Abstract
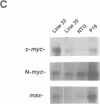
Although myc family genes are differentially expressed during development, their expression frequently overlaps, suggesting that they may serve both distinct and common biological functions. In addition, alterations in their expression occur at major developmental transitions in many cell lineages. For example, during mouse lens maturation, the growth arrest and differentiation of epithelial cells into lens fiber cells is associated with a decrease in L- and c-myc expression and a reciprocal rise in N-myc levels. To determine whether the down-regulation of L- and c-myc are required for mitotic arrest and/or completion of differentiation and whether these genes have distinct or similar activities in the same cell type, we have studied the consequences of forced L- and c-myc expression in the lens fiber cell compartment using the alpha A-crystallin promoter in transgenic mice (alpha A/L-myc and alpha A/c-myc mice). With respect to morphological and molecular differentiation, alpha A/L-myc lenses were characterized by a severely disorganized lens fiber cell compartment and a significant decrease in the expression of a late-stage differentiation marker (MIP26); in contrast, differentiation appeared to be unaffected in alpha A/c-myc mice. Furthermore, an analysis of proliferation indicated that while alpha A/L-myc fiber cells withdrew properly from the cell cycle, inappropriate cell cycle progression occurred in the lens fiber cell compartment of alpha A/c-myc mice. These observations indicate that continued late-stage expression of L-myc affected differentiation processes directly, rather than indirectly through deregulated growth control, whereas constitutive c-myc expression inhibited proliferative arrest, but did not appear to disturb differentiation. As a direct corollary, our data indicate that L-Myc and c-Myc are involved in distinct physiological processes in the same cell type.
Full text
PDF

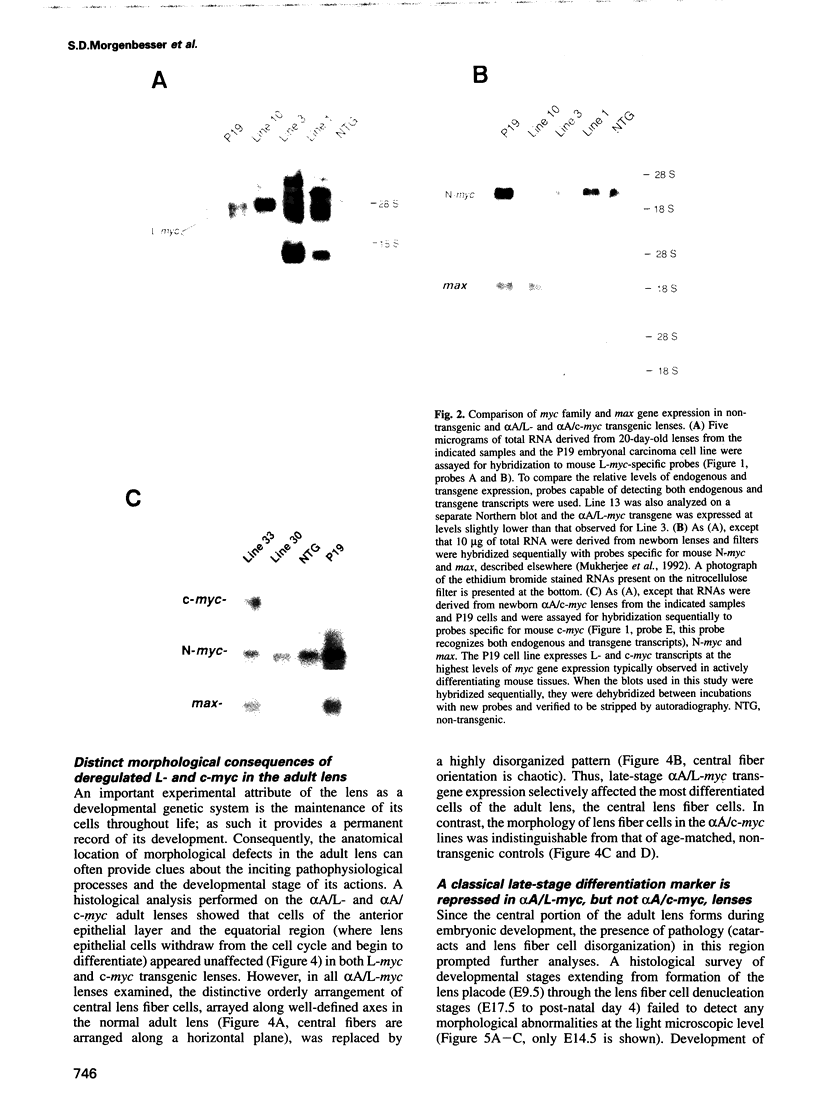
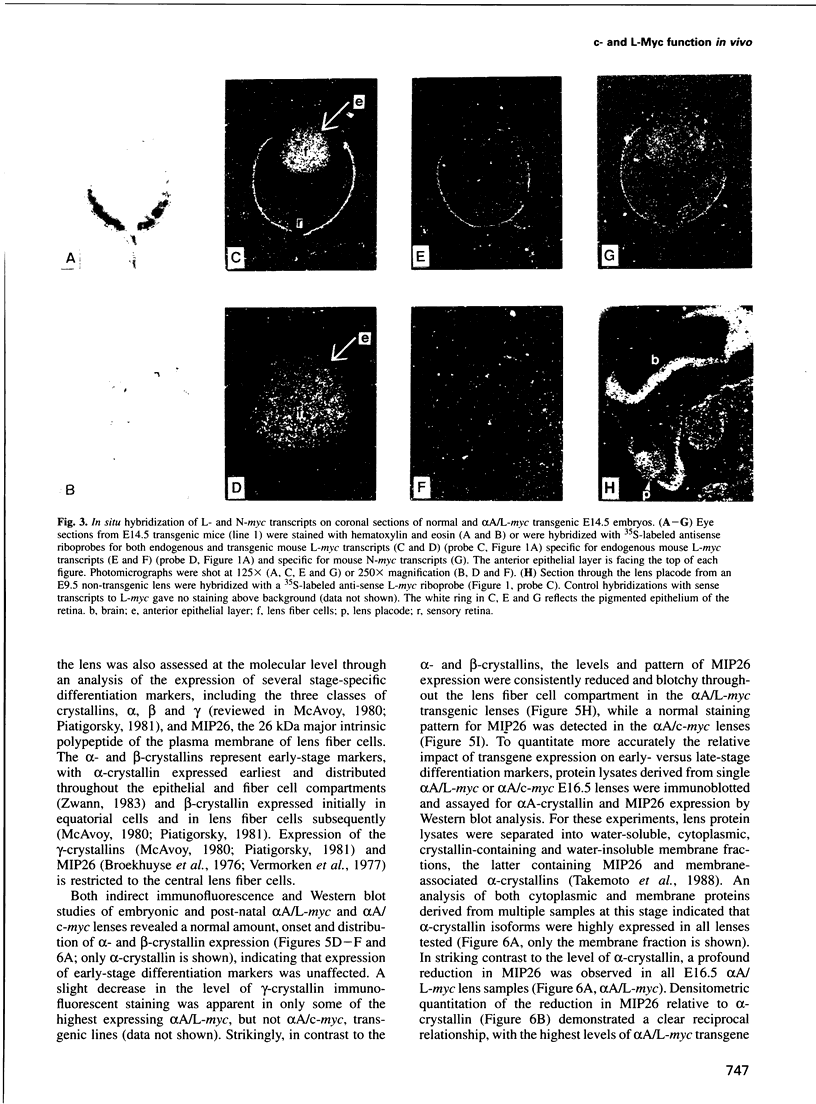
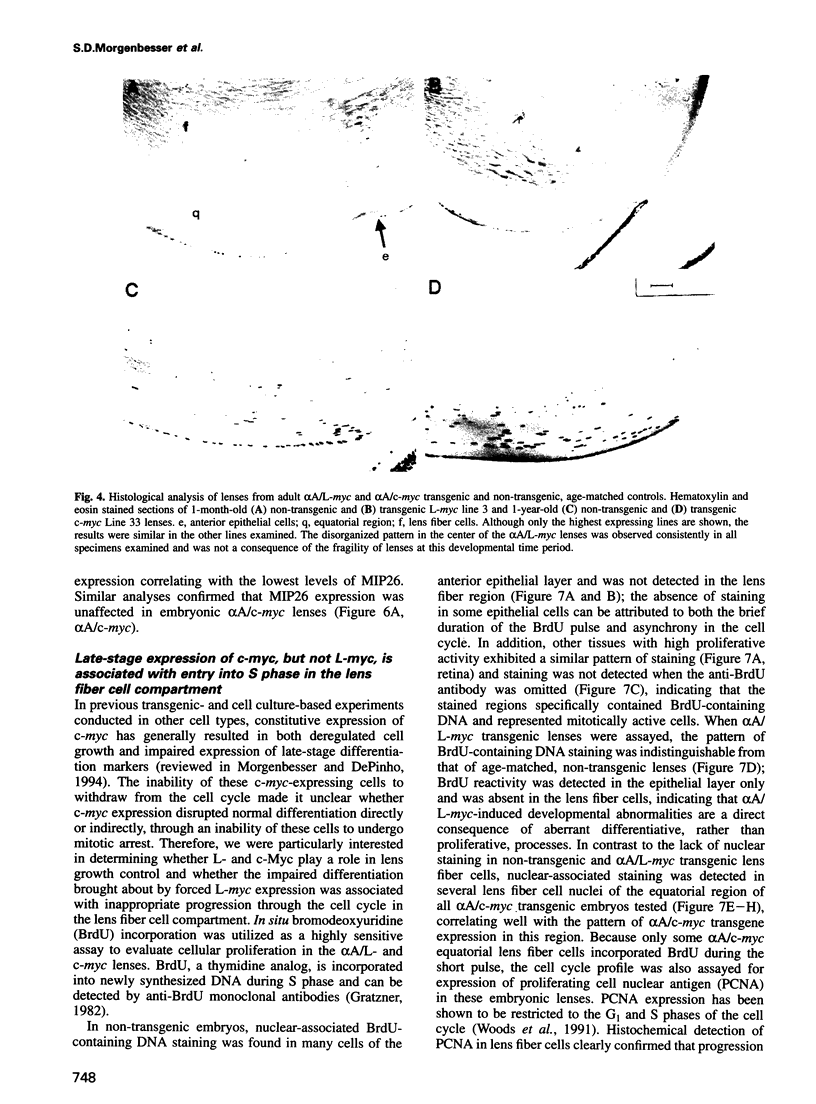
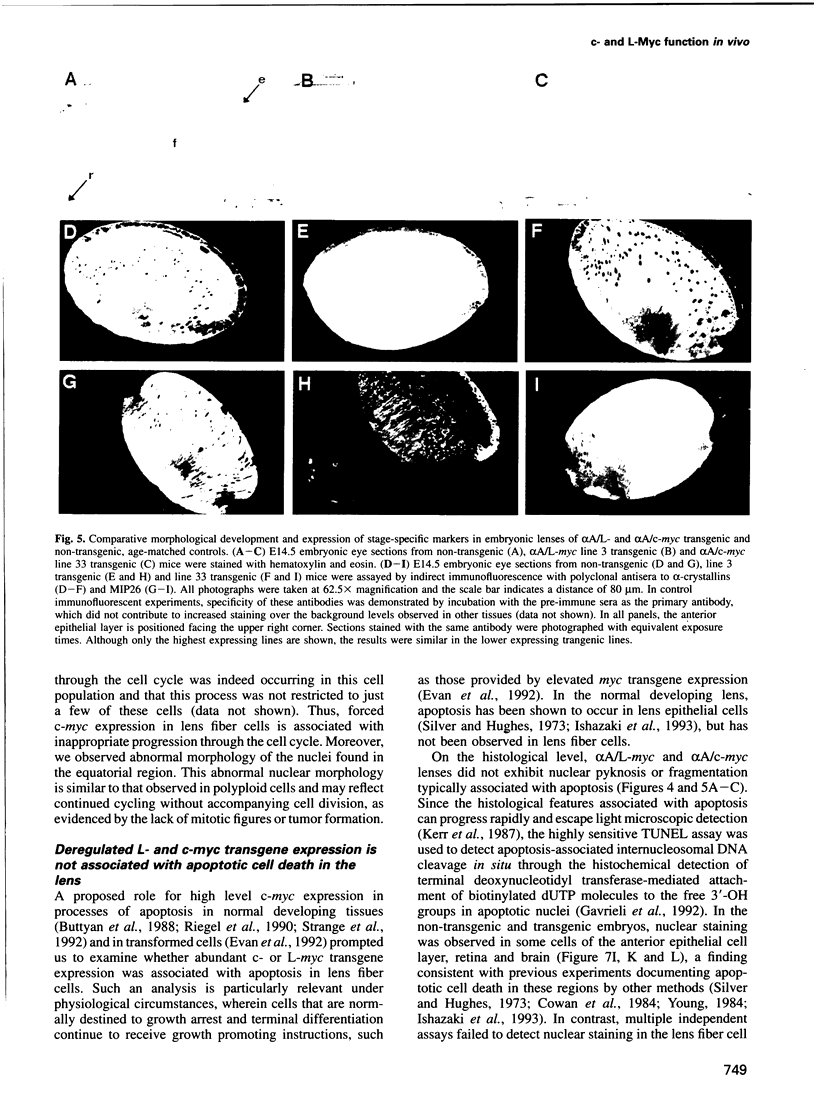




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alex R., Sözeri O., Meyer S., Dildrop R. Determination of the DNA sequence recognized by the bHLH-zip domain of the N-Myc protein. Nucleic Acids Res. 1992 May 11;20(9):2257–2263. doi: 10.1093/nar/20.9.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Eisenman R. N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993 Nov;7(11):2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Kretzner L., Eisenman R. N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993 Jan 29;72(2):211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- Barrett J., Birrer M. J., Kato G. J., Dosaka-Akita H., Dang C. V. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol Cell Biol. 1992 Jul;12(7):3130–3137. doi: 10.1128/mcb.12.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrer M. J., Raveh L., Dosaka H., Segal S. A transfected L-myc gene can substitute for c-myc in blocking murine erythroleukemia differentiation. Mol Cell Biol. 1989 Jun;9(6):2734–2737. doi: 10.1128/mcb.9.6.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrer M. J., Segal S., DeGreve J. S., Kaye F., Sausville E. A., Minna J. D. L-myc cooperates with ras to transform primary rat embryo fibroblasts. Mol Cell Biol. 1988 Jun;8(6):2668–2673. doi: 10.1128/mcb.8.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Lüscher B., Eisenman R. N. Myc and Max associate in vivo. Genes Dev. 1992 Jan;6(1):71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- Bok D., Dockstader J., Horwitz J. Immunocytochemical localization of the lens main intrinsic polypeptide (MIP26) in communicating junctions. J Cell Biol. 1982 Jan;92(1):213–220. doi: 10.1083/jcb.92.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekhuyse R. M., Kuhlmann E. D., Stols A. L. Lens membranes II. Isolation and characterization of the main intrinsic polypeptide (MIP) of bovine lens fiber membranes. Exp Eye Res. 1976 Sep;23(3):365–371. doi: 10.1016/0014-4835(76)90135-4. [DOI] [PubMed] [Google Scholar]
- Buttyan R., Zakeri Z., Lockshin R., Wolgemuth D. Cascade induction of c-fos, c-myc, and heat shock 70K transcripts during regression of the rat ventral prostate gland. Mol Endocrinol. 1988 Jul;2(7):650–657. doi: 10.1210/mend-2-7-650. [DOI] [PubMed] [Google Scholar]
- Campbell G. R., Zimmerman K., Blank R. D., Alt F. W., D'Eustachio P. Chromosomal location of N-myc and L-myc genes in the mouse. Oncogene Res. 1989;4(1):47–54. [PubMed] [Google Scholar]
- Charron J., Malynn B. A., Fisher P., Stewart V., Jeannotte L., Goff S. P., Robertson E. J., Alt F. W. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992 Dec;6(12A):2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- Chepelinsky A. B., King C. R., Zelenka P. S., Piatigorsky J. Lens-specific expression of the chloramphenicol acetyltransferase gene promoted by 5' flanking sequences of the murine alpha A-crystallin gene in explanted chicken lens epithelia. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2334–2338. doi: 10.1073/pnas.82.8.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola J. A., Cole M. D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986 Apr 24;320(6064):760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- Cowan W. M., Fawcett J. W., O'Leary D. D., Stanfield B. B. Regressive events in neurogenesis. Science. 1984 Sep 21;225(4668):1258–1265. doi: 10.1126/science.6474175. [DOI] [PubMed] [Google Scholar]
- Crews S., Barth R., Hood L., Prehn J., Calame K. Mouse c-myc oncogene is located on chromosome 15 and translocated to chromosome 12 in plasmacytomas. Science. 1982 Dec 24;218(4579):1319–1321. doi: 10.1126/science.7146913. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R., Bregni M., Erikson J., Patterson D., Gallo R. C., Croce C. M. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. C., Wims M., Spotts G. D., Hann S. R., Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993 Apr;7(4):671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- De Greve J., Battey J., Fedorko J., Birrer M., Evan G., Kaye F., Sausville E., Minna J. The human L-myc gene encodes multiple nuclear phosphoproteins from alternatively processed mRNAs. Mol Cell Biol. 1988 Oct;8(10):4381–4388. doi: 10.1128/mcb.8.10.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePinho R. A., Hatton K. S., Tesfaye A., Yancopoulos G. D., Alt F. W. The human myc gene family: structure and activity of L-myc and an L-myc pseudogene. Genes Dev. 1987 Dec;1(10):1311–1326. doi: 10.1101/gad.1.10.1311. [DOI] [PubMed] [Google Scholar]
- DePinho R. A., Legouy E., Feldman L. B., Kohl N. E., Yancopoulos G. D., Alt F. W. Structure and expression of the murine N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1827–1831. doi: 10.1073/pnas.83.6.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePinho R. A., Schreiber-Agus N., Alt F. W. myc family oncogenes in the development of normal and neoplastic cells. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- Dmitrovsky E., Kuehl W. M., Hollis G. F., Kirsch I. R., Bender T. P., Segal S. Expression of a transfected human c-myc oncogene inhibits differentiation of a mouse erythroleukaemia cell line. Nature. 1986 Aug 21;322(6081):748–750. doi: 10.1038/322748a0. [DOI] [PubMed] [Google Scholar]
- Downs K. M., Martin G. R., Bishop J. M. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 1989 Jun;3(6):860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- Evan G. I., Littlewood T. D. The role of c-myc in cell growth. Curr Opin Genet Dev. 1993 Feb;3(1):44–49. doi: 10.1016/s0959-437x(05)80339-9. [DOI] [PubMed] [Google Scholar]
- Evan G. I., Wyllie A. H., Gilbert C. S., Littlewood T. D., Land H., Brooks M., Waters C. M., Penn L. Z., Hancock D. C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992 Apr 3;69(1):119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y., Sherman Y., Ben-Sasson S. A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992 Nov;119(3):493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokkel E., Grossman Z., Ramot B., Yarden Y., Rechavi G., Givol D. Structural organization of the murine c-kit proto-oncogene. Oncogene. 1992 Jul;7(7):1423–1429. [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady E. F., Schwab M., Rosenau W. Expression of N-myc and c-src during the development of fetal human brain. Cancer Res. 1987 Jun 1;47(11):2931–2936. [PubMed] [Google Scholar]
- Gratzner H. G. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science. 1982 Oct 29;218(4571):474–475. doi: 10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]
- Griep A. E., Herber R., Jeon S., Lohse J. K., Dubielzig R. R., Lambert P. F. Tumorigenicity by human papillomavirus type 16 E6 and E7 in transgenic mice correlates with alterations in epithelial cell growth and differentiation. J Virol. 1993 Mar;67(3):1373–1384. doi: 10.1128/jvi.67.3.1373-1384.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griep A. E., Kuwabara T., Lee E. J., Westphal H. Perturbed development of the mouse lens by polyomavirus large T antigen does not lead to tumor formation. Genes Dev. 1989 Jul;3(7):1075–1085. doi: 10.1101/gad.3.7.1075. [DOI] [PubMed] [Google Scholar]
- Gu W., Bhatia K., Magrath I. T., Dang C. V., Dalla-Favera R. Binding and suppression of the Myc transcriptional activation domain by p107. Science. 1994 Apr 8;264(5156):251–254. doi: 10.1126/science.8146655. [DOI] [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Determination of the c-MYC DNA-binding site. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6162–6166. doi: 10.1073/pnas.88.14.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hateboer G., Timmers H. T., Rustgi A. K., Billaud M., van 't Veer L. J., Bernards R. TATA-binding protein and the retinoblastoma gene product bind to overlapping epitopes on c-Myc and adenovirus E1A protein. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8489–8493. doi: 10.1073/pnas.90.18.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Koike M., Matsutani M., Hoshino T. Effects of fixation time and enzymatic digestion on immunohistochemical demonstration of bromodeoxyuridine in formalin-fixed, paraffin-embedded tissue. J Histochem Cytochem. 1988 May;36(5):511–514. doi: 10.1177/36.5.3282006. [DOI] [PubMed] [Google Scholar]
- Hirning U., Schmid P., Schulz W. A., Rettenberger G., Hameister H. A comparative analysis of N-myc and c-myc expression and cellular proliferation in mouse organogenesis. Mech Dev. 1991 Feb;33(2):119–125. doi: 10.1016/0925-4773(91)90078-k. [DOI] [PubMed] [Google Scholar]
- Ishizaki Y., Voyvodic J. T., Burne J. F., Raff M. C. Control of lens epithelial cell survival. J Cell Biol. 1993 May;121(4):899–908. doi: 10.1083/jcb.121.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T., Allard M. F., Sreenan C. M., Doss L. K., Bishop S. P., Swain J. L. The c-myc proto-oncogene regulates cardiac development in transgenic mice. Mol Cell Biol. 1990 Jul;10(7):3709–3716. doi: 10.1128/mcb.10.7.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobovits A., Schwab M., Bishop J. M., Martin G. R. Expression of N-myc in teratocarcinoma stem cells and mouse embryos. Nature. 1985 Nov 14;318(6042):188–191. doi: 10.1038/318188a0. [DOI] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Lee W. M., Chen L. L., Dang C. V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992 Jan;6(1):81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- Kaye F., Battey J., Nau M., Brooks B., Seifter E., De Greve J., Birrer M., Sausville E., Minna J. Structure and expression of the human L-myc gene reveal a complex pattern of alternative mRNA processing. Mol Cell Biol. 1988 Jan;8(1):186–195. doi: 10.1128/mcb.8.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoff E., Bister K., Klempnauer K. H. Sequence-specific DNA binding by Myc proteins. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4323–4327. doi: 10.1073/pnas.88.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl N. E., Kanda N., Schreck R. R., Bruns G., Latt S. A., Gilbert F., Alt F. W. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983 Dec;35(2 Pt 1):359–367. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- Kratzke R. A., Otterson G. A., Lin A. Y., Shimizu E., Alexandrova N., Zajac-Kaye M., Horowitz J. M., Kaye F. J. Functional analysis at the Cys706 residue of the retinoblastoma protein. J Biol Chem. 1992 Dec 25;267(36):25998–26003. [PubMed] [Google Scholar]
- Lahoz E. G., Xu L., Schreiber-Agus N., DePinho R. A. Suppression of Myc, but not E1a, transformation activity by Max-associated proteins, Mad and Mxi1. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5503–5507. doi: 10.1073/pnas.91.12.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder A., Pattengale P. K., Kuo A., Stewart T. A., Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986 May 23;45(4):485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- Legouy E., DePinho R., Zimmerman K., Collum R., Yancopoulos G., Mitsock L., Kriz R., Alt F. W. Structure and expression of the murine L-myc gene. EMBO J. 1987 Nov;6(11):3359–3366. doi: 10.1002/j.1460-2075.1987.tb02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A., Moroy T., Collum R., Weintraub H., Alt F. W., Blackwell T. K. DNA binding by N- and L-Myc proteins. Oncogene. 1993 Apr;8(4):1093–1098. [PubMed] [Google Scholar]
- Ma A., Smith R. K., Tesfaye A., Achacoso P., Dildrop R., Rosenberg N., Alt F. W. Mechanism of endogenous myc gene down-regulation in E mu-N-myc tumors. Mol Cell Biol. 1991 Jan;11(1):440–444. doi: 10.1128/mcb.11.1.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon K. A., Chepelinsky A. B., Khillan J. S., Overbeek P. A., Piatigorsky J., Westphal H. Oncogenesis of the lens in transgenic mice. Science. 1987 Mar 27;235(4796):1622–1628. doi: 10.1126/science.3029873. [DOI] [PubMed] [Google Scholar]
- McAvoy J. W. Induction of the eye lens. Differentiation. 1980;17(3):137–149. doi: 10.1111/j.1432-0436.1980.tb01091.x. [DOI] [PubMed] [Google Scholar]
- McBurney M. W., Jones-Villeneuve E. M., Edwards M. K., Anderson P. J. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982 Sep 9;299(5879):165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- Miller M. W., Nowakowski R. S. Use of bromodeoxyuridine-immunohistochemistry to examine the proliferation, migration and time of origin of cells in the central nervous system. Brain Res. 1988 Aug 2;457(1):44–52. doi: 10.1016/0006-8993(88)90055-8. [DOI] [PubMed] [Google Scholar]
- Morgenbesser S. D., DePinho R. A. Use of transgenic mice to study myc family gene function in normal mammalian development and in cancer. Semin Cancer Biol. 1994 Feb;5(1):21–36. [PubMed] [Google Scholar]
- Morgenbesser S. D., Williams B. O., Jacks T., DePinho R. A. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature. 1994 Sep 1;371(6492):72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- Mugrauer G., Alt F. W., Ekblom P. N-myc proto-oncogene expression during organogenesis in the developing mouse as revealed by in situ hybridization. J Cell Biol. 1988 Oct;107(4):1325–1335. doi: 10.1083/jcb.107.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugrauer G., Ekblom P. Contrasting expression patterns of three members of the myc family of protooncogenes in the developing and adult mouse kidney. J Cell Biol. 1991 Jan;112(1):13–25. doi: 10.1083/jcb.112.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B., Morgenbesser S. D., DePinho R. A. Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants. Genes Dev. 1992 Aug;6(8):1480–1492. doi: 10.1101/gad.6.8.1480. [DOI] [PubMed] [Google Scholar]
- Möröy T., Fisher P., Guidos C., Ma A., Zimmerman K., Tesfaye A., DePinho R., Weissman I., Alt F. W. IgH enhancer deregulated expression of L-myc: abnormal T lymphocyte development and T cell lymphomagenesis. EMBO J. 1990 Nov;9(11):3659–3666. doi: 10.1002/j.1460-2075.1990.tb07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Mahon K. A., Miskin R., Dey A., Kuwabara T., Westphal H. Differentiation and oncogenesis: phenotypically distinct lens tumors in transgenic mice. New Biol. 1989 Nov;1(2):193–204. [PubMed] [Google Scholar]
- Nath P., Getzenberg R., Beebe D., Pallansch L., Zelenka P. c-myc mRNA is elevated as differentiating lens cells withdraw from the cell cycle. Exp Cell Res. 1987 Mar;169(1):215–222. doi: 10.1016/0014-4827(87)90239-4. [DOI] [PubMed] [Google Scholar]
- Nau M. M., Brooks B. J., Battey J., Sausville E., Gazdar A. F., Kirsch I. R., McBride O. W., Bertness V., Hollis G. F., Minna J. D. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985 Nov 7;318(6041):69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
- Neel B. G., Jhanwar S. C., Chaganti R. S., Hayward W. S. Two human c-onc genes are located on the long arm of chromosome 8. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7842–7846. doi: 10.1073/pnas.79.24.7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisen P. D., Zimmerman K. A., Cotter S. V., Gilbert F., Alt F. W. Enhanced expression of the N-myc gene in Wilms' tumors. Cancer Res. 1986 Dec;46(12 Pt 1):6217–6222. [PubMed] [Google Scholar]
- Overbeek P. A., Chepelinsky A. B., Khillan J. S., Piatigorsky J., Westphal H. Lens-specific expression and developmental regulation of the bacterial chloramphenicol acetyltransferase gene driven by the murine alpha A-crystallin promoter in transgenic mice. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7815–7819. doi: 10.1073/pnas.82.23.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatigorsky J. Lens differentiation in vertebrates. A review of cellular and molecular features. Differentiation. 1981;19(3):134–153. doi: 10.1111/j.1432-0436.1981.tb01141.x. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Prochownik E. V., Kukowska J. Deregulated expression of c-myc by murine erythroleukaemia cells prevents differentiation. 1986 Aug 28-Sep 3Nature. 322(6082):848–850. doi: 10.1038/322848a0. [DOI] [PubMed] [Google Scholar]
- Riegel J. S., Richie E. R., Allison J. P. Nuclear events after activation of CD4+8+ thymocytes. J Immunol. 1990 May 1;144(9):3611–3618. [PubMed] [Google Scholar]
- Rustgi A. K., Dyson N., Bernards R. Amino-terminal domains of c-myc and N-myc proteins mediate binding to the retinoblastoma gene product. Nature. 1991 Aug 8;352(6335):541–544. doi: 10.1038/352541a0. [DOI] [PubMed] [Google Scholar]
- Sandgren E. P., Quaife C. J., Pinkert C. A., Palmiter R. D., Brinster R. L. Oncogene-induced liver neoplasia in transgenic mice. Oncogene. 1989 Jun;4(6):715–724. [PubMed] [Google Scholar]
- Sawai S., Shimono A., Wakamatsu Y., Palmes C., Hanaoka K., Kondoh H. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development. 1993 Apr;117(4):1445–1455. doi: 10.1242/dev.117.4.1445. [DOI] [PubMed] [Google Scholar]
- Schmid P., Schulz W. A., Hameister H. Dynamic expression pattern of the myc protooncogene in midgestation mouse embryos. Science. 1989 Jan 13;243(4888):226–229. doi: 10.1126/science.2911736. [DOI] [PubMed] [Google Scholar]
- Schoenenberger C. A., Andres A. C., Groner B., van der Valk M., LeMeur M., Gerlinger P. Targeted c-myc gene expression in mammary glands of transgenic mice induces mammary tumours with constitutive milk protein gene transcription. EMBO J. 1988 Jan;7(1):169–175. doi: 10.1002/j.1460-2075.1988.tb02797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber-Agus N., Horner J., Torres R., Chiu F. C., DePinho R. A. Zebra fish myc family and max genes: differential expression and oncogenic activity throughout vertebrate evolution. Mol Cell Biol. 1993 May;13(5):2765–2775. doi: 10.1128/mcb.13.5.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Alitalo K., Klempnauer K. H., Varmus H. E., Bishop J. M., Gilbert F., Brodeur G., Goldstein M., Trent J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983 Sep 15;305(5931):245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- Silver J., Hughes A. F. The role of cell death during morphogenesis of the mammalian eye. J Morphol. 1973 Jun;140(2):159–170. doi: 10.1002/jmor.1051400204. [DOI] [PubMed] [Google Scholar]
- Spanopoulou E., Early A., Elliott J., Crispe N., Ladyman H., Ritter M., Watt S., Grosveld F., Kioussis D. Complex lymphoid and epithelial thymic tumours in Thy1-myc transgenic mice. Nature. 1989 Nov 9;342(6246):185–189. doi: 10.1038/342185a0. [DOI] [PubMed] [Google Scholar]
- Stanton B. R., Perkins A. S., Tessarollo L., Sassoon D. A., Parada L. F. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 1992 Dec;6(12A):2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Strange R., Li F., Saurer S., Burkhardt A., Friis R. R. Apoptotic cell death and tissue remodelling during mouse mammary gland involution. Development. 1992 May;115(1):49–58. doi: 10.1242/dev.115.1.49. [DOI] [PubMed] [Google Scholar]
- Takemoto L., Kuck J., Kuck K. Changes in the major intrinsic polypeptide (MIP26K) during opacification of the Emory mouse lens. Exp Eye Res. 1988 Aug;47(2):329–336. doi: 10.1016/0014-4835(88)90015-2. [DOI] [PubMed] [Google Scholar]
- Taub R., Kirsch I., Morton C., Lenoir G., Swan D., Tronick S., Aaronson S., Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudel M., D'Agati V., Costantini F. C-myc as an inducer of polycystic kidney disease in transgenic mice. Kidney Int. 1991 Apr;39(4):665–671. doi: 10.1038/ki.1991.80. [DOI] [PubMed] [Google Scholar]
- Vermorken A. J., Hilderink J. M., Dunia I., Benedetti E. L., Bloemendal H. Changes in membrane protein pattern in relation to lens cell differentiation. FEBS Lett. 1977 Nov 15;83(2):301–306. doi: 10.1016/0014-5793(77)81028-4. [DOI] [PubMed] [Google Scholar]
- Wenzel A., Cziepluch C., Hamann U., Schürmann J., Schwab M. The N-Myc oncoprotein is associated in vivo with the phosphoprotein Max(p20/22) in human neuroblastoma cells. EMBO J. 1991 Dec;10(12):3703–3712. doi: 10.1002/j.1460-2075.1991.tb04938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. M., Robertson N. P., Horwitz J. Heat induced aggregation of the sodium dodecyl sulfate-solubilized main intrinsic polypeptide isolated from bovine lens plasma membrane. Biochem Biophys Res Commun. 1978 Sep 14;84(1):158–165. doi: 10.1016/0006-291x(78)90277-2. [DOI] [PubMed] [Google Scholar]
- Woods A. L., Hall P. A., Shepherd N. A., Hanby A. M., Waseem N. H., Lane D. P., Levison D. A. The assessment of proliferating cell nuclear antigen (PCNA) immunostaining in primary gastrointestinal lymphomas and its relationship to histological grade, S+G2+M phase fraction (flow cytometric analysis) and prognosis. Histopathology. 1991 Jul;19(1):21–27. doi: 10.1111/j.1365-2559.1991.tb00890.x. [DOI] [PubMed] [Google Scholar]
- Wyllie A. H. Apoptosis and the regulation of cell numbers in normal and neoplastic tissues: an overview. Cancer Metastasis Rev. 1992 Sep;11(2):95–103. doi: 10.1007/BF00048057. [DOI] [PubMed] [Google Scholar]
- Xu L., Morgenbesser S. D., DePinho R. A. Complex transcriptional regulation of myc family gene expression in the developing mouse brain and liver. Mol Cell Biol. 1991 Dec;11(12):6007–6015. doi: 10.1128/mcb.11.12.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. W. Cell death during differentiation of the retina in the mouse. J Comp Neurol. 1984 Nov 1;229(3):362–373. doi: 10.1002/cne.902290307. [DOI] [PubMed] [Google Scholar]
- Zervos A. S., Gyuris J., Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993 Jan 29;72(2):223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- Zimmerman K. A., Yancopoulos G. D., Collum R. G., Smith R. K., Kohl N. E., Denis K. A., Nau M. M., Witte O. N., Toran-Allerand D., Gee C. E. Differential expression of myc family genes during murine development. 1986 Feb 27-Mar 5Nature. 319(6056):780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]