instructions. Anti-miR-203: 5′-CUAGUGGUCCUAAACAUUUCAC-3′; Anti-GFP: 5′-AAGGCAAGCUGACCCUGAAGU-3′.
Transfection of plasmid DNA and miRNA mimic/anti-miRNA
HCT116 p53(+/+), p53-null, and Cos7 cells were transfected with FuGENE 6 (Roche or Promega) for plasmid DNA and RNAiMax for miRNA mimics or anti-miRNAs. miRNA mimics and anti-miRNAs were transfected at a concentration of 20 μM. Transfection was performed according to the manufacturer’s specifications.
Stable p53 cell lines
Flag epitope-tagged cDNA encoding WT or K120R p53 was cloned into pcDNA6.2/V5/GW/D-TOPO (Life Technologies). HCT116 p53-null cells were transfected with these plasmids or empty vector (control), and multiple stable clones were selected in the media containing 10 μg/ml of Blasticidin S (Life Technologies).
Quantitative real-time PCR assay
Quantitative real-time PCR (qRT–PCR) assays were performed for measurement of the expression levels of pri- and pre-miRNAs as described previously (Kruse and Gu, 2009). In brief, total RNA was extracted with Trizol (Invitrogen) and subjected to reverse transcription with the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s instructions. Quantitative analysis of expression levels was performed with the iQ5 real-time PCR machine (Bio-Rad). Expression levels of mature miRNAs were quantified with the TaqMan MicroRNA Assay Kit (Applied Biosystems) following the manufacturer’s instructions. An average of three experiments was performed in triplicate, with standard errors presented.
PCR primer sets: Human GAPDH: 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′; Human Bcl-w: 5′-TGTCAACAAGGAGATGGAAC-3′ and 5′-CCCGTATAGAGCTGTGAACT-3′; Human pri-miR-143: 5′-CCCTCTAACACCCCTTCT-3′ and 5′-TGGAGTCTGGAAACACTCTG-3′; Human pre-miR-143: 5′-CAGTGCTGCATCTCTGGT-3′ and 5′-CAGAACAACTTCTCTCTTCCTG-3′; Human pri-miR-16-1: 5′-ACAAAAACAAAGGAAAAGGA-3′ and 5′-TCGTTTTATGTTTGGATGAA-3′; Human pri-miR-214: 5′-CTGCTTTCTTTCAATGGCTGGTTGT-3′ and 5′-CTGATTGTATCTGTCTATGAGCAAA-3′; Human pri-miR-107: 5′-TTGTATGTCACCAGCTCCAC-3′ and 5′-GTGTCCACTGAAATGTGAGG-3′; Human pri-miR-34a: 5′-ATCTCTCGCTTCATCTTCC-3′ and 5′-CCCACATTTCCTTCTTATCA-3′; Human miR-pri-miR-199a: 5′-GCCAACCCAGTGTTCAGACTA-3′ and 5′-CCTAACCAATGTGCAGACTA-3′; Human p21/CIP: 5′-CATGTGGACCTGTCACTGTC-3′ and 5′-TTCCTCTTGGAGAAGATCAG-3′; Human pre-miR-16-1: 5′-AGCAGCACAGTTAATACTGGA-3′ and 5′-GCAGCACGTAAATATTGG-3′; Human pre-miR-103: 5′-TCTTTACAGTGCTGCCTTG-3′ and 5′-TCATAGCCCTGTACAATGCT-3′; Human pre-miR-203: 5′-TGGTCCTAAACATTTCACAA-3′ and 5′-TCCAGTGGTTCTTAACAGTTC-3′; Human pre-miR-206: 5′-GCTTCTTTATATCCCCATA-3′ and 5′-CACACACTTCCTTACATTCCA-3′; Human pre-miR-21: 5′-TGTCGGGTAGCTTATCAGAC-3′ and 5′-TGTCAGACAGCCCATCGACT-3′; Human pri-miR-421: 5′-ATCATTGTCCGTGTCTATGG-3′ and 5′-CATTCTGAAGAGAGCTTGGA-3; Human pri-miR-21: 5′-TTTTGTTTTGCTTGGGAGGA-3′ and 5′-AGCAGACAGTCAGGCAGGAT-3′.
Plasmid constructs
p53 K120R and K164R single lysine mutants were provided by the Gu laboratory. The K120/164R double lysine mutant and K120Q mutant were generated using the Single-Site Mutagenesis Kit (Agilent). The Bcl-w expression construct was generated by cloning a full-length Bcl-w cDNA without the 3′UTR into a pCMV5 construct. Bcl-w 3′UTR luciferase constructs were synthesized by cloning Bcl-w full-length 3′UTR (2794 nucleotides) into the firefly luciferase pISO vector, which was purchased from Addgene. Human pri-miR-203 expression construct was generated by amplifying ∼400 bp genomic fragments encoding the pri-miR-203 by following primers: 5′-CTAGGATCCGGCGGTCCCAAGGGC-3′ and 5′-CTAGAATTCGCGCACCCCTGACTGT-3′. The genomic fragment was digested with BamHI and EcoRI and ligated into the pCDNA3.1(+) vector.
In vitro miRNA processing assay
Pri-miR-203 expression plasmid DNA was linearized with XhoI. In vitro transcription reaction was incubated for 2 h at 37°C using the Biotin RNA labelling mix (Roche) according to the manufacturer’s protocol. In vitro transcribed RNAs are treated with DNase I 40U (Roche 10U/μl) at 37°C for 30 min, followed by inhibition of DNAase I by addition of EDTA (final concentration 25 mM). In vitro transcribed RNAs were purified by ethanol/lithium chloride precipitation. In vitro processing was performed with total cell lysate from HCT-116 cells with or without stimulation with 4 μM CPT for 8 h. Cells were washed with ice-cold phosphate saline buffer, harvested and lysed by sonication in the lysis buffer (20 mM Tris–HCl pH 8.0, 100 mM KCl and 0.2 mM EDTA), followed by centrifugation for 15 min at 4°C. Protein concentration of the supernatant was determined by Bradford assay. In vitro processing was performed by incubating 800 ng of in vitro transcribed RNA in the processing buffer (20 mM Tris–HCl pH7.9, 0.1 M KCl, 10% glycerol, 5 mM DTT, 0.2 mM PMSF) supplemented with ATP buffer containing 6.5 mM MgCl2, 10 mM ATP, 200 mM Creatine Phosphate and 20U RNasin Plus RNase inhibitor (Promega) at 37°C for 90 min. In vitro processing products were purified with Phenol-Chloroform extraction, followed by ethanol precipitation. RNA samples were resuspended in 5 μl RNA Loading Buffer. After heat denaturing at 90°C for 10 min and chilled on ice for 10 min, RNA samples were separated by polyacrylamide gel electrophoresis (PAGE). After PAGE, the 7 M Urea-6% Polyacrylamide gel was soaked in SYBR Gold (1:20 000 dilution) in 1 × TBE for 15 min at room temperature, followed by a transfer onto Nylon membrane. Biotinylated-RNA was UV cross-linked to the Nylon membrane for 30 s at 120 000 mJ/cm2 and incubated with a LI-COR blocking buffer with 0.1% SDS and 0.01% Tween for 30 min. Streptavidin staining was performed for 1 h incubation in a blocking buffer, followed by visualization by Odssey (LI-COR).
Luciferase reporter assay
After a 24-h transfection of the reporter construct together with LacZ plasmid as an internal control, cells were supplied with fresh media (DMEM/10% FBS) for 4 h. Cells were then transfected with miR-203 mimic, anti-miR-203, or their corresponding controls. After 24 h of transfection with mimic, anti-miRNAs, or controls, cells were reseeded onto 12-well plates and allowed to attach for 24 h. Luciferase assays were carried out using the Promega’s Luciferase assay system. Luciferase activity was normalized to LacZ activity.
Apoptosis assay
HCT116 cells were treated with 50 μM CPT, 10 μM, Nutlin-3, or vehicle for 16 h followed by counting trypan blue-positive cells. Briefly, both attached and unattached cells were collected, and a minimum of 200 cells for each condition were examined for trypan blue uptake. RNA was extracted from the remaining cells using Trizol (Invitrogen). Cleaved caspase-3 levels were also assessed by western blotting. Activity of caspase-3/7 was measured using Caspase-Glo 3/7 assay (Promega) according to the manufacturer’s protocol.
RNA immunoprecipitation
RNA-IP was performed as described previously (Furuta et al, 2010). All buffers were supplemented with 0.5 U/μl Superase-In (Ambion). HCT116(p53(+)) or HCT116(p53(−)) cells were first fixed with 1% formaldehyde for 10 min. Formaldehyde-fixed cells were then collected and sonicated for chromatin fragmentation and cell lysis. After IP with anti-p53 or anti-c-kit antibody (as a negative control), washing, and elution, RNA was purified with Trizol (Invitrogen). Purified RNA was resuspended in 88 μl of nuclease-free water, 1 μl of Superase-In (Ambion), 1 μl of DNase I (Roche), and 10 μl of 10 × DNase I Reaction Buffer (Roche). The mixture was incubated at 37°C for 25 min, and the RNA was purified again by phenol extraction and isopropanol precipitation (Invitrogen). One microgram RNA was used for a 20-μl cDNA synthesis reaction. Quantitative PCRs were then performed in the iQ5 real-time PCR machine (Bio-Rad). Primer sets used for RNA-IP were Human miR-203: 5′-TCAGAGTCACAGTCAGGGGT-3′ and 5′-GTCTTCCCTGGCAGCAGG-3′.
Statistical analysis
The results presented are the average of at least three experiments performed in triplicate with standard errors. Statistical analyses were performed by ANOVA, followed by Tukey’s multiple comparison test or by Student’s t-test as appropriate, using Prism 4 (GraphPAD Software Inc.). P-values of <0.05 were considered as significant and are indicated with asterisks.
Supplementary Material
Acknowledgments
We thank Dr Vogelstein and Dr Lieberman for sharing HCT116 cells and Dr Roeder for hMOF expression plasmid. We thank Ms Gruttadauria for technical support and all members of the Hata laboratory for helpful suggestions and critical discussion, in particular Dr Blahna for critical reading of the manuscript. This work was supported by grants from the National Institute of Health: HL093154 and HL108317, and the American Heart Association: 0940095N to AH and CA85533 and CA080058 to WG.
Author contributions: JC, BND-D, RK, XJ, JL, RS, and NM designed and performed experiments, and interpreted data. WG provided reagents. GL edited the manuscript. AH designed experiments, interpreted data, and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arbely E, Natan E, Brandt T, Allen MD, Veprintsev DB, Robinson CV, Chin JW, Joerger AC, Fersht AR (2011) Acetylation of lysine 120 of p53 endows DNA-binding specificity at effective physiological salt concentration. Proc Natl Acad Sci USA 108: 8251–8256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo J, Yang G, Huo K, Jiang H, Zhang L, Liu D, Huang Y (2011) microRNA-203 suppresses bladder cancer development by repressing bcl-2 expression. FEBS J 278: 786–792 [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W (2011) The impact of acetylation and deacetylation on the p53 pathway. Protein Cell 2: 456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernandez-Piqueras J, Malumbres M (2008) Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 13: 496–506 [DOI] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282: 1497–1501 [DOI] [PubMed] [Google Scholar]
- Chen HY, Han ZB, Fan JW, Xia J, Wu JY, Qiu GQ, Tang HM, Peng ZH (2012) miR-203 expression predicts outcome after liver transplantation for hepatocellular carcinoma in cirrhotic liver. Med Oncol 29: 1859–1865 [DOI] [PubMed] [Google Scholar]
- Craig VJ, Cogliatti SB, Rehrauer H, Wundisch T, Muller A (2011) Epigenetic silencing of microRNA-203 dysregulates ABL1 expression and drives Helicobacter-associated gastric lymphomagenesis. Cancer Res 71: 3616–3624 [DOI] [PubMed] [Google Scholar]
- Dai C, Gu W (2010) p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med 16: 528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A (2008) SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454: 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A (2010) Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell 39: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Dusenbery BN, Hata A (2010) Mechanisms of control of microRNA biogenesis. J Biochem 148: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Hoover JM, Szostek SA, Espinosa JM (2007) Stimulus-specific transcriptional regulation within the p53 network. Cell Cycle 6: 2594–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J (2010) miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 31: 766–776 [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M (1995) Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592 [DOI] [PubMed] [Google Scholar]
- Hermeking H (2007) p53 enters the microRNA world. Cancer cell 12: 414–418 [DOI] [PubMed] [Google Scholar]
- Junttila MR, Evan GI (2009) p53—a1 Jack of all trades but master of none. Nat Rev Cancer 9: 821–829 [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W (2009) Modes of p53 regulation. Cell 137: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultz D (2005) Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol 67: 225–257 [DOI] [PubMed] [Google Scholar]
- Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B (2009) MicroRNA-125b is a novel negative regulator of p53. Genes Dev 23: 862–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, Candi E (2008) miR-203 represses 'stemness' by repressing DeltaNp63. Cell Death Differ 15: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Li J, Chen Y, Zhao J, Kong F, Zhang Y (2011) miR-203 reverses chemoresistance in p53-mutated colon cancer cells through downregulation of Akt2 expression. Cancer Lett 304: 52–59 [DOI] [PubMed] [Google Scholar]
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W (2012) Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149: 1269–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W (2004) Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci USA 101: 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzna P, Gregar J, Uberall I, Radova L, Prochazka V, Ehrmann J Jr (2011) Changes of microRNAs-192, 196a and 203 correlate with Barrett's esophagus diagnosis and its progression compared to normal healthy individuals. Diagn Pathol 6: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna DJ, McDade SS, Patel D, McCance DJ (2010) MicroRNA 203 expression in keratinocytes is dependent on regulation of p53 levels by E6. J Virol 84: 10644–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Olson EN (2012) MicroRNAs in stress signaling and human disease. Cell 148: 1172–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard DM, Print C, O'Reilly L, Adams JM, Potten CS, Hickman JA (2000) Bcl-w is an important determinant of damage-induced apoptosis in epithelia of small and large intestine. Oncogene 19: 3955–3959 [DOI] [PubMed] [Google Scholar]
- Ru P, Steele R, Hsueh EC, Ray RB (2011) Anti-miR-203 upregulates SOCS3 expression in breast cancer cells and enhances cisplatin chemosensitivity. Genes Cancer 2: 720–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S, Arora S, Majid S, Shahryari V, Chen Y, Deng G, Yamamura S, Ueno K, Dahiya R (2011) Curcumin modulates microRNA-203-mediated regulation of the Src-Akt axis in bladder cancer. Cancer Prev Res (Phila) 4: 1698–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC (2010) Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell 38: 323–332 [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K (2009) Modulation of microRNA processing by p53. Nature 460: 529–533 [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB (2006) Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 24: 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W (2006) Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 24: 827–839 [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM (2006) Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923 [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q et al. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219 [DOI] [PubMed] [Google Scholar]
- Xu Y, Brenn T, Brown ER, Doherty V, Melton DW (2012) Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer 106: 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Samson M, Jegou B, Toppari J (2000) Bcl-w forms complexes with Bax and Bak, and elevated ratios of Bax/Bcl-w and Bak/Bcl-w correspond to spermatogonial and spermatocyte apoptosis in the testis. Mol Endocrinol 14: 682–699 [DOI] [PubMed] [Google Scholar]
- Yu H, Lu J, Zuo L, Yan Q, Yu Z, Li X, Huang J, Zhao L, Tang H, Luo Z, Liao Q, Zeng Z, Zhang J, Li G (2012) Epstein-Barr virus downregulates microRNA 203 through the oncoprotein latent membrane protein 1: a contribution to increased tumor incidence in epithelial cells. J Virol 86: 3088–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.