Abstract
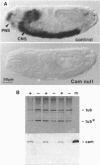
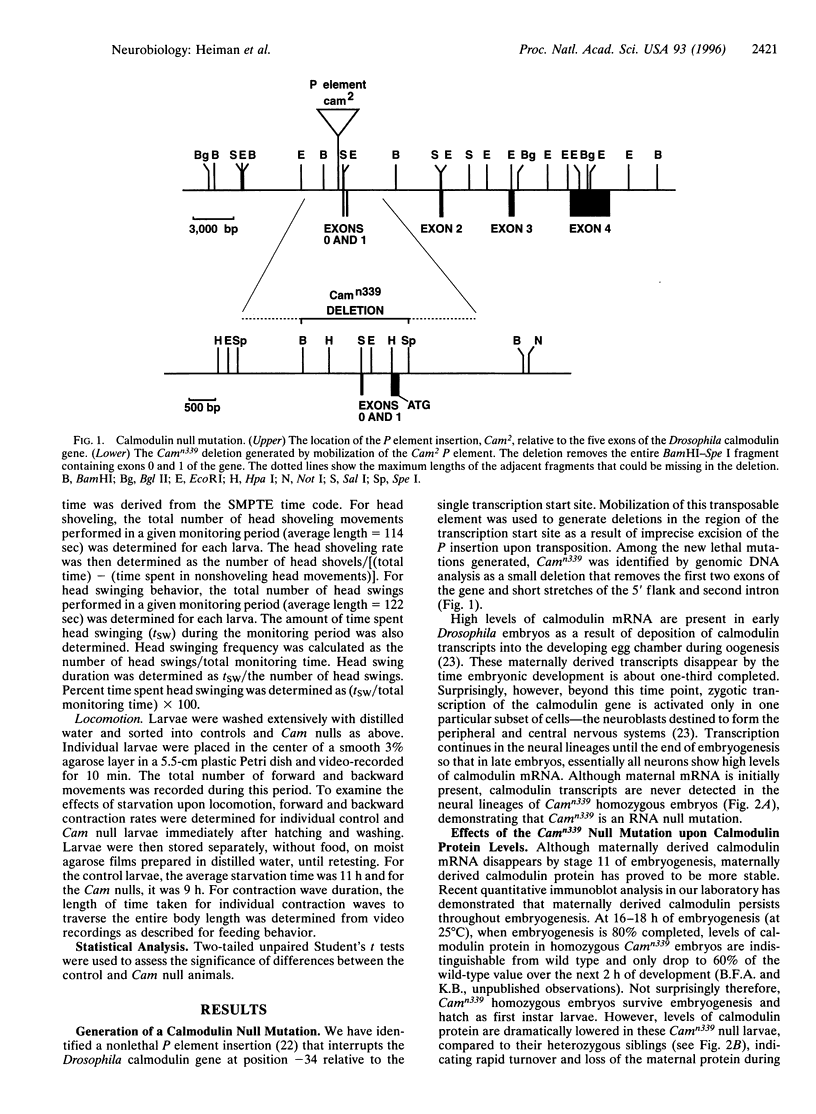
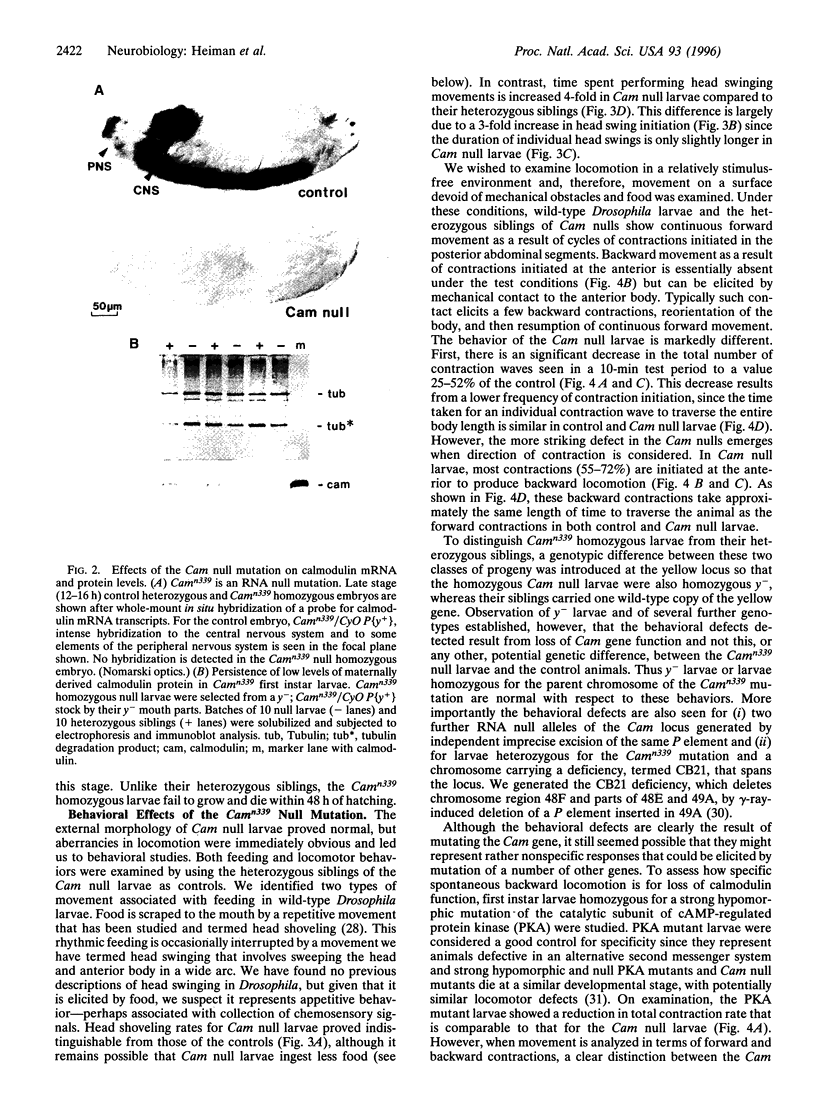
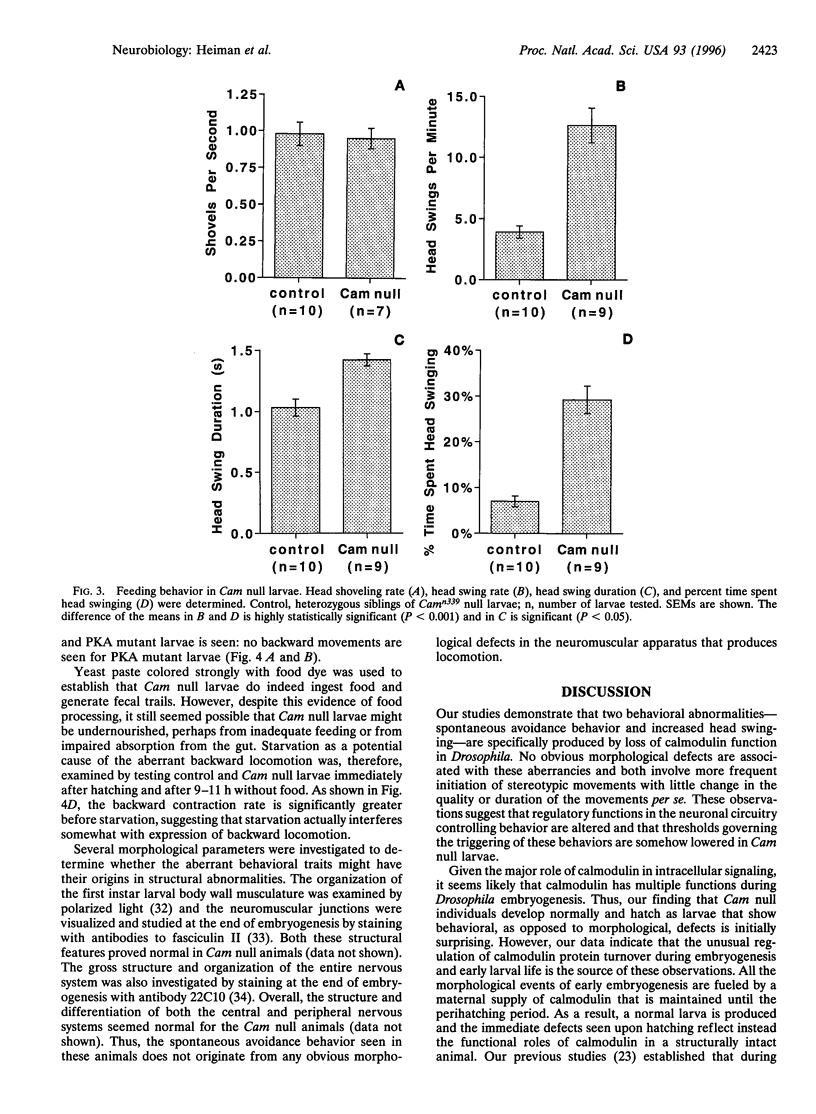
The regulatory protein calmodulin is a major mediator of calcium-induced changes in cellular activity. To analyze the roles of calmodulin in an intact animal, we have generated a calmodulin null mutation in Drosophila melanogaster. Maternal calmodulin supports calmodulin null individuals throughout embryogenesis, but they die within 2 days of hatching as first instar larvae. We have detected two pronounced behavioral abnormalities specific to the loss of calmodulin in these larvae. Swinging of the head and anterior body, which occurs in the presence of food, is three times more frequent in the null animals. More strikingly, most locomotion in calmodulin null larvae is spontaneous backward movement. This is in marked contrast to the wild-type situation where backward locomotion is seen only as a stimulus-elicited avoidance response. Our finding of spontaneous avoidance behavior has striking similarities to the enhanced avoidance responses produced by some calmodulin mutations in Paramecium. Thus our results suggest evolutionary conservation of a role for calmodulin in membrane excitability and linked behavioral responses.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelman J. P., Shen K. Z., Kavanaugh M. P., Warren R. A., Wu Y. N., Lagrutta A., Bond C. T., North R. A. Calcium-activated potassium channels expressed from cloned complementary DNAs. Neuron. 1992 Aug;9(2):209–216. doi: 10.1016/0896-6273(92)90160-f. [DOI] [PubMed] [Google Scholar]
- Brockerhoff S. E., Stevens R. C., Davis T. N. The unconventional myosin, Myo2p, is a calmodulin target at sites of cell growth in Saccharomyces cerevisiae. J Cell Biol. 1994 Feb;124(3):315–323. doi: 10.1083/jcb.124.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivici A., Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- Davis T. N., Urdea M. S., Masiarz F. R., Thorner J. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell. 1986 Nov 7;47(3):423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- Doyle K. E., Kovalick G. E., Lee E., Beckingham K. Drosophila melanogaster contains a single calmodulin gene. Further structure and expression studies. J Mol Biol. 1990 Jun 20;213(4):599–605. doi: 10.1016/S0022-2836(05)80245-1. [DOI] [PubMed] [Google Scholar]
- Epstein P. N., Overbeek P. A., Means A. R. Calmodulin-induced early-onset diabetes in transgenic mice. Cell. 1989 Sep 22;58(6):1067–1073. doi: 10.1016/0092-8674(89)90505-9. [DOI] [PubMed] [Google Scholar]
- Geiser J. R., Sundberg H. A., Chang B. H., Muller E. G., Davis T. N. The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Dec;13(12):7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith L. C., Wang J., Zhong Y., Wu C. F., Greenspan R. J. Calcium/calmodulin-dependent protein kinase II and potassium channel subunit eag similarly affect plasticity in Drosophila. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):10044–10048. doi: 10.1073/pnas.91.21.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruver C. L., DeMayo F., Goldstein M. A., Means A. R. Targeted developmental overexpression of calmodulin induces proliferative and hypertrophic growth of cardiomyocytes in transgenic mice. Endocrinology. 1993 Jul;133(1):376–388. doi: 10.1210/endo.133.1.8319584. [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Schulman H. Neuronal Ca2+/calmodulin-dependent protein kinases. Annu Rev Biochem. 1992;61:559–601. doi: 10.1146/annurev.bi.61.070192.003015. [DOI] [PubMed] [Google Scholar]
- Hinrichsen R. D., Burgess-Cassler A., Soltvedt B. C., Hennessey T., Kung C. Restoration by calmodulin of a Ca2+-dependent K+ current missing in a mutant of Paramecium. Science. 1986 Apr 25;232(4749):503–506. doi: 10.1126/science.2421410. [DOI] [PubMed] [Google Scholar]
- Hinrichsen R. D., Fraga D., Reed M. W. 3'-modified antisense oligodeoxyribonucleotides complementary to calmodulin mRNA alter behavioral responses in Paramecium. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8601–8605. doi: 10.1073/pnas.89.18.8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichsen R. D., Fraga D., Russell C. The regulation of calcium in Paramecium. Adv Second Messenger Phosphoprotein Res. 1995;30:311–338. doi: 10.1016/s1040-7952(05)80013-8. [DOI] [PubMed] [Google Scholar]
- Hulen D., Baron A., Salisbury J., Clarke M. Production and specificity of monoclonal antibodies against calmodulin from Dictyostelium discoideum. Cell Motil Cytoskeleton. 1991;18(2):113–122. doi: 10.1002/cm.970180206. [DOI] [PubMed] [Google Scholar]
- Ikeshima H., Yuasa S., Matsuo K., Kawamura K., Hata J., Takano T. Expression of three nonallelic genes coding calmodulin exhibits similar localization on the central nervous system of adult rats. J Neurosci Res. 1993 Sep 1;36(1):111–119. doi: 10.1002/jnr.490360112. [DOI] [PubMed] [Google Scholar]
- James P., Vorherr T., Carafoli E. Calmodulin-binding domains: just two faced or multi-faceted? Trends Biochem Sci. 1995 Jan;20(1):38–42. doi: 10.1016/s0968-0004(00)88949-5. [DOI] [PubMed] [Google Scholar]
- Kink J. A., Maley M. E., Preston R. R., Ling K. Y., Wallen-Friedman M. A., Saimi Y., Kung C. Mutations in paramecium calmodulin indicate functional differences between the C-terminal and N-terminal lobes in vivo. Cell. 1990 Jul 13;62(1):165–174. doi: 10.1016/0092-8674(90)90250-i. [DOI] [PubMed] [Google Scholar]
- Klee C. B. Concerted regulation of protein phosphorylation and dephosphorylation by calmodulin. Neurochem Res. 1991 Sep;16(9):1059–1065. doi: 10.1007/BF00965851. [DOI] [PubMed] [Google Scholar]
- Klee C. B., Vanaman T. C. Calmodulin. Adv Protein Chem. 1982;35:213–321. doi: 10.1016/s0065-3233(08)60470-2. [DOI] [PubMed] [Google Scholar]
- Kovalick G. E., Beckingham K. Calmodulin transcription is limited to the nervous system during Drosophila embryogenesis. Dev Biol. 1992 Mar;150(1):33–46. doi: 10.1016/0012-1606(92)90005-2. [DOI] [PubMed] [Google Scholar]
- Lane M. E., Kalderon D. Genetic investigation of cAMP-dependent protein kinase function in Drosophila development. Genes Dev. 1993 Jul;7(7A):1229–1243. doi: 10.1101/gad.7.7a.1229. [DOI] [PubMed] [Google Scholar]
- Lee J. K., Coyne R. S., Dubreuil R. R., Goldstein L. S., Branton D. Cell shape and interaction defects in alpha-spectrin mutants of Drosophila melanogaster. J Cell Biol. 1993 Dec;123(6 Pt 2):1797–1809. doi: 10.1083/jcb.123.6.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K. P., Osmani S. A., Osmani A. H., Means A. R. Essential roles for calcium and calmodulin in G2/M progression in Aspergillus nidulans. J Cell Biol. 1993 May;121(3):621–630. doi: 10.1083/jcb.121.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K. P., Rasmussen C. D., May G. S., Means A. R. Cooperative regulation of cell proliferation by calcium and calmodulin in Aspergillus nidulans. Mol Endocrinol. 1992 Mar;6(3):365–374. doi: 10.1210/mend.6.3.1584213. [DOI] [PubMed] [Google Scholar]
- Maune J. F., Klee C. B., Beckingham K. Ca2+ binding and conformational change in two series of point mutations to the individual Ca(2+)-binding sites of calmodulin. J Biol Chem. 1992 Mar 15;267(8):5286–5295. [PubMed] [Google Scholar]
- Means A. R. Calcium, calmodulin and cell cycle regulation. FEBS Lett. 1994 Jun 20;347(1):1–4. doi: 10.1016/0014-5793(94)00492-7. [DOI] [PubMed] [Google Scholar]
- Nojima H. Structural organization of multiple rat calmodulin genes. J Mol Biol. 1989 Jul 20;208(2):269–282. doi: 10.1016/0022-2836(89)90388-4. [DOI] [PubMed] [Google Scholar]
- Ohya Y., Botstein D. Diverse essential functions revealed by complementing yeast calmodulin mutants. Science. 1994 Feb 18;263(5149):963–966. doi: 10.1126/science.8310294. [DOI] [PubMed] [Google Scholar]
- Saimi Y., Kung C. Behavioral genetics of Paramecium. Annu Rev Genet. 1987;21:47–65. doi: 10.1146/annurev.ge.21.120187.000403. [DOI] [PubMed] [Google Scholar]
- Saimi Y., Ling K. Y. Calmodulin activation of calcium-dependent sodium channels in excised membrane patches of Paramecium. Science. 1990 Sep 21;249(4975):1441–1444. doi: 10.1126/science.2169650. [DOI] [PubMed] [Google Scholar]
- Skoulakis E. M., Kalderon D., Davis R. L. Preferential expression in mushroom bodies of the catalytic subunit of protein kinase A and its role in learning and memory. Neuron. 1993 Aug;11(2):197–208. doi: 10.1016/0896-6273(93)90178-t. [DOI] [PubMed] [Google Scholar]
- Sokolowski M. B. Foraging strategies of Drosophila melanogaster: a chromosomal analysis. Behav Genet. 1980 May;10(3):291–302. doi: 10.1007/BF01067774. [DOI] [PubMed] [Google Scholar]
- Takeda T., Yamamoto M. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3580–3584. doi: 10.1073/pnas.84.11.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vactor D. V., Sink H., Fambrough D., Tsoo R., Goodman C. S. Genes that control neuromuscular specificity in Drosophila. Cell. 1993 Jun 18;73(6):1137–1153. doi: 10.1016/0092-8674(93)90643-5. [DOI] [PubMed] [Google Scholar]
- Yamanaka M. K., Saugstad J. A., Hanson-Painton O., McCarthy B. J., Tobin S. L. Structure and expression of the Drosophila calmodulin gene. Nucleic Acids Res. 1987 Apr 24;15(8):3335–3348. doi: 10.1093/nar/15.8.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink B., Engström Y., Gehring W. J., Paro R. Direct interaction of the Polycomb protein with Antennapedia regulatory sequences in polytene chromosomes of Drosophila melanogaster. EMBO J. 1991 Jan;10(1):153–162. doi: 10.1002/j.1460-2075.1991.tb07931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipursky S. L., Venkatesh T. R., Teplow D. B., Benzer S. Neuronal development in the Drosophila retina: monoclonal antibodies as molecular probes. Cell. 1984 Jan;36(1):15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]