Abstract
EMBO J 32 13, 1855–1868 doi:; DOI: 10.1038/emboj.2013.135; published online June 11 2013
EMBO J 32 13, 1842–1854 doi:; DOI: 10.1038/emboj.2013.63; published online March 15 2013
Nature 497: 244–248 doi:; DOI: 10.1038/nature12119
Regulated degradation plays a major role in determining the levels of both non-coding (miRNA) and coding (mRNA) transcripts. Thus, insights into the factors and pathways that influence this process have broad, interdisciplinary implications. New findings by Malecki et al (2013), Lubas et al (2013), and Chang et al (2013) identify the protein Dis3L2 as a major player in the 3′–5′ exonucleolytic decay of transcripts. Furthermore, they demonstrate a strong connection between terminal uridylation of the RNA substrate and enzymatic activity.
Looking for a set of truly high impact, paradigm-shifting papers for your Molecular Biology Journal Club this week that will cause your audience to put down their snack and drink and really take notice? Try the recently published studies by Malecki et al (2013), Lubas et al (2013), and/or Chang et al (2013). If these do not turn heads during your discussions, track one of us down at a meeting this summer and we will buy you a cold beverage.
The level of RNAs in the cell is not simply a product of transcription, but is also heavily influenced by the relative stability of the transcript. For the past 15 years, the enzymes involved in general mRNA decay have been firmly entrenched in the field. In general, mRNAs are targeted for decay in the cytoplasm as soon as they are no longer needed. Once an RNA undergoes deadenylation or endonucleolytic cleavage, it becomes subjected to decapping and Xrn1-mediated 5′–3′ decay and/or 3–5′ exonucleolytic decay by the Dis3 subunit of the exosome (Schoenberg and Maquat, 2012). A few years ago, interesting work started a ripple in the placid waters of the mRNA decay pool by indicating that non-polyadenylated histone mRNA decay (Mullen and Marzluff, 2008), mRNA decay in S. pombe (Rissland and Norbury, 2009), and the turnover of miRNAs (Heo et al, 2009) are associated with the addition of one or more uridylate residues to the 3′ end of targeted transcripts. Three new studies (Chang et al, 2013; Lubas et al, 2013 and Malecki et al, 2013) have now turned this ripple into quite a splash by identifying both a major new disease-associated exonuclease, Dis3L2, involved in 3′–5′ RNA decay as well as a possible widespread role for terminal uridylation as a signal for the RNA decay in multiple eukaryotes.
Dis3 has been well-studied as the ribonuclease of the cytoplasmic exosome in the yeast S. cerevisiae. Interestingly, many other eukaryotes express two additional Dis3 isoforms. One of these is Dis3L2 (SOV in plants; Zhang et al, 2010), an isoform containing an intact RNB exonuclease-signature domain but lacking a PIN endonuclease domain and having an extended CR3 RNA-binding region near its N terminus. Dis3L2 has now been clearly shown to possess processive 3′–5′ exonuclease activity. Dis3L2 is more effective than its Dis3 relative in degrading dsRNAs with a short 3′ single-stranded landing pad and gives a slightly shorter final decay product. Dis3L2 is located in the cytoplasm of cells, is curiously not found associated with the exosome, and can be associated with P bodies and perhaps polysomes under select conditions. Since P bodies have previously been known to only harbour components of the 5′–3′ mRNA decay pathway, this is the first report of an association of a 3′–5′ decay factor with these RNA granules. Thus, the 3′–5′ exonuclease Dis3L2 may be in close communication with the 5′–3′ arm of the mRNA decay machinery to coordinate RNA decay. Dis3L2 is likely to be very important for general mRNA decay for three main reasons. First, mutations in Dis3L2 are synthetically lethal with Xrn1 mutations in S. pombe. This observation indicates that Dis3L2, rather than the exosome per se, may be the major player in 3′–5′ mRNA decay in fission yeast and perhaps other metazoans. Second, an effective block in mRNA decay is observed in double mutants of Dis3L2 and the 5′–3′ decay factor Lsm1, directly implicating Dis3L2 in the general mechanism of mRNA degradation. Finally, knockdown of Dis3L2 in mammalian cells results in the stabilization of a reporter mRNA containing a well-studied instability element from the TNF-α 3′-untranslated region.
In addition to its role in mRNA decay, Chang et al (2013) also demonstrated that Dis3L2, via its ribonuclease activity, helps maintain the pluripotency of stem cells by acting as an effector of Lin28-mediated inhibition of let-7 miRNA expression. Dis3L2 mediates this effect by binding to cytoplasmic precursors of let-7 (pre-let7) miRNAs that contain a 3′ terminal uridylate tail. The enzymatic activity of Dis3L2 is dramatically (10X) stimulated by the addition of 10–14 uridylates onto the 3′ end of miRNA targets. In the absence of Dis3L2, uridylated pre-let7 miRNAs accumulate in the cell. Collectively these observations establish a major role for the Dis3L2 enzyme in cellular RNA decay, changing our dogmatic picture of mRNA exonucleolytic decay enzymes and pathways (Figure 1).
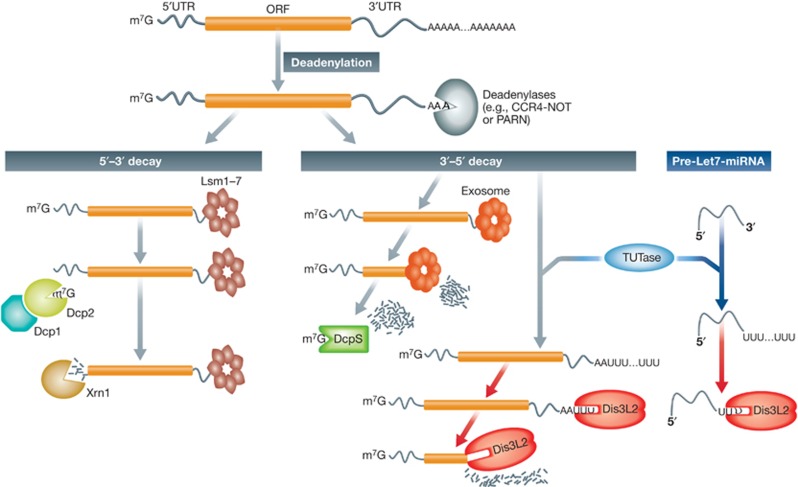
Figure 1.
Dis3L2 is a new player in a major pathway of RNA decay. Many mRNAs normally undergo poly(A) tail removal by deadenylases as a first step prior to their decay by an exonucleolytic mechanism. Recruitment of the Lsm1-7 complex to the 3′ end of a mRNA triggers decapping by the Dcp1–Dcp2 complex, making the RNA accessible to the 5′–3′ exoribonuclease Xrn1. Degradation can also occur in the 3′→5′ direction if the exosome complex is recruited to the 3′ end of deadenylated transcript. In this pathway, short-capped RNA oligomers generated by the exosome are acted on by the scavenger-decapping enzyme DcpS. The recent observations of Malecki et al (2013) and Lubas et al (2013) indicate that the 3′–5′ decay of mRNAs can also occur in an exosome-independent and uridylation-dependent manner. A TUTase enzyme adds a short poly(U) tract to a deadenylated mRNA that becomes an ideal landing pad for the 3′–5′ exoribonuclease Dis3L2. Chang et al (2013) demonstrated that a similar uridylation/Dis3L2-mediated 3′–5′ decay pathway also acts on let7 pre-miRNAs.
Not surprising, these paradigm-shifting papers lead to numerous interesting discussion points and areas for future investigation. From a purely technical perspective, the work by Chang et al nicely points out the importance of not solely relying on PCR-based assessments of RNA abundances to determine the impact of a factor on gene expression. A direct assessment of RNA decay rates and an analysis of the actual species of RNA that accumulated was required to determine the influence of Dis3L2 on miRNA biogenesis. At a fundamental level, it will be interesting to understand structurally how Dis3L2 differs from other Dis3 family members in terms of its ability to degrade dsRNA and its preference for substrates with terminal uridylates. Next, what’s the relationship of Dis3L2 to general RNA decay in specific organisms? It will be curious to see if Dis3L2 plays as large a role in mRNA decay in humans as it appears to in S. pombe. Past experience from studies in S. cerevisiae indicate that it may be dangerous to assume that mRNA decay pathways/enzymes are created equal in all eukaryotes. It will also be interesting to determine the contributions of Dis3L2 to RNA quality control pathways in addition to its role in regulating mRNA levels. One also cannot overlook the fact that these studies provide the first evidence for a 3′–5′ decay factor in P bodies, an observation that may help expand our mechanistic understanding of these rather enigmatic structures. Furthermore, how does the cell regulate the highly processive Dis3L2 exonuclease activity? Terminal uridylation is likely part of the answer to this question, and undoubtedly research is currently being undertaken to determine the extent of terminal uridylation on a variety of types of RNA under Dis3L2 knockdown conditions. While Dis3L2 is not part of the exosome, it may be in direct communication with TUTases or perhaps regulatory protein(s) that function in a similar fashion as Lin28 in regulating the processing/decay of miRNAs. Given the fact that Lin28 plays a key role in the generation of induced pluripotent stem cells and stem cell biology (Shyh-Chang and Daley, 2013), it will be interesting to determine the contribution of Dis3L2 to this fascinating population of cells.
Finally, the fact that a mutation in Dis3L2 gene has been associated with Perlman’s fetal overgrowth syndrome and a propensity for Wilm’s tumour development (Astuti et al, 2012) confirms the importance of this protein in maintaining normal cell metabolism and development. We still do not know, however, how alterations in an mRNA decay pathway can lead to the overgrowth characteristics of Perlman’s syndrome and contribute to cancer. Perhaps some clues to this puzzle can be drawn from mining the global analyses presented in the Lubas et al (2013) study. Clearly, the functional and biological implications of these studies are far-reaching and may very well lay the foundation for many more interesting papers for journal club in the future.
Footnotes
The authors declare that they have no conflict of interest.
References
- Astuti D, Morris MR, Cooper WN, Staals RH, Wake NC, Fews GA, Gill H, Gentle D, Shuib S, Ricketts CJ, Cole T, van Essen AJ, van Lingen RA, Neri G, Opitz JM, Rump P, Stolte-Dijkstra I, Müller F, Pruijn GJ, Latif F et al. (2012) Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet 44: 277–284 [DOI] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI (2013) A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 497: 244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN (2009) TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Nat Cell Biol 11: 1157–1163 [DOI] [PubMed] [Google Scholar]
- Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A (2013) Exonuclease hDIS3L2 specifies an exosome-independent 3′-5′ degradation pathway of human cytoplasmic mRNA. EMBO J 32: 1855–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM (2013) The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J 32: 1842–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF (2008) Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5' to 3' and 3' to 5'. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ (2009) Decapping is preceded by 3' uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 16: 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE (2012) Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13: 246–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ (2013) Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12: 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Murphy C, Sieburth LE (2010) Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc Natl Acad Sci USA 107: 15981–15985 [DOI] [PMC free article] [PubMed] [Google Scholar]