Abstract
The sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) couples ATP hydrolysis to transport of Ca2+. This directed energy transfer requires cross-talk between the two Ca2+ sites and the phosphorylation site over 50 Å distance. We have addressed the mechano-structural basis for this intramolecular signal by analysing the structure and the functional properties of SERCA mutant E309Q. Glu309 contributes to Ca2+ coordination at site II, and a consensus has been that E309Q only binds Ca2+ at site I. The crystal structure of E309Q in the presence of Ca2+ and an ATP analogue, however, reveals two occupied Ca2+ sites of a non-catalytic Ca2E1 state. Ca2+ is bound with micromolar affinity by both Ca2+ sites in E309Q, but without cooperativity. The Ca2+-bound mutant does phosphorylate from ATP, but at a very low maximal rate. Phosphorylation depends on the correct positioning of the A-domain, requiring a shift of transmembrane segment M1 into an ‘up and kinked position’. This transition is impaired in the E309Q mutant, most likely due to a lack of charge neutralization and altered hydrogen binding capacities at Ca2+ site II.
Keywords: Ca2+-ATPase, Ca2+ binding, intramolecular signalling, membrane protein crystallography, phosphorylation
Introduction
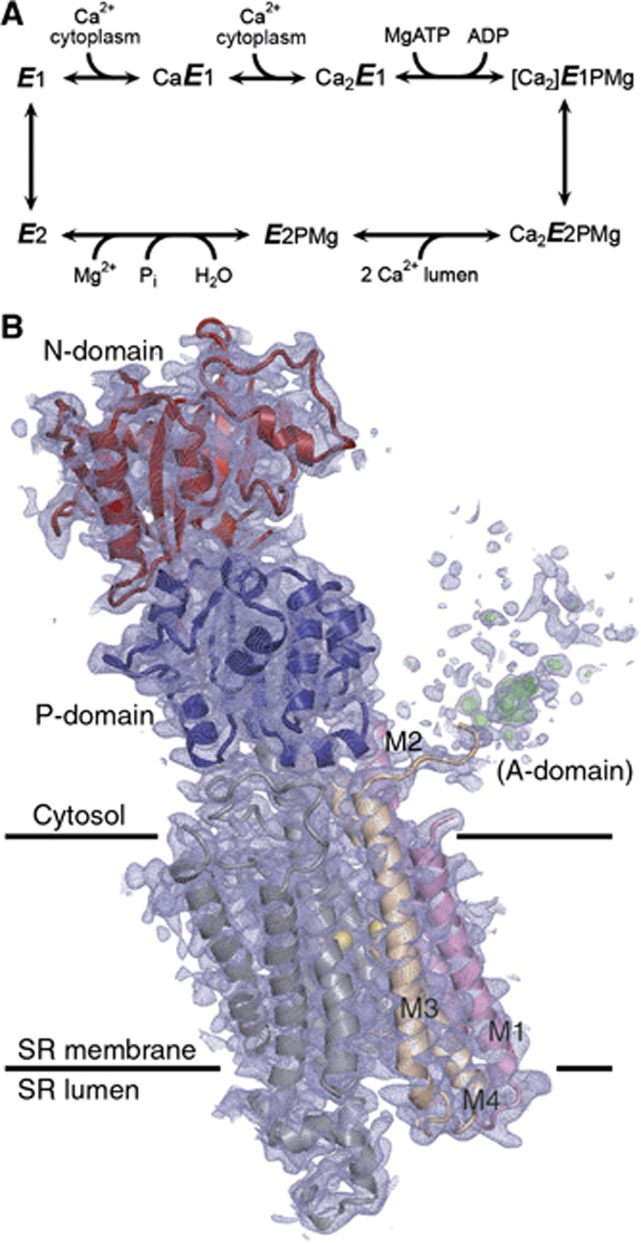
The sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA1a) is a P-type cation pump that couples ATP hydrolysis to the active transport of Ca2+ across the membrane by a Ca2+/H+ exchange mechanism. The ion translocation is achieved by well-defined steps of a reaction cycle (Figure 1A) where the enzyme alternates between E1 and E2 and phosphorylated and dephosphorylated states with inward- and outward-facing ion binding sites of varying ion affinities (de Meis and Vianna, 1979; Inesi, 1985; Mintz and Guillain, 1997). A major conformational change of the enzyme from the E2 to the E1 state allows cooperative binding of two Ca2+ ions in exchange for 2–3 protons at the ion binding sites exposed to the cytosol. Binding of both Ca2+ ions is required for activation of the enzyme for autophosphorylation by ATP (Petithory and Jencks, 1988) to form the [Ca2]E1P state, in which the two Ca2+ ions are tightly occluded at their binding sites. A conformational change of [Ca2]E1P to the Ca2E2P state is associated with the exposure of the Ca2+ sites towards the lumen, with the Ca2+ affinity now dramatically lowered. Ca2+ release to the lumen is accompanied by protonation of the ion binding sites, which activates the autophosphatase activity and leads to dephosphorylation (E2 state) and subsequent translocation of protons to the cytosolic side in the E1 state.
Figure 1.
SERCA reaction cycle and crystal structure of the mutant E309Q in the Ca2E1 state. (A) Ca2+-ATPase reaction cycle. Major conformational changes and substrate binding and dissociation steps are shown. (B) The crystal structure of E309Q. Final electron density after refinement is shown as blue (2mFo-DFc, 0.7 σ) and green (mFo-DFc, 2.7 σ, for clarity only shown for A-domain region) mesh. N-domain depicted in red and P-domain in blue. M1–M2 is shown in pink, M3–M4 in beige, M5–M10 in grey, and Ca2+ ions as yellow spheres.
Structural analysis has demonstrated that SERCA consists of a bundle of 10 transmembrane helices (M1–M10) connected to a cytosolic headpiece comprising three distinct and flexibly connected domains named N (nucleotide binding), P (phosphorylation), and A (actuator) (Toyoshima et al, 2000) (Figure 1B). Side chain oxygens and main chain carbonyls in transmembrane helices M4, M5, M6, and M8 contribute to the coordination of the two Ca2+ ions in the Ca2E1 and [Ca2]E1P states. Crystal structures of SERCA stabilized in well-defined intermediate states of the transport cycle have allowed the major functional events to be described also in structural terms (reviewed in Toyoshima, 2009; Bublitz et al, 2010; and Møller et al, 2010). Central to the energy interchange between ATP hydrolysis and Ca2+ transport are successive displacements of the cytosolic domains relative to one another, which—by their connection to the transmembrane helices—affect the disposition of the Ca2+ binding sites and vice versa. Along with the E2→E1 transition, the A-domain rotates ∼90° around an axis roughly perpendicular to the membrane. During this transition, the M1–M2 segment is displaced downwards and sidewards with respect to the lipid membrane, thereby opening up a cytosolic entrance pathway to the Ca2+ sites (Toyoshima et al, 2013; Winther et al, 2013). The [Ca2]E1P→Ca2E2P transition, a rate-limiting step in the transport cycle (Champeil et al, 1986), structurally corresponds to the displacement of the N-domain away from the P-domain and a back-rotation of the A-domain; a movement, which is transmitted further down to M1–M4, leading to the disruption of the high affinity Ca2+ binding sites and the opening up of a luminal Ca2+ exit pathway (Olesen et al, 2007).
A central unresolved issue of the SERCA mechanism is how the signal is transmitted from the Ca2+ sites in the transmembrane domain to activate the phosphorylation by ATP at the catalytic site in the cytosolic region—sites that are separated by ∼50 Å in the structure. Glu309 in M4 is a conserved and important residue in the transmembrane ion-binding pocket. Glu309 is involved in Ca2+ coordination at site II, the second Ca2+ site to be occupied in a sequential and cooperative mechanism (Inesi, 1985; Andersen and Vilsen, 1994; Toyoshima et al, 2000). Mutation of Glu309 to glutamine abolishes Ca2+ transport activity, and it was reported that the E309Q mutant is unable to undergo the Ca2+-activated phosphorylation from ATP, which requires both Ca2+ sites to be occupied, whereas phosphorylation from Pi, occurring in the absence of Ca2+, is possible (Clarke et al, 1989; Andersen and Vilsen, 1992). Experiments demonstrating a defective Ca2+ occlusion after mutation of Glu309 indicated that Glu309 acts as a gating residue at the cytosolic entrance of the Ca2+ sites (Vilsen and Andersen, 1992, 1998; Inesi et al, 2004). Equilibrium Ca2+ binding measurements performed at low, micromolar Ca2+ concentrations, as well as studies of the Ca2+ dependencies of intrinsic tryptophan fluorescence and inhibition of phosphorylation from Pi led to the conclusion that Ca2+ site II is unable to bind Ca2+ in E309Q, while site I is almost unaffected. The defective Ca2+ binding at site II has widely been assumed to account for the apparent inability of E309Q to undergo phosphorylation from ATP (Andersen and Vilsen, 1992; Skerjanc et al, 1993; Zhang et al, 2000; Inesi et al, 2002, 2004; Lenoir et al, 2006; Montigny et al, 2008).
Mutation of Glu309 furthermore blocks the dephosphorylation of E2P as formed from Pi (Andersen and Vilsen, 1992; Vilsen and Andersen, 1998; Clausen and Andersen, 2010). During the transition from [Ca2]E1P to E2P, Glu309 is displaced towards the luminal side (Olesen et al, 2007), suggesting a possible link between the importance of Glu309 for dephosphorylation of E2P and a function of Glu309 as a ligand at a luminal Ca2+/H+ site in E2P, thus being directly involved also in proton countertransport (Andersen and Vilsen, 1992; Vilsen and Andersen, 1998; Clausen and Andersen, 2010; Musgaard et al, 2011).
To understand the structural and mechanistic basis for the multiple roles of Glu309 in the binding and translocation of Ca2+ as well as in the mediation of long-range effects of Ca2+ binding in the transmembrane region on the phosphorylation and dephosphorylation at the catalytic site, we have determined the crystal structure of the SERCA mutant E309Q in the presence of Ca2+. The structure was obtained from the recombinant enzyme expressed in Saccharomyces cerevisiae and was refined at 3.5 Å resolution. To our surprise, we find that the crystal structure of E309Q contains not only one, but two bound Ca2+ ions at the transmembrane ion sites. Our biochemical assays furthermore show that both Ca2+ ions are bound with reasonable affinity, albeit without cooperativity, and that a phosphorylated state of E309Q accumulates in the presence of Ca2+ and ATP at 25°C. The maximal rate of phosphorylation is, however, strongly reduced as compared with the wild-type enzyme. The structural features of the mutant explain the impaired signal transmission between the Ca2+ sites and the catalytic site and point to the structural determinants of the coupling between the two sites.
Results
Purification and crystallization of SERCA E309Q
Milligram amounts of rabbit SERCA1a E309Q were obtained by overexpression in S. cerevisiae followed by purification by streptavidin affinity chromatography, affinity tag cleavage, and gel filtration (Supplementary Figure S1; Jidenko et al, 2005; Marchand et al, 2008; Cardi et al, 2010). The purified protein was relipidated with 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC). The yield and purity of the E309Q protein was similar to that obtained previously with the recombinant wild-type protein (Jidenko et al, 2005), as evaluated by gel electrophoresis and gel filtration analysis (compare Supplementary Figures S1A and B with figures 15.5 and 15.4, respectively, in Cardi et al, 2010).
The Ca2+-bound E309Q enzyme was crystallized in the presence of Ca2+ and the non-hydrolysable ATP-analogue β,γ-methyleneadenosine 5′-triphosphate (AMPPCP) and using polyethylene glycol (PEG) 6000 and MgCl2 in the crystallization buffer (‘high MgCl2’ condition; Supplementary Figure S2A). Data were collected and scaled at 3.5 Å resolution in space group C2 (Table I). Crystals of the same form were also obtained with CaCl2 and PEG 2000 mono-methyl ether (MME) (‘high CaCl2’ condition; Supplementary Figure S2B). Importantly, the unit cell parameters and the crystal packing of this crystal form differ significantly from those found previously for the wild-type enzyme (Table I; Supplementary Figures S3A and B), and no crystals were obtained with E309Q under the standard conditions used for the wild-type enzyme (see Materials and methods).
Table I. Data collection and refinement statistics.
aNumbers in parentheses refer to the highest resolution shells as indicated.
bWeighted scaling R-factor against published wild-type data (PDB 1T5S) as calculated with Scaleit for the range of 50–7.5 Å (Howell and Smith, 1992).
To examine whether the novel crystal form is dependent on the E309Q mutation, we prepared and crystallized the wild-type enzyme under similar conditions as those used for E309Q. Only the PEG2000-MME/CaCl2 conditions resulted in three-dimensional crystals, but distinctly different in morphology (rhombic, Supplementary Figure S2C). We also reproduced crystals under the previously published conditions for the wild-type enzyme (Sørensen et al, 2004; Jidenko et al, 2005) yielding again rhombic crystals (Supplementary Figure S2D). X-ray diffraction analysis of the wild-type crystals confirmed that the same, previously reported form was reproduced (Sørensen et al, 2004; Jidenko et al, 2005) as also observed for the D351A and P312A mutant forms (Marchand et al, 2008). The E309Q crystals therefore stand out as being specific to the E309Q mutant form (Table I).
The structure of SERCA E309Q
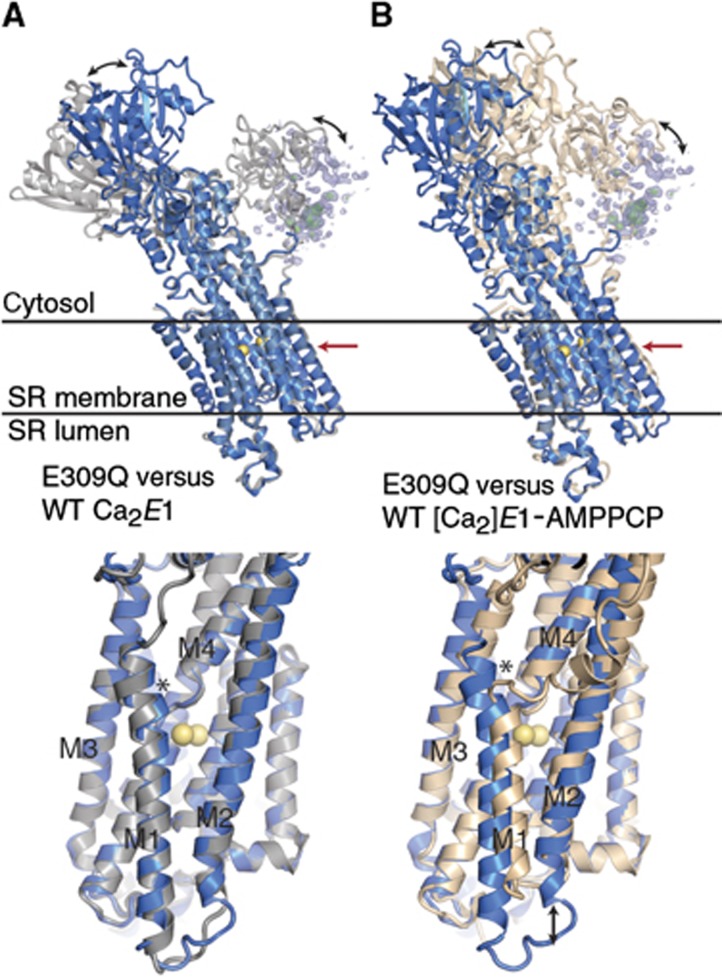
The E309Q structure (Figure 1B) was determined by molecular replacement using individual domains of previously published structures of the wild-type Ca2+-ATPase in the [Ca2]E1P-like state with bound AMPPCP (PDB 1T5S; Sørensen et al, 2004) or the Ca2E1 state without nucleotide (PDB 1SU4; Toyoshima et al, 2000) as search models. Refinement yielded a final model with an Rfree of 26.7% and with electron density maps of relatively high quality for large parts of the structure. Surprisingly, the configuration of the transmembrane helices in the E309Q mutant is similar to that seen in the nucleotide-free Ca2E1 structure of the native enzyme (Figure 2A). Hence, in E309Q, M1 adopts a straight, unkinked position, albeit slightly more shifted towards M4 than that of wild-type Ca2E1 (see asterisk in the bottom part of Figure 2A). In contrast, in the occluded [Ca2]E1P wild-type states (as represented by the [Ca2]E1-AMPPCP, [Ca2]E1-AlF4−-ADP, and [Ca2]E1P-AMPPN structures), the M1/M2 bundle is shifted vertically towards the cytosolic side by roughly two helix turns, and the cytosolic part of M1 is kinked (near Leu61, see arrow and asterisk in the bottom part of Figure 2B) in an ∼90° angle.
Figure 2.
Superposition of the E309Q structure with the structures of wild type in Ca2E1 and [Ca2]E1-AMPPCP states. E309Q depicted in blue, wild-type Ca2E1 (PDB 1SU4; Toyoshima et al, 2000) in grey (A), and wild-type Ca2E1-AMPPCP (PDB 1T5S; Sørensen et al, 2004) in wheat (B). E309Q A-domain electron density is shown in blue (2mFo-DFc, 0.7 σ) and green (mFo-DFc, 2.7 σ) mesh. Black arrows in the top and lower panels indicate significant differences in cytosolic domain and M1/M2 helice bundle positions, respectively. The lower panels show details of the transmembrane domains, viewed in the direction of the red arrows in the top panels. The asterisk in (A) indicates the position shift of M1 and in (B) the position of the M1 kink near Leu61 present in the wild-type[Ca2]E1-AMPPCP state, but absent in the E309Q Ca2E1 structure.
In the E309Q mutant, the position of the N-domain is roughly halfway between that seen in wild-type Ca2E1 and [Ca2]E1P (Figure 2). The electron density for the A-domain was only weakly defined, indicating that the domain assumes a flexible position in the crystals (Figure 1B; Supplementary Figure 3A). Although this led us to omit the A-domain from the final structural model, we were able to determine the overall core position of the domain (indicated by the electron density meshes in Figures 1B and 2) by fitting the published structure of the A-domain from the wild-type Ca2+-ATPase as a rigid body into the contours of the electron density map. The resulting position of the A-domain is some 10 Å closer to the membrane surface relative to its position in the Ca2E1 wild-type structures (Figure 2).
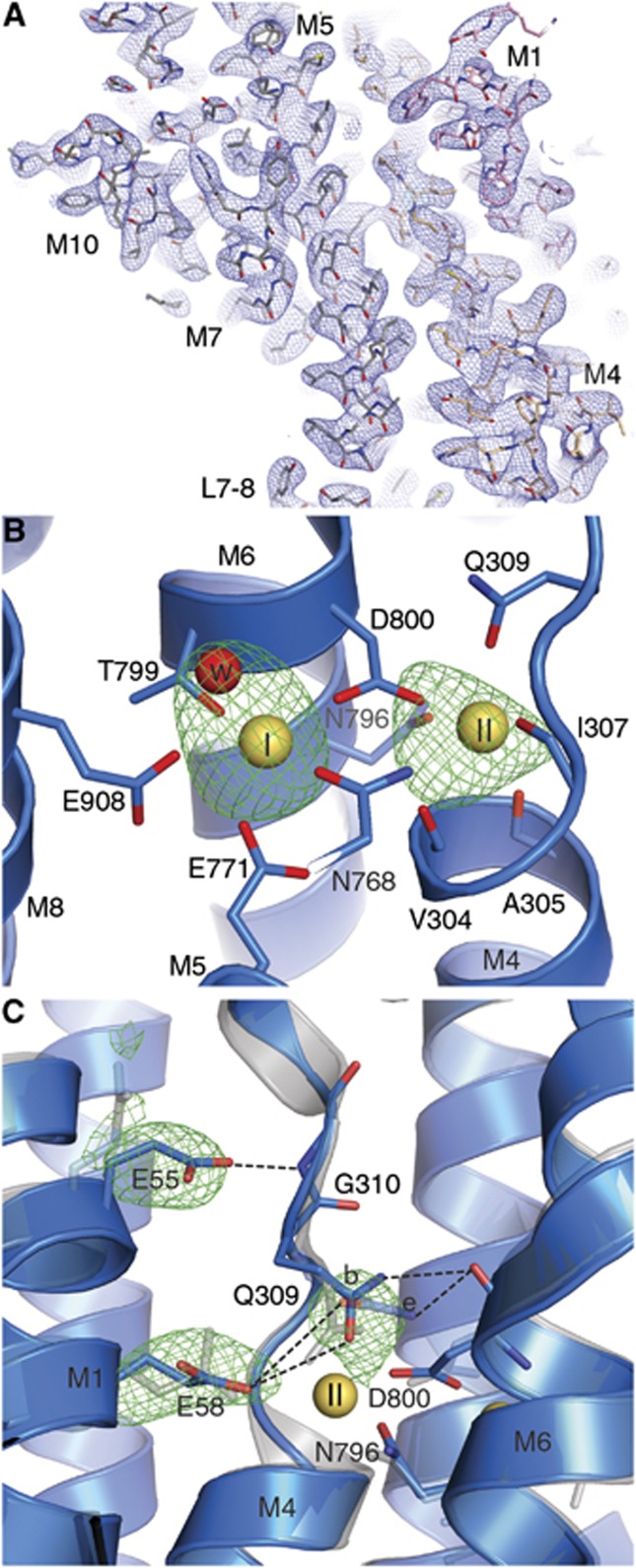
The good quality of the model phases (which were derived from the higher resolution SERCA structures used in molecular replacement), and two strong peaks (⩾8 σ) in Ca2+ omit electron density maps (Figure 3A) allowed us to unambiguously determine that both Ca2+ sites are occupied in the E309Q structure (Figure 3B), and not just site I. Placing Ca2+ ions at the sites, the refined B-factors were 50 Å2 (site I) and 69 Å2 (site II), comparable to the coordinating protein atoms (∼43–107 Å2), whereas refinement of a model with Mg2+ ions converged to unrealistically low values (∼30 Å2). We conclude that the structure contains Ca2+ ions at both sites and at full occupancy.
Figure 3.
Details of the Ca2+ binding sites in the E309Q structure. (A) Representative section of electron density (blue mesh, 2mFo-DFc, 1.0 σ) in the transmembrane region of the E309Q structure. (B) Simulated annealing mFo-DFc map calculated with empty ion binding sites (green mesh, 3.5 σ). The two Ca2+ ions are depicted as yellow spheres, and relevant side chains and main chain carbonyls are shown as stick model. Ca2+-site I contains a water molecule (red sphere), as also found in the wild-type Ca2E1 structure (PDB 1SU4; Toyoshima et al, 2000). (C) Superposition of E309Q (blue) with the Ca2E1 wild-type structure (grey; PDB 1SU4). The green mesh represents mFo-DFc average kicked omit maps for the respective side chains at 3.2 σ. Two possible side chain conformations (denoted as b for bent and e for extended) are modelled for Gln309, dashed lines indicate possible hydrogen bond networks (distance ?4 Å) in E309Q.
The side chain atoms of Gln309 have considerably higher B-factors (∼107 Å2) than those of the other Ca2+ coordinating residues (∼43–73 Å2), a feature not observed for the wild-type glutamate residue. This indicates a higher mobility of this side chain in the mutant, reflecting the fact that it can only coordinate the Ca2+ ion in site II through the amide oxygen, not in a bidentate manner as observed for the wild-type glutamate. Due to the high flexibility of the Gln309 side chain and the limited resolution of the data, it was not possible to unambiguously determine its conformation. However, assuming a flexible side chain and modelling possible side chain conformations, we find a hydrogen bond to be possible between the Gln309 amide group and the main-chain carbonyl oxygen of Asp800, an interaction not likely to occur with the wild-type Gln309 (Figure 3C). Furthermore, Gln309 may form a hydrogen bond to Glu58. Although an accurate distribution of Gln309 side chain conformations cannot be assigned at the present resolution, a test refinement with two side chain conformations at 50% occupancy gave a more reasonable fit to the data (as judged from difference maps) than each of the single solutions. The previously mentioned slight shift in M1 of the mutant, relative to the wild-type structure (Figure 2A, lower panel), furthermore brings Glu55 into hydrogen bonding distance with the main chain amide of Gly310, allowing another unique hydrogen bond in close vicinity to Ca2+ binding site II (Figure 3C).
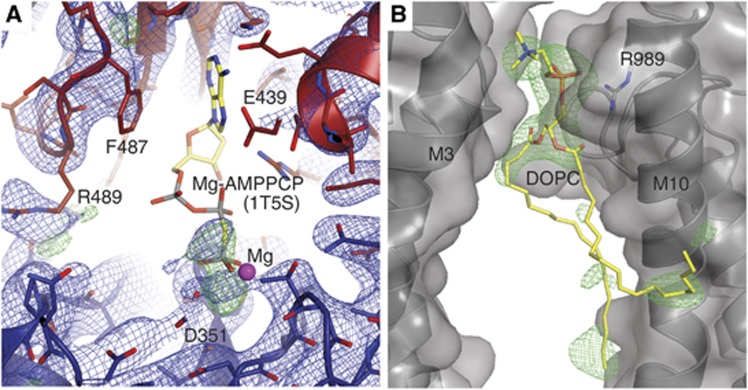
Another important finding is that the nucleotide ligand in the E309Q structure is not ordered (Figure 4A). We observe an electron density peak roughly overlapping with the position of the β- and γ-phosphate in the published [Ca2]E1-AMPPCP structures (when superposed on the P-domain), but the adenosine moiety is invisible in the electron density, unlike the binding pocket itself, indicating disorder. In the [Ca2]E1-AMPPCP structures of native and expressed wild-type SERCA (Sørensen et al, 2004; Jidenko et al, 2005) the N- and P-domains come together and both contribute to sandwich the nucleotide at the Asp351 phosphorylation site. The E309Q structure, on the other hand, displays an open arrangement of the cytosolic domains, with the N-domain still at a distance from the P-domain. Thus, judging from the position of the observed electron density peak, the nucleotide only establishes interactions to the P-domain—a phenomenon that has been observed before in crystals obtained with AMPPCP in high Mg2+ conditions (Winther et al, 2013). In this connection, it should also be noted that the wild-type Ca2+-ATPase has a much lower affinity for AMPPCP than for ATP in the presence of Mg2+ (see also data below in Figure 6), which is known to stimulate ATP binding and inhibit AMPPCP binding (Pang and Briggs, 1977; Picard et al, 2005).
Figure 4.
Details of the nucleotide and lipid binding sites in the E309Q structure. (A) N-domain depicted in red, P-domain in blue, and electron density as blue (2mFo-DFc, 1.0 σ) and green (mFo-DFc, 3.0 σ) mesh (the refined model does not include a nucleotide). The AMPPCP β- and γ-phosphates of the superposed wild-type [Ca2]E1-AMPPCP structure (PDB 1T5S) roughly overlap with the electron density peaks in E309Q. (B) The DOPC binding site, positioned in a groove between two ATPase molecules, with the phosphate moiety of the lipid head group being coordinated by Arg989. Green mesh: mFo-DFc SA-omit map at 3.0 σ. Note that the two labelled helices are from adjacent SERCA molecules in the crystal.
Figure 6.
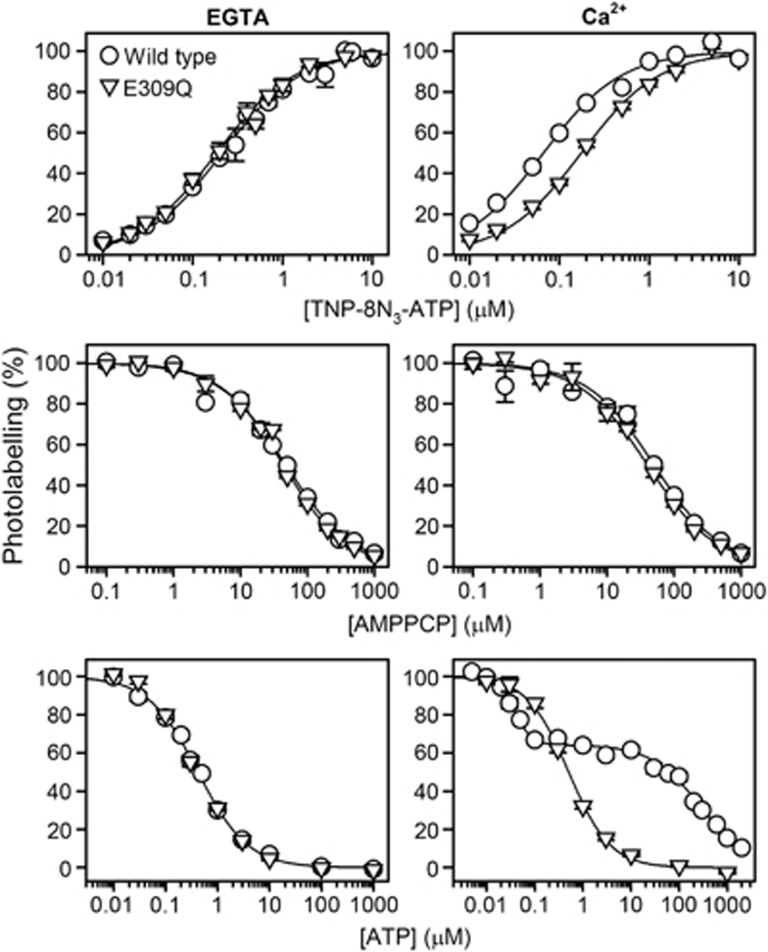
Affinity of the E1 state for TNP-8N3-ATP, AMPPCP and ATP in the absence (left panels) or presence (right panels) of Ca2+. Wild type Ca2+-ATPase (circles) or E309Q (triangles) expressed in COS-1 cells was subjected to TNP-8N3-ATP photolabelling at the indicated concentrations of TNP-8N3-ATP without ATP and AMPPCP (upper panels), or at 3 × the K0.5 for TNP-8N3-ATP with the indicated concentrations of AMPPCP (middle panels) or ATP (lower panels). The affinity constants extracted from the analysis are listed in Table II.
We observe also a lipid molecule (DOPC) between the transmembrane domains of two crystallographically related SERCA molecules of the unit cell. The phosphate moiety of the lipid head group is coordinated by Arg989 in M10 (Figure 4B).
Phosphorylation of E309Q by ATP
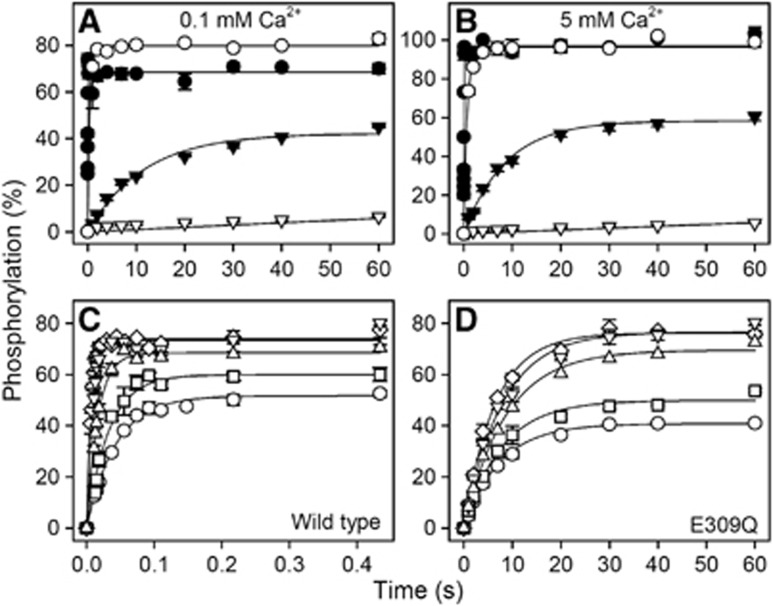
Phosphorylation studies were undertaken with wild type and E309Q expressed in COS-1 cells. Figure 5A shows the time course of phosphorylation in the presence of 5 μM [γ-32P]ATP, 0.1 mM Ca2+, and 25 mM Mg2+ (corresponding to the high Mg2+ concentration used for crystallization) at both 0 and 25°C. The phosphorylation level is indicated relative to the ‘active site concentration’ as determined by the stoichiometric phosphorylation of E2 with 32Pi (Sørensen et al, 1997; Clausen and Andersen, 2010). First, our data confirmed the previous findings that no significant phosphorylation of E309Q from ATP can be detected at 0°C in buffer conditions where the wild-type enzyme phosphorylates rapidly (Clarke et al, 1989; Andersen and Vilsen, 1992; Strock et al, 1998; Zhang et al, 2000). However, when the temperature was raised to 25°C, a considerable amount of phosphoenzyme accumulated for the mutant, albeit at an ∼600-fold lower phosphorylation rate as compared with the wild-type enzyme. Steady-state accumulation of phosphoenzyme at such a low rate is only possible because of the concomitant block of dephosphorylation (E2P→E2) characteristic of E309Q (Andersen and Vilsen, 1992; Clausen and Andersen, 2010). A rather similar time course was observed when the Mg2+ concentration was reduced to a more physiological concentration of 5 mM at otherwise similar buffer conditions (Figures 5C and D). To examine whether the slow phosphorylation of E309Q is caused by insufficient saturation of the Ca2+ site(s), we determined the phosphorylation rate also in 5 mM Ca2+. The 50-fold increase in Ca2+ concentration only led to a moderate 1.4-fold increase in phosphorylation rate for E309Q and a slight increase in the steady-state phosphoenzyme level (Figure 5B). Hence, the reduced phosphorylation rate of E309Q does not appear to result from impaired Ca2+ binding at site II.
Figure 5.
Time course and ATP concentration dependence of phosphorylation by [γ-32P]ATP. (A, B) The time course of phosphorylation of wild-type Ca2+-ATPase (circles) or mutant E309Q (triangles) expressed in COS-1 cells was studied at 0°C (open symbols) or 25°C (closed symbols) by incubation in a medium containing 40 mM MOPS/Tris (pH 7.0), 80 mM KCl, 25 mM MgCl2, 5 μM [γ-32P]ATP, and 0.1 mM CaCl2 (A) or 5 mM CaCl2 (B), followed by acid quench at varying time intervals. (C, D) Phosphorylation of the wild-type Ca2+-ATPase (C) or mutant E309Q (D) was carried out for varying time intervals at 25°C at various [γ-32P]ATP concentrations. The medium contained 40 mM MOPS/Tris (pH 7.0), 80 mM KCl, 5 mM MgCl2, 0.1 mM CaCl2, and the following concentrations of [γ-32P]ATP: circles, 1 μM; squares, 2 μM; triangles pointing upward, 5 μM; triangles pointing downward, 10 μM; diamonds, 20 μM. In each case, the level of phosphorylation is shown relative to the total concentration of Ca2+-ATPase active sites present, as determined by phosphorylation with 32Pi. The lines show the best fits of a monoexponential function, EP=EPmax·(1−e−kt), to the data, giving the rate constants shown in Supplementary Table S1 (for panels A and B) and Supplementary Table S2 (for panels C and D).
Nucleotide binding
Because a possible explanation for the very low phosphorylation rate of E309Q depicted in Figures 5A and B could be a deficiency of [γ-32P]ATP binding, we examined the ATP concentration dependence of phosphorylation (Figures 5C and D). For the wild-type Ca2+-ATPase, the phosphorylation rate increased 6.5-fold upon increasing the ATP concentration from 1 to 20 μM ATP, the Km (ATP) being ∼10 μM (see also Clausen et al, 2001). In comparison, the phosphorylation rate of E309Q increased only 1.2-fold over the same ATP concentration range, thus indicating that the mutant is, in fact, closer to saturation with ATP than the wild type at 1 μM ATP. The observed difference between wild-type and mutant phosphorylation rates was as large as 1000-fold at the nearly saturating ATP concentration of 20 μM. A low saturation level of the ATP binding site of the E309Q mutant can therefore not explain the low phosphorylation rate, either.
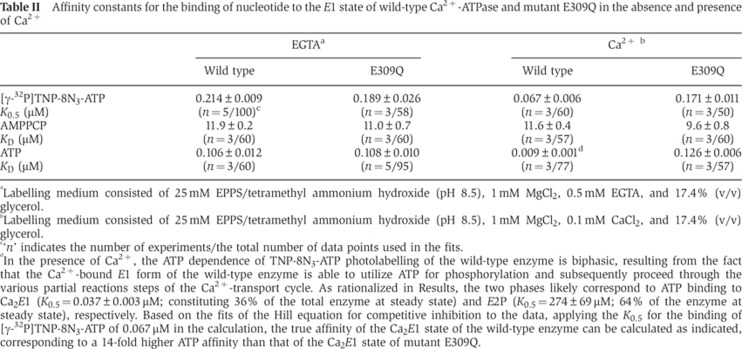
The nucleotide binding properties of the expressed wild-type and mutant enzymes were further examined by studying the nucleotide concentration dependence of [γ-32P]2′,3′-O-(2,4,6-trinitrophenyl)-8-azido-ATP ([γ-32P]TNP-8N3-ATP) photolabelling of Lys492 and the ATP/AMPPCP competitive inhibition thereof using a previously validated method (McIntosh et al, 1996, 1999; Clausen et al, 2011). The experiments were carried out at 1 mM MgCl2 and pH 8.5, either with or without 0.1 mM Ca2+ present. Even without Ca2+ the high pH ensures accumulation of the E1 state in the wild-type enzyme (Forge et al, 1993). It can be assumed that E309Q, like the wild type, resides preferentially in the E1 state at pH 8.5, because the E2↔E1 equilibrium is shifted towards E1 in E309Q as observed previously in proteolysis experiments (Menguy et al, 1998; Inesi et al, 2008). As seen in Figure 6 (left panels) and Table II, there was only a marginal difference between the wild-type enzyme and E309Q with respect to the affinity of the Ca2+-free E1 conformation for [γ-32P]TNP-8N3-ATP, AMPPCP, or ATP. In the presence of 0.1 mM Ca2+ (Figure 6, right panels, and Table II), E309Q displayed a 2.6-fold reduced affinity for [γ-32P]TNP-8N3-ATP relative to the wild type, whereas the affinity of E309Q for AMPPCP was wild type-like. The inhibition of [γ-32P]TNP-8N3-ATP photolabelling by ATP was also studied in medium containing 0.1 mM Ca2+ (lower right panel of Figure 6), although the wild type undergoes a significant phosphorylation when Ca2+ and ATP are present simultaneously. The wild-type enzyme therefore cycles between the various intermediate states shown in Figure 1A, resulting in a biphasic ATP inhibition profile with high- and low-affinity components that likely reflect ATP binding to Ca2E1 and E2P, respectively (Champeil et al, 1988). Although the highest ATP affinity assigned to Ca2E1 is an apparent affinity, influenced by the rate constant of phosphorylation, it is worth noting that it is as much as 14-fold higher than the ATP affinity of E309Q in the presence of Ca2+. It is also remarkable that the ATP affinity of E309Q, unlike that of the wild type, is rather independent of the presence of Ca2+, being ∼0.1 μM both with and without Ca2+. Hence, there appears to be a significant difference between the wild type and E309Q with respect to the ability to respond to Ca2+ binding by a change in the conformation of the nucleotide binding region.
Table II. Affinity constants for the binding of nucleotide to the E1 state of wild-type Ca2+-ATPase and mutant E309Q in the absence and presence of Ca2+.
aLabelling medium consisted of 25 mM EPPS/tetramethyl ammonium hydroxide (pH 8.5), 1 mM MgCl2, 0.5 mM EGTA, and 17.4% (v/v) glycerol.
bLabelling medium consisted of 25 mM EPPS/tetramethyl ammonium hydroxide (pH 8.5), 1 mM MgCl2, 0.1 mM CaCl2, and 17.4% (v/v) glycerol.
c‘n’ indicates the number of experiments/the total number of data points used in the fits.
dIn the presence of Ca2+, the ATP dependence of TNP-8N3-ATP photolabelling of the wild-type enzyme is biphasic, resulting from the fact that the Ca2+-bound E1 form of the wild-type enzyme is able to utilize ATP for phosphorylation and subsequently proceed through the various partial reactions steps of the Ca2+-transport cycle. As rationalized in Results, the two phases likely correspond to ATP binding to Ca2E1 (K0.5=0.037±0.003 μM; constituting 36% of the total enzyme at steady state) and E2P (K0.5=274±69 μM; 64% of the enzyme at steady state), respectively. Based on the fits of the Hill equation for competitive inhibition to the data, applying the K0.5 for the binding of [γ-32P]TNP-8N3-ATP of 0.067 μM in the calculation, the true affinity of the Ca2E1 state of the wild-type enzyme can be calculated as indicated, corresponding to a 14-fold higher ATP affinity than that of the Ca2E1 state of mutant E309Q.
Equilibrium 45Ca2+ binding measurements
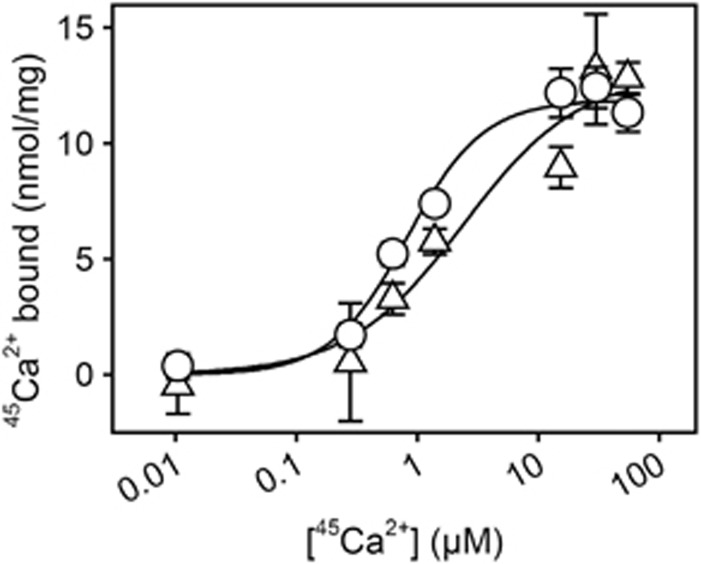
Figure 7 shows the results of equilibrium 45Ca2+ binding measurements carried out with the purified yeast-expressed E309Q mutant reconstituted with DOPC. For comparison, binding of 45Ca2+ by wild-type SERCA from deoxycholate-purified native membranes was also examined. Reliable binding data could be obtained by perfusion of the proteoliposomes deposited on Millipore filters up to a Ca2+ concentration of 55 μM, where E309Q was found to bind 12.8±0.7 nmol Ca2+/mg protein and wild-type SERCA 11.3±0.8 nmol/mg. With respect to wild type, this corresponds to the binding of two Ca2+ per active enzyme monomer, since even in the best preparations at most 60% (corresponding to 5–6 nmol ATPase/mg) is functional, the remainder being present in an inactive and aggregated state as evidenced by measurements of phosphorylation capacity and HPLC analysis of detergent solubilized ATPase (Andersen et al, 1986; le Maire et al, 2008). For the E309Q mutant expressed in yeast, the HPLC data also indicate the presence of inactive, aggregated ATPase (Supplementary Figure S1A). Because one binding site would correspond to 9.1 nmol/mg protein in a 100% pure and active enzyme preparation (the molecular mass is 110 kDa), and there is inactive protein present in significant amounts, the finding of 12.8±0.7 nmol bound Ca2+/mg is in good accordance with a similar binding stoichiometry of two Ca2+ per active Ca2+-ATPase for the wild type and the mutant. By fitting a Hill function to the binding data in Figure 7, the apparent affinity for Ca2+ of E309Q was found to be ∼3-fold lower than that of the wild type. The Hill coefficient for Ca2+ binding was ∼1.4 for the wild type, in accordance with cooperative binding (Inesi, 1985; Mintz and Guillain, 1997), while for the E309Q mutant cooperativity seems to be lost as indicated by a Hill coefficient close to one (Figure 7).
Figure 7.
Equilibrium 45Ca2+ binding measurements. 45Ca2+ binding to deoxycholate-purified native sarcoplasmic reticulum vesicles and to DOPC-reconstituted yeast-expressed E309Q was measured by filtration. Filters, deposited with the protein samples, were perfused with 20 ml medium containing 100 mM MOPS/Tris (pH 7.2), 100 mM KCl, 1 mM MgCl2, 55 μM CaCl2 with radiolabelled 45Ca2+, and various concentrations of EGTA, giving the free 45Ca2+ concentrations indicated on the abscissa. Circles, wild type; triangles, E309Q. The lines show the best fits of the Hill equation, Y=Ymax · [Ca2+]h/(K0.5h+[Ca2+]h), to the data giving the following affinity constants (K0.5) and Hill coefficients (h): wild type, K0.5=0.82±0.16 μM, h=1.38 (n=2, 28 data points); E309Q, K0.5=2.42±1.96 μM, h=0.91 (n=2, 28 data points). In each Hill fit, the Ymax was defined by the amount of 45Ca2+ bound following perfusion with EGTA-free medium (hence, with 55 μM free Ca2+).
Discussion
Although ATP binds to SERCA throughout the whole reaction cycle, only the Ca2+-bound form undergoes phosphorylation from ATP. In wild-type SERCA, the occupied transmembrane Ca2+ sites thus communicate with the catalytic site to enable the transfer of the γ-phosphate of ATP to Asp351, and the E309Q mutation disrupts this coupled activity. By structural analysis, we found that the mutant has both transport sites occupied by Ca2+ (Figure 3B) and the transmembrane domain possesses a conformation similar to that observed with the wild type in the absence of nucleotide (PDB 1SU4; Toyoshima et al, 2000) (Figure 2), despite the presence of nucleotide during crystallization and a partial electron density signal in the nucleotide binding site. On the other hand, the cytosolic domains are arranged in positions different from those seen in the wild-type enzyme (Figure 2), as considered further below. Our 45Ca2+ equilibrium binding measurements confirm that the mutant is able to bind two Ca2+ ions, and the affinity is only three-fold lower than that of the wild type, but with a lost cooperativity (Figure 7). This contrasts with previous analyses suggesting that E309Q binds only one Ca2+ ion, at site I (Skerjanc et al, 1993; Strock et al, 1998; Zhang et al, 2000). Furthermore, we find that E309Q is able to slowly form a stable phosphoenzyme by phosphorylation with ATP (Figure 5); again in contrast to conclusions of previous studies (Clarke et al, 1989; Andersen and Vilsen, 1992; Strock et al,1998; Zhang et al, 2000), but consistent with the retention of a residual communication with the phosphorylation site, as a consequence of the occupation of both Ca2+ sites (Petithory and Jencks, 1988). The reason why the previous phosphorylation studies failed to detect phosphorylation of E309Q from ATP is that they only tested phosphorylation under ice-cold buffer conditions, where the reaction is too slow to allow the accumulation of measureable amounts of phosphoenzyme. We find that the maximum phosphorylation rate of the mutant, as detected at saturating Ca2+ and ATP concentrations, is three orders of magnitude lower than that of the wild-type enzyme (Figure 5), thus clearly attesting to a defective signal transmission between the Ca2+ sites and the catalytic site. The severe inhibition of ATP utilization in E309Q appears to relate to the straight M1 helix and the disordered nature of the A-domain in the E309Q structure, reflecting an increased flexibility of the A-domain. Thus, the formation of a compact structure of the cytoplasmic A-, P-, and N-domains, enclosing the phosphorylation site and shielding it from the bulk solvent, as displayed by the wild-type enzyme in the Ca2E1/Ca2E1P states (Sørensen et al, 2004; Olesen et al, 2007), is likely impeded in the mutant. This leads to a diminished ability to attain and/or stabilize the catalytic [Ca2]E1P-ADP transition state, thus lowering the Vmax for phosphorylation.
In the E309Q crystal structure, the Gln309 side chain can only take part in coordinating Ca2+ II with the amide oxygen. This is reflected in a higher degree of flexibility than in the wild-type glutamate, and with several alternative conformations of the mutated side chain being likely, as indicated by high B-factors and a test model encompassing two alternating conformations (Figure 3C). These features probably cause the incompetence of the mutant with respect to Ca2+ occlusion (Vilsen and Andersen, 1992; Inesi et al, 2004). Our analysis of the immediate surroundings of the Gln309 side chain reveals that the possible side chain conformations would allow hydrogen bonding interactions with the main chain carbonyl oxygen of Asp800 in M6, another central Ca2+ liganding residue, and Glu58 in M1 (Figures 3B and C). Also, the shifted position of M1 in E309Q would allow a hydrogen bonding interaction between Glu55 (M1) and the carbonyl oxygen of Gly310 (Figure 3C); an interaction absent in the wild-type structure.
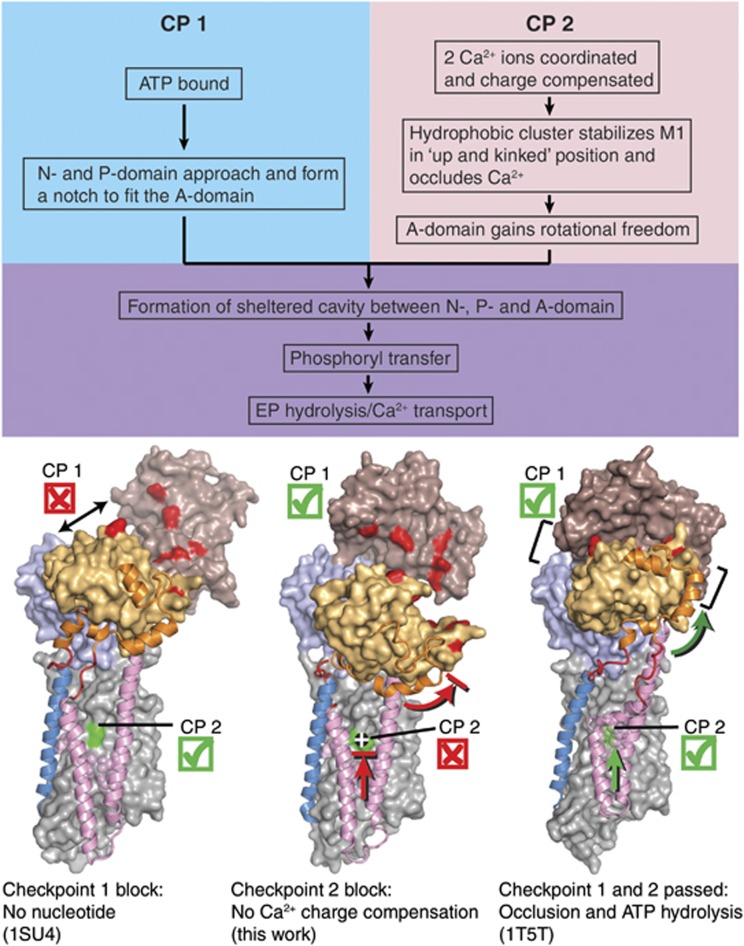
We can now devise a schematic model (Figure 8) to address the key question of our study: how is the Ca2+-binding signal transmitted across the ∼50 Å distance from the Ca2+ sites in the transmembrane domain to the catalytic site in the cytosolic region? First we note that in the absence of nucleotide, the N-, P-, and A-domains are not interconnected, neither in the wild-type enzyme nor in the E309Q mutant (Figure 8, ‘Checkpoint 1’). For the formation of a solvent shielded catalytically competent site after ATP binding, the A-domain has to be properly positioned at the N-P domain interface, and the M1–M2 segment must undergo an ‘upward’ translation (in the direction towards the cytosol). This involves the formation of a kink in M1 and leads to the bound Ca2+ becoming occluded via a hydrophobic cluster of Phe57, Val62, Leu65, Ile94, Ile97, and Leu98 that pack against the bent Cβ and Cγ atoms of Glu309 (Sørensen et al, 2004; Toyoshima and Mizutani, 2004). Furthermore, for stable occlusion to occur at the Ca2+ sites near the M1 kink, overall neutralization of charges is required, which is ensured by the negatively charged Glu309 side chain (‘Checkpoint 2’ in Figure 8). The latter is not fulfilled in E309Q due to the residual positive charge of the Ca2+ ion at site II. In addition, an alternative hydrogen-bonding pattern in the mutant, involving Gln309, Gly310, Glu55, Glu58, and Asp800, could counteract the M1–M2 movements necessary for Ca2+ occlusion and cooperative interaction. Asp800 may be instrumental for the cooperative binding pattern of the wild type, because it coordinates both Ca2+ ions, whereas for E309Q it may instead be affected by engaging in a hydrogen bonding interaction with Gln309, which could negatively influence cooperativity. The rotational freedom necessary for proper positioning of the A-domain lateral to the nucleotide binding site is also compromised, rendering catalytically competent nucleotide coordination and solvent shielding a less favoured and infrequent event. The result is a strongly reduced phosphorylation rate, no increase in affinity for ATP in the presence of Ca2+ (Figure 6), and, as indicated by our data with AMPPCP, a disorder of the adenosine moiety of the nucleotide in E309Q (Figure 4A). ATP binding occurs but is not coupled to Ca2+ binding, being rather reminiscent of the ATP binding to E2-thapsigargin (Jensen et al, 2006).
Figure 8.
Scheme for the structural requirements of SERCA for Ca2+-dependent activation of phosphorylation by ATP. The three structures shown are (from left to right): wild-type Ca2E1 (PDB 1SU4; Toyoshima et al. 2000), E309Q Ca2E1 (AMPPCP) (present work), and wild-type [Ca2]E1-AlF4−-ADP (PDB 1T5T; Sørensen et al, 2004). Requirements at both ‘checkpoints’, CP1 and CP2, have to be met; otherwise, the enzyme activity will be strongly lowered or completely abolished. In each structure, M1 and M2 are shown in pink cartoon, M3 in blue cartoon, the A-M1 and A-M3 linker regions in red cartoon, and the N-terminal part of the A-domain (amino acids 1–46) in orange cartoon, to aid in visualizing the upward motion of the M1/M2 bundle as well as the rotational motion of the A-domain upon nucleotide binding to the wild type. The remainder of the ATPase molecule is shown in surface representation, with the N-domain in brown, the P-domain in light blue, the A-domain in light orange, and the transmembrane domain in light grey. Some of the amino acids involved in interdomain contacts and Ca2+ binding are highlighted in the surface representation by red and green colouring, respectively.
Additionally, it is of interest to consider the central core of interacting, conserved residues that connect the Ca2+ binding sites with the phosphorylation site (Scarborough, 2002; Møller et al, 2010). These include the residues in M4 stretching from Ca2+ site II (Glu309) to the phosphorylated residue in the P-domain, Asp351, as well as the residues in M5 stretching from Ca2+ site I (Glu771) to the C-terminal part of the P-domain containing the long (699–721) signature motif of P-type ATPases. The central core forms what appears to be a plastic structure that can be moulded in various ways during transport. Via this direct connection between the transport sites and the phosphorylation site, the central core may enable the cooperatively interacting Ca2+ ions to transmit the long-distance signal that triggers the reaction of Asp351 with ATP. In line with this view, molecular dynamics simulations have indicated that the critical kinetic factor involved in N- and P-domain movements closing the nucleotide binding pocket of the wild type (after addition of Ca2+ and ATP to the E1 state) is Ca2+ binding rather than nucleotide binding (Espinoza-Fonseca and Thomas, 2011). Thus, it is likely that the half-open N-domain position seen in E309Q relative to the [Ca2]E1-AMPPCP state of native SERCA reflects a defective signal transmission along the central core in response to binding at Ca2+ site II, with Glu309 being involved as a critical mediator of the signalling.
The positioning of the A-domain is also an essential aspect of the dephosphorylation of E2P, where the conserved T181GES motif of the A-domain participates in catalysis of aspartyl phosphate hydrolysis (Clausen et al, 2004; Olesen et al, 2004). Therefore, the A-domain displacement towards the membrane seen in the E309Q structure is likely the reason why the E2P dephosphorylation, as well, is a severely retarded reaction step in the E309Q mutant (Andersen and Vilsen, 1992; Clausen and Andersen, 2010). Analogous to our model for the realization of a phosphorylation- and occlusion-competent E1 conformation presented in Figure 8, one can imagine a scenario in E2P where, akin to checkpoint 1 (ATP binding), the proper positioning of the T181GES motif of the A-domain is required not only to mediate the dephosphorylation by placing the water molecule in the correct position for the phosphoenzyme hydrolysis, but also to bring the cytosolic domains together and carry out the necessary rearrangement of transmembrane helices M1–M3 (connected directly to the A-domain) leading to the release of Ca2+ from Ca2E2P and subsequent protonation of the carboxylate groups at the transport sites (including also that of Glu309). In turn (akin to checkpoint 2, Ca2+ binding/occlusion), the luminal protonation of Glu309 in the E2P state is a prerequisite for charge neutralization at the transport sites, acting as the instigator of the signalling from the transport sites to the catalytic site, these events being blocked in the E309Q mutant, as mirrored by the severely retarded dephosphorylation.
To sum up, we find that the structural features agree very well with the functional properties of the E309Q mutant. Thereby, our study also provides information on the basis for the critical role that Glu309 plays for the mechanistic aspects of the signal transmission between the transmembrane ion binding sites and the catalytic domains in wild-type SERCA. The signalling depends critically on the range of movement of the A-domain, which is in turn determined by the hydrogen bonding pattern and charge neutralization around Glu309.
Materials and methods
Purification of protein for crystallization
The purification of the E309Q mutant Ca2+-ATPase (SERCA1a isoform) expressed in Saccharomyces cerevisiae was performed as described for the recombinant wild-type enzyme in Cardi et al (2010), made feasible by the presence of a biotin acceptor domain (biotinylated in vivo in the yeast) linked to the C-terminus of the Ca2+-ATPase by a thrombin cleavage site. For each preparation, a typical yield of ∼1 mg pure E309Q protein was available for crystallization trials, and was prepared contained in ∼100 μl of a buffer composed of 100 mM MOPS/Tris (pH 6.8), 80 mM KCl, 20% (v/v) glycerol, 1 mM CaCl2, 1 mM MgCl2, and ∼36 mg/ml octaethylene glycol monododecyl ether (C12E8). To relipidate the protein, the sample was added to a thin layer of N2-dried DOPC to reach a C12E8/DOPC ratio of 3:1 (w/w), vortexed gently, and equilibrated overnight at 4°C. Prior to crystallization trials, the relipidated and C12E8-solubilized Ca2+-ATPase was supplemented with CaCl2, MgCl2, and AMPPCP to final concentrations of 10, 3, and 1 mM, respectively, to accumulate a Ca2+-saturated nucleotide-bound E1 state. The sample was centrifuged at 20 000 g and 4°C for 30 min and the supernatant used for crystallization.
Protein crystallization
Crystallization trials were performed using the vapour diffusion technique. Each crystallization drop contained a mixture of the concentrated, DOPC-relipidated, and C12E8-solubilized Ca2+-ATPase solution and the reservoir solution, typically in a 1:1 ratio. Initial broad spectrum screening in low-volume (0.28 μl) crystallization drops was carried out using a liquid-handling Mosquito robot (TTP Labtech, Melbourn, Hertfordshire, UK), testing various concentrations of different PEGs, salts, and organic solvents. Promising hits were subjected to grid screens using higher volume (1–4 μl) hanging crystallization drops manually pipetted onto siliconized glass cover slides (Hampton Research, Aliso Viejo, CA) sealed to the reservoir with microscopy immersion oil. The crystals were cryoprotected by addition of an extra 10–20% glycerol to the reservoir solution and pre-cooling to 4°C for 24–48 h and were then mounted in either LithoLoops (Molecular Dimensions, Newmarket, Suffolk, UK) or nylon CryoLoops (Hampton Research, Aliso Viejo, CA) before flash-cooling in liquid nitrogen.
For E309Q, the final content of the crystallization drops during set-up was as follows: 4–5 mg/ml mutant protein, ∼18 mg/ml C12E8, ∼6 mg/ml DOPC, 50 mM MOPS/Tris (pH 6.8), 40 mM KCl, 15% (v/v) glycerol, 0.5 mM AMPPCP, and either 5.5% (w/v) PEG 6000, 5 mM CaCl2, 27 mM MgCl2, and 3% (v/v) 2-methyl-2,4-pentanediol (‘high MgCl2’ condition), or 7.5% (w/v) PEG 2000 MME, 55 mM CaCl2, 1.5 mM MgCl2, and 1.5% (v/v) t-butanol (‘high CaCl2’ condition).
For SERCA1a wild type, the final content of the crystallization drops during set-up was as follows: ∼5 mg/ml protein, ∼18 mg/ml C12E8, ∼6 mg/ml DOPC, 0.5 mM AMPPCP, 50 mM MOPS/Tris (pH 6.8), 40 mM KCl, 15% (v/v) glycerol, and either 4.5% (w/v) PEG 2000 MME, 55 mM CaCl2, 1.5 mM MgCl2, and 1% (v/v) t-butanol (‘high CaCl2’ condition), or 3.5% (w/v) PEG 6000, 5 mM CaCl2, 100 mM sodium acetate, and 2% (v/v) t-butanol (wild-type [Ca2]E1 AMPPCP crystallization conditions similar to those reported in Sørensen et al, 2004 and Jidenko et al, 2005).
Diffraction data collection and structure refinement
X-ray diffraction data were collected at the Swiss Light Source beamline X06SA (Villigen, Switzerland) and at the European Synchrotron Radiation Facility beamline ID23-1 (Grenoble, France). The data were processed and scaled using the XDS package (Kabsch, 1993), and structure determination by molecular replacement was carried out with PHASER, using the PHENIX AutoMR function (McCoy et al, 2007; Adams et al, 2010). Low Z-scores for the rotation (RFZ) and translation (TFZ) searches (5.9 and 4.0, respectively) and a high number of packing clashes clearly ruled out the nucleotide-bound Ca2E1 state search model (PDB 1T5S). The nucleotide-free Ca2E1 model (PDB 1SU4) yielded higher Z-scores (RFZ=7.4, TFZ=10.6), but was still unable to be packed without clashes into the new unit cell. We then proceeded to delete individual domains, and deletion of both the N- and the A-domain yielded the highest Z-scores (RFZ=8.4, TFZ=14.3) and a log-likelihood gain (LLG) value of 161. The N-domain was then placed manually into the extra electron density obtained from the molecular replacement procedure, while the A-domain never appeared unambiguously from the electron density maps. The model was built in Coot (Emsley et al, 2010) and refinement was performed with PHENIX (Adams et al, 2010) using rigid body refinement in initial rounds, followed by refinement of atomic coordinates, B-factors and TLS motion (five TLS groups: N-domain, P-domain, M1/M2-, M3/M4-, and M5-M10 helix bundles). Even though the exact position of the A-domain could not be deduced from the electron density maps, including it in to the model (derived from a previously solved structure and placed manually as a rigid body to match the electron density cloud) during refinement (rigid body and B-factors only) and omit map calculation, led to a considerable improvement of the model phases (most likely for low-resolution structure factors in particular) and thereby of the resulting maps. For this reason, all electron density maps, except for those in Figures 1 and 2, were calculated from models including the A-domain. Unbiased difference Fourier maps (simulated annealing omit maps or average kicked omit maps) were calculated with PHENIX to reveal the presence or absence of expected ligands or to evaluate the position of side chains. The atomic coordinates and experimental data (code 4NAB) have been deposited in the Protein Data Bank ( www.pdb.org).
Protein expression in mammalian cell culture
For expression of the Ca2+-ATPase in COS-1 cells to be used in the phosphorylation and photolabelling experiments, cDNA encoding the wild-type or mutant enzyme was inserted into the expression vector pMT2 (Kaufman et al, 1989). COS-1 cells were transfected using the calcium phosphate precipitation method (Chen and Okayama, 1987), and microsomal vesicles containing the expressed Ca2+-ATPase were isolated by differential centrifugation (Maruyama and MacLennan, 1988). The concentration of the expressed Ca2+-ATPase was determined by measurement of the maximum capacity for phosphorylation with Pi in the presence of dimethyl sulfoxide (‘active site concentration’; Sørensen et al, 1997).
Measurements of phosphorylation by [γ-32P]ATP
The phosphorylation by [γ-32P]ATP at various buffer and temperature conditions (detailed in the figure legends) of wild type and mutant Ca2+-ATPase expressed in COS-1 cells was carried out either by manual mixing or using a Bio-Logic quench-flow module QFM-5, as described (Sørensen et al, 2000). Acid quenching of the phosphorylated enzyme was performed with 0.5–2 volumes of 25% (w/v) trichloroacetic acid containing 100 mM H3PO4. The acid-precipitated protein was washed by centrifugation and subjected to SDS-polyacrylamide gel electrophoresis in a 7% polyacrylamide gel at pH 6.0 (Weber and Osborn, 1969), and the radioactivity associated with the separated Ca2+-ATPase band was quantified by imaging with a Cyclone Storage Phosphor System (Perkin Elmer, Waltham, MA). Background phosphorylation levels, determined in parallel experiments by adding an excess of EGTA, were subtracted from all data points.
Nucleotide affinity determination by [γ-32P]TNP-8N3-ATP photolabelling
The synthesis of the [γ-32P]TNP-8N3-ATP photolabel, its application as a specific photolabel of the recombinant Ca2+-ATPase expressed in COS-1 cells, the competitive inhibition by ATP of [γ-32P]TNP-8N3-ATP photolabelling, the quantification of 32P-labelled bands by electronic autoradiography following SDS–PAGE, and the analysis of the results were carried out using previously established procedures (Seebregts and McIntosh, 1989; McIntosh et al, 1996) with the modifications to the photolabelling set-up detailed in Clausen et al (2011). The medium used for the photolabelling contained 25 mM EPPS/tetramethyl ammonium hydroxide (pH 8.5), 1 mM MgCl2, either 0.5 mM EGTA or 0.1 mM CaCl2, 17.4% (v/v) glycerol, 4–5 nM Ca2+-ATPase and [γ-32P]TNP-8N3-ATP without or with ATP or AMPPCP at the concentrations indicated in the figures. The analysis of the data obtained from the ATP or AMPPCP inhibition of TNP-8N3-ATP photolabelling was generally based on the Hill equation for inhibition, Y=Ymax·(1−[ATP]h/(K0.5h+[ATP]h)), in which Y and Ymax are the amount of photolabelled Ca2+-ATPase and the maximal value, respectively, K0.5 is the concentration of ATP giving half-maximum effect, and h is the Hill coefficient (varying between 0.86 and 1.01 for the present data). The ‘true’ dissociation constant, KD, for ATP binding was calculated from the measured K0.5 values using the validated equation for competitive inhibition (McIntosh et al, 1996). In one case, namely the wild-type data for the inhibition by ATP of TNP-8N3-ATP photolabelling in the presence of Ca2+, the data displayed a biphasic inhibition profile, and the analysis was therefore based on a two-component Hill equation for inhibition, Y=Ymax1·(1−[ATP]h1/(K0.5,1h1+[ATP]h1))+Ymax2·(1−[ATP]h2/(K0.5,2h2+[ATP]h2)), in which the parameters are defined as above but assuming two reaction intermediates.
Equilibrium 45Ca2+ binding measurements
The yeast-expressed E309Q mutant, eluted in 50 mM Tris (pH 7.0), 150 mM NaCl, 20% (v/v) glycerol, 2.5 mM CaCl2, 1 mM β-mercaptoethanol, and 0.5 mg/ml n-dodecyl-β-D-maltopyranoside (DDM), was concentrated to ∼1 mg/ml, followed by reconstitution into proteoliposomes by addition of 2.5 mg/ml DOPC, solubilized by 5 mg/ml DDM. The detergent was then removed by addition of 200 mg Bio-beads per mg detergent and gentle stirring for 3 h, followed by resuspension of the reconstituted sample in 100 mM MOPS/Tris (pH 7.2), 100 mM KCl, and 1 mM MgCl2. For the 45Ca2+ binding experiments, 220 μg samples of either deoxycholate-purified native SERCA vesicles or reconstituted E309Q were deposited on double layers of 0.45 μM Millipore filters (Merck Millipore, Darmstadt, Germany). The filters were perfused with 20 ml of 100 mM MOPS/Tris (pH 7.2), 100 mM KCl, and 1 mM MgCl2, containing in addition 55 μM CaCl2 with radiolabelled 45Ca2+ and various concentrations (0, 25, 40, 70, and 1000 μM) of EGTA. Ca2+ binding was calculated from the difference between the radioactive counts in the upper and lower Millipore filters.
The validity of the reconstitution procedure was checked by subjecting native SERCA in sarcoplasmic reticulum vesicles, isolated from rabbit skeletal muscle, to the same DOPC/DDM reconstitution procedure, with DOPC:protein ratios varying from 2.5:1 to 10:1 (w/w). The same Ca2+ binding level as with the non-reconstituted sarcoplasmic reticulum vesicles was only obtained at the 2.5:1 reconstitution ratio, presumably because the Millipore filters became leaky to proteoliposomes with higher lipid contents. Furthermore, the addition of ATP did not give rise to vesicular 45Ca2+ uptake, consistent with the absence of sealed vesicles which, as a result of asymmetric ATPase incorporation, could have masked some of the binding sites.
Analysis of functional data and statistics
The functional experiments were conducted at least three times on independent microsomal preparations, unless otherwise stated in the figure legends and data tables. Average values are shown in Figures 5, 6, 7, error bars are seen when larger than the size of the symbols and correspond to SEM. The complete data sets (including all experimental data points before averaging) were analysed by non-linear regression using the SigmaPlot program (SPSS, Inc.), giving the lines in the figures and the rate constants, affinity constants, and SE listed in the figure legends. The equations used and the total number of data points included in the fits, as well as the number of independent experiments (n), are detailed in the figure legends.
Supplementary Material
Acknowledgments
We are very grateful to Dr Marie Jidenko who initiated this work when she was a PhD student at University Paris-Sud. A special appreciation goes to the late Bitten Holm for her expert technical assistance given in this and other projects performed in the JVM laboratory. We thank Karin Kracht and Lene Jacobsen (Department of Biomedicine, Aarhus University, Denmark) and Jesper L Karlsen and Anna Marie Nielsen (Department of Molecular Biology and Genetics, Aarhus University, Denmark) for expert technical assistance, and Dr Joseph Lyons and Dr Claus Olesen (Department of Molecular Biology and Genetics, Aarhus University, Denmark) for discussions. Beam time and support at the Swiss Light Source (Villigen, Switzerland) and the European Synchrotron Radiation Facility (Grenoble, France) are greatly acknowledged. This work was supported by grants from the Lundbeck Foundation (to JDC), the Danish Medical Research Council (to JPA), the Novo Nordisk/Vilhelm Pedersen Foundation (to JPA and JVM), the Danish Council for Strategic Research (to MB, JVM, and PN), the Agence Nationale de la Recherche (to MlM, CJ, and CM), the Domaines d'Intérêt Majeur Maladies Infectieuses Région Ile de France (to MlM, CJ, and CM), and the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INSB-05-01 (to MlM, CJ, and CM).
Author contributions: JDC, MB, BA, and CM performed the experiments. JDC, MB, BA, CM, CJ, JVM, PN, JPA, and MlM participated in the experimental design. JDC, JPA, and MB wrote the first draft of the manuscript, which was revised by all authors in collaboration.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JP, Vilsen B, Nielsen H, Møller JV (1986) Characterization of detergent-solubilized sarcoplasmic reticulum Ca2+-ATPase by high-performance liquid chromatography. Biochemistry 25: 6439–6447 [DOI] [PubMed] [Google Scholar]
- Andersen JP, Vilsen B (1992) Functional consequences of alterations to Glu309, Glu771, and Asp800 in the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem 267: 19383–19387 [PubMed] [Google Scholar]
- Andersen JP, Vilsen B (1994) Amino acids Asn796 and Thr799 of the Ca2+-ATPase of sarcoplasmic reticulum bind Ca2+ at different sites. J Biol Chem 269: 15931–15936 [PubMed] [Google Scholar]
- Bublitz M, Poulsen H, Morth JP, Nissen P (2010) In and out of the cation pumps: P-Type ATPase structure revisited. Curr Opin Struct Biol 20: 431–439 [DOI] [PubMed] [Google Scholar]
- Cardi D, Montigny C, Arnou B, Jidenko M, Marchal E, le Maire M, Jaxel C (2010) Heterologous expression and affinity purification of eukaryotic membrane proteins in view of functional and structural studies: The example of the sarcoplasmic reticulum Ca2+-ATPase. Methods Mol Biol 601: 247–267 [DOI] [PubMed] [Google Scholar]
- Champeil P, le Maire M, Andersen JP, Guillain F, Gingold M, Lund S, Møller JV (1986) Kinetic characterization of the normal and detergent-perturbed reaction cycles of the sarcoplasmic reticulum calcium pump. Rate-limiting step(s) under different conditions. J Biol Chem 261: 16372–16384 [PubMed] [Google Scholar]
- Champeil P, Riollet S, Orlowski S, Guillain F, Seebregts CJ, McIntosh DB (1988) ATP regulation of sarcoplasmic reticulum Ca2+-ATPase. Metal-free ATP and 8-bromo-ATP bind with high affinity to the catalytic site of phosphorylated ATPase and accelerate dephosphorylation. J Biol Chem 263: 12288–12294 [PubMed] [Google Scholar]
- Chen C, Okayama H (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol 7: 2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DM, Loo TW, Inesi G, MacLennan DH (1989) Location of high affinity Ca2+-binding sites within the predicted transmembrane domain of the sarcoplasmic reticulum Ca2+-ATPase. Nature 339: 476–478 [DOI] [PubMed] [Google Scholar]
- Clausen JD, Andersen JP (2010) Glutamate 90 at the luminal ion gate of sarcoplasmic reticulum Ca2+-ATPase is critical for Ca2+ binding on both sides of the membrane. J Biol Chem 285: 20780–20792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen JD, McIntosh DB, Woolley DG, Andersen JP (2001) Importance of Thr-353 of the conserved phosphorylation loop of the sarcoplasmic reticulum Ca2+-ATPase in MgATP binding and catalytic activity. J Biol Chem 276: 35741–35750 [DOI] [PubMed] [Google Scholar]
- Clausen JD, McIntosh DB, Woolley DG, Andersen JP (2011) Modulatory ATP binding affinity in intermediate states of E2P dephosphorylation of sarcoplasmic reticulum Ca2+-ATPase. J Biol Chem 286: 11792–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen JD, Vilsen B, McIntosh DB, Einholm AP, Andersen JP (2004) Glutamate-183 in the conserved TGES motif of domain A of sarcoplasmic reticulum Ca2+-ATPase assists in catalysis of E2/E2P partial reactions. Proc Natl Acad Sci USA 101: 2776–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meis L, Vianna al (1979) Energy interconversion by the Ca2+-dependent ATPase of the sarcoplasmic reticulum. Annu Rev Biochem 48: 275–292 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Fonseca LM, Thomas DD (2011) Atomic-level characterization of the activation mechanism of SERCA by calcium. PLoS One 6: e26936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forge V, Mintz E, Guillain F (1993) Ca2+ binding to sarcoplasmic reticulum ATPase revisited. I. Mechanism of affinity and cooperativity modulation by H+ and Mg2+. J Biol Chem 268: 10953–10960 [PubMed] [Google Scholar]
- Howell PL, Smith GD (1992) Identification of heavy-atom derivatives by normal probability methods. J Appl Cryst 25: 81–86 [Google Scholar]
- Inesi G (1985) Mechanism of calcium transport. Annu Rev Physiol 47: 573–601 [DOI] [PubMed] [Google Scholar]
- Inesi G, Lewis D, Toyoshima C, Hirata A, de Meis L (2008) Conformational fluctuations of the Ca2+-ATPase in the native membrane environment. Effects of pH, temperature, catalytic substrates, and thapsigargin. J Biol Chem 283: 1189–1196 [DOI] [PubMed] [Google Scholar]
- Inesi G, Ma H, Lewis D, Xu C (2004) Ca2+ occlusion and gating function of Glu309 in the ADP-fluoroaluminate analog of the Ca2+-ATPase phosphoenzyme intermediate. J Biol Chem 279: 31629–31637 [DOI] [PubMed] [Google Scholar]
- Inesi G, Zhang Z, Lewis D (2002) Cooperative setting for long-range linkage of Ca2+ binding and ATP synthesis in the Ca2+ ATPase. Biophys J 83: 2327–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AM, Sørensen TL, Olesen C, Møller JV, Nissen P (2006) Modulatory and catalytic modes of ATP binding by the calcium pump. EMBO J 25: 2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jidenko M, Nielsen RC, Sørensen TL, Møller JV, le Maire M, Nissen P, Jaxel C (2005) Crystallization of a mammalian membrane protein overexpressed in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 102: 11687–11691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crys 26: 795–800 [Google Scholar]
- Kaufman RJ, Davies MV, Pathak VK, Hershey JW (1989) The phosphorylation state of eucaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Mol Cell Biol 9: 946–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Maire M, Arnou B, Olesen C, Georgin D, Ebel C, Møller JV (2008) Gel chromatography and analytical ultracentrifugation to determine the extent of detergent binding and aggregation, and Stokes radius of membrane proteins using sarcoplasmic reticulum Ca2+-ATPase as an example. Nat Protoc 3: 1782–1795 [DOI] [PubMed] [Google Scholar]
- Lenoir G, Jaxel C, Picard M, le Maire M, Champeil P, Falson P (2006) Conformational changes in sarcoplasmic reticulum Ca2+-ATPase mutants: effect of mutations either at Ca2+-binding site II or at tryptophan 552 in the cytosolic domain. Biochemistry 45: 5261–5270 [DOI] [PubMed] [Google Scholar]
- Marchand A, Winther AM, Holm PJ, Olesen C, Montigny C, Arnou B, Champeil P, Clausen JD, Vilsen B, Andersen JP, Nissen P, Jaxel C, Møller JV, le Maire M (2008) Crystal structure of D351A and P312A mutant forms of the mammalian sarcoplasmic reticulum Ca2+-ATPase reveals key events in phosphorylation and Ca2+ release. J Biol Chem 283: 14867–14882 [DOI] [PubMed] [Google Scholar]
- Maruyama K, MacLennan DH (1988) Mutation of aspartic acid-351, lysine-352, and lysine-515 alters the Ca2+ transport activity of the Ca2+-ATPase expressed in COS-1 cells. Proc Natl Acad Sci USA 85: 3314–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh DB, Woolley DG, MacLennan DH, Vilsen B, Andersen JP (1999) Interaction of nucleotides with Asp351 and the conserved phosphorylation loop of sarcoplasmic reticulum Ca2+-ATPase. J Biol Chem 274: 25227–25236 [DOI] [PubMed] [Google Scholar]
- McIntosh DB, Woolley DG, Vilsen B, Andersen JP (1996) Mutagenesis of segment 487Phe-Ser-Arg-Asp-Arg-Lys492 of sarcoplasmic reticulum Ca2+-ATPase produces pumps defective in ATP binding. J Biol Chem 271: 25778–25789 [DOI] [PubMed] [Google Scholar]
- Menguy T, Corre F, Bouneau L, Deschamps S, Møller JV, Champeil P, le Maire M, Falson P (1998) The cytoplasmic loop located between transmembrane segments 6 and 7 controls activation by Ca2+ of sarcoplasmic reticulum Ca2+-ATPase. J Biol Chem 273: 20134–20143 [DOI] [PubMed] [Google Scholar]
- Mintz E, Guillain F (1997) Ca2+ transport by the sarcoplasmic reticulum ATPase. Biochim Biophys Acta 1318: 52–70 [DOI] [PubMed] [Google Scholar]
- Møller JV, Olesen C, Winther AM, Nissen P (2010) The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Q Rev Biophys 43: 1–66 [DOI] [PubMed] [Google Scholar]
- Montigny C, Arnou B, Marchal E, Champeil P (2008) Use of glycerol-containing media to study the intrinsic fluorescence properties of detergent-solubilized native or expressed SERCA1a. Biochemistry 47: 12159–12174 [DOI] [PubMed] [Google Scholar]
- Musgaard M, Thogersen L, Schiøtt B (2011) Protonation states of important acidic residues in the central Ca2+ ion binding sites of the Ca2+-ATPase: a molecular modeling study. Biochemistry 50: 11109–11120 [DOI] [PubMed] [Google Scholar]
- Olesen C, Picard M, Winther AM, Gyrup C, Morth JP, Oxvig C, Møller JV, Nissen P (2007) The structural basis of calcium transport by the calcium pump. Nature 450: 1036–1042 [DOI] [PubMed] [Google Scholar]
- Olesen C, Sørensen TL, Nielsen RC, Møller JV, Nissen P (2004) Dephosphorylation of the calcium pump coupled to counterion occlusion. Science 306: 2251–2255 [DOI] [PubMed] [Google Scholar]
- Pang DC, Briggs FN (1977) Effect of calcium and magnesium on binding of beta, gamma-methylene ATP to sarcoplasmic reticulum. J Biol Chem 252: 3262–3266 [PubMed] [Google Scholar]
- Petithory JR, Jencks WP (1988) Sequential dissociation of Ca2+ from the calcium adenosinetriphosphatase of sarcoplasmic reticulum and the calcium requirement for its phosphorylation by ATP. Biochemistry 27: 5553–5564 [DOI] [PubMed] [Google Scholar]
- Picard M, Toyoshima C, Champeil P (2005) The average conformation at micromolar [Ca2+] of Ca2+-ATPase with bound nucleotide differs from that adopted with the transition state analog ADP.AlFx or with AMPPCP under crystallization conditions at millimolar [Ca2+]. J Biol Chem 280: 18745–18754 [DOI] [PubMed] [Google Scholar]
- Scarborough GA (2002) Molecular mechanism of the P-type ATPases. J Bioenerg Biomembr 34: 235–250 [DOI] [PubMed] [Google Scholar]
- Seebregts CJ, McIntosh DB (1989) 2′,3′-O-(2,4,6-trinitrophenyl)-8-azido-adenosine mono-, di-, and triphosphates as photoaffinity probes of the Ca2+-ATPase of sarcoplasmic reticulum. Regulatory/superfluorescent nucleotides label the catalytic site with high efficiency. J Biol Chem 264: 2043–2052 [PubMed] [Google Scholar]
- Skerjanc IS, Toyofuku T, Richardson C, MacLennan DH (1993) Mutation of glutamate 309 to glutamine alters one Ca2+-binding site in the Ca2+-ATPase of sarcoplasmic reticulum expressed in Sf9 cells. J Biol Chem 268: 15944–15950 [PubMed] [Google Scholar]
- Strock C, Cavagna M, Peiffer WE, Sumbilla C, Lewis D, Inesi G (1998) Direct demonstration of Ca2+ binding defects in sarco-endoplasmic reticulum Ca2+ ATPase mutants overexpressed in COS-1 cells transfected with adenovirus vectors. J Biol Chem 273: 15104–15109 [DOI] [PubMed] [Google Scholar]
- Sørensen T, Vilsen B, Andersen JP (1997) Mutation Lys758→Ile of the sarcoplasmic reticulum Ca2+-ATPase enhances dephosphorylation of E2P and inhibits the E2 to E1Ca2 transition. J Biol Chem 272: 30244–30253 [DOI] [PubMed] [Google Scholar]
- Sørensen TL, Dupont Y, Vilsen B, Andersen JP (2000) Fast kinetic analysis of conformational changes in mutants of the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem 275: 5400–5408 [DOI] [PubMed] [Google Scholar]
- Sørensen TL, Møller JV, Nissen P (2004) Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science 304: 1672–1675 [DOI] [PubMed] [Google Scholar]
- Toyoshima C (2009) How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim Biophys Acta 1793: 941–946 [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Iwasawa S, Ogawa H, Hirata A, Tsueda J, Inesi G (2013) Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state. Nature 495: 260–264 [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Mizutani T (2004) Crystal structure of the calcium pump with a bound ATP analogue. Nature 430: 529–535 [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Nakasako M, Nomura H, Ogawa H (2000) Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6A resolution. Nature 405: 647–655 [DOI] [PubMed] [Google Scholar]
- Vilsen B, Andersen JP (1992) CrATP-induced Ca2+ occlusion in mutants of the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem 267: 25739–25743 [PubMed] [Google Scholar]
- Vilsen B, Andersen JP (1998) Mutation to the glutamate in the fourth membrane segment of Na+,K+- ATPase and Ca2+-ATPase affects cation binding from both sides of the membrane and destabilizes the occluded enzyme forms. Biochemistry 37: 10961–10971 [DOI] [PubMed] [Google Scholar]
- Weber K, Osborn M (1969) The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem 244: 4406–4412 [PubMed] [Google Scholar]
- Winther AM, Bublitz M, Karlsen JL, Møller JV, Hansen JB, Nissen P, Buch-Pedersen MJ (2013) The sarcolipin-bound calcium pump stabilizes calcium sites exposed to the cytoplasm. Nature 495: 265–269 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lewis D, Strock C, Inesi G, Nakasako M, Nomura H, Toyoshima C (2000) Detailed characterization of the cooperative mechanism of Ca2+ binding and catalytic activation in the Ca2+ transport (SERCA) ATPase. Biochemistry 39: 8758–8767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.