Abstract
Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) is one of the most abundant RNA binding proteins. hnRNP A1 is localized prevalently in the nucleus but it can relocate to the cytoplasm in response to specific stimuli shuttling between nuclear and cytoplasmic compartments. The cellular localization of this protein is regulated by a short C-terminus motif (M9) and other less defined sequences. The RNA binding specificity of this protein is dependent on multiple RNA binding domains (RBDs), which regulate its role in RNA processing and expression. hnRNP A1 plays multiple roles in gene expression by regulating the biogenesis and translation of messengers RNAs, the processing of miRNAs, affecting transcription and controlling telomere maintenance. The multiple functions of this protein correlate with diverse roles in genetic disease, cancer and the replication of viral pathogens. Utilizing a tagged hnRNP A1 deletion library we have shown that the three hnRNP A1 RBDs contribute to the prevalent nuclear distribution of the protein. Our data also indicate that a truncated form of the protein, lacking one of the RBDs, the RGG-box, can regulate splicing of a splicing reporter minigene and down-regulate replication of the HIV-1 virus with efficiency comparable to the wild type protein. This functional hnRNP A1 deletion mutant is similar to a predicted hnRNP A1 isoform, which had not been previously experimentally characterized.
Keywords: hnRNP A1, RRM, RGG-box, alternative splicing, HIV-1
1. Introduction
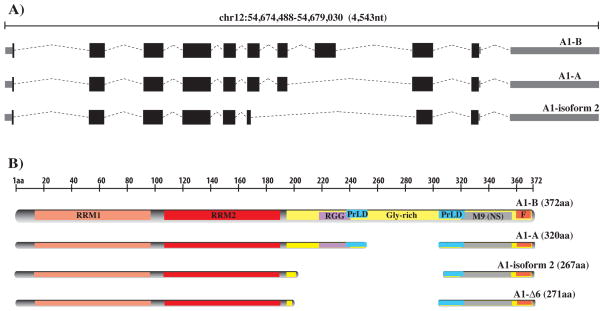
hnRNP A1 is one of the most abundant and ubiquitously expressed nuclear proteins and plays a major role in the biogenesis and expression of messenger RNAs. The characterization of specific RNA sequences recognized by this protein and evidence of its functions in both DNA and RNA metabolism quickly defined its role as a major regulator of gene expression. hnRNP A1 contributes to the processing of microRNAs, telomere maintenance and transcription regulation although it is its function as a constitutive and alternative splicing factor that is the most studied and better understood (reviewed in [1]). Several coding and non-coding transcripts have been predicted for hnRNP A1, nevertheless only two alternatively spliced isoforms have been characterized; A1-B (372 aa, 38 kDa, ENSEMBL ID ENST00000340913), and A1-A (320 aa, 34 kDa, ENSEMBL ID ENST00000546500), which, in most tissues, is over 20-times more abundant than the larger isoform [2].
hnRNP A1 is composed by structurally and functionally independent domains. The N-terminus contains two closely-related RNA binding domains (RBDs) of the RRM type (RRM1 and RRM2), followed by a highly flexible glycine-rich (Gly-rich) C-terminal region (Fig. 1A) [3]. The Gly-rich domain has been shown to have both protein and RNA binding properties, the latter mostly due to the presence of a third RBD characterized by closely spaced clusters of Arg-Gly-Gly tripeptide repeats with interspersed aromatic (Phe, Tyr) residues, named the RGG-box [4]. The M9 nucleo-cytoplasmic shuttling sequence [5], which is required for the protein shuttle between the nucleus and the cytoplasm [6, 7], is located within the C-terminal Gly-rich domain. Furthermore, a recent report showed that the Gly-rich region also harbors a prion-like domain (PrLD), which, if mutated, might induce the aggregation of hnRNP A1 into fibrils [8].
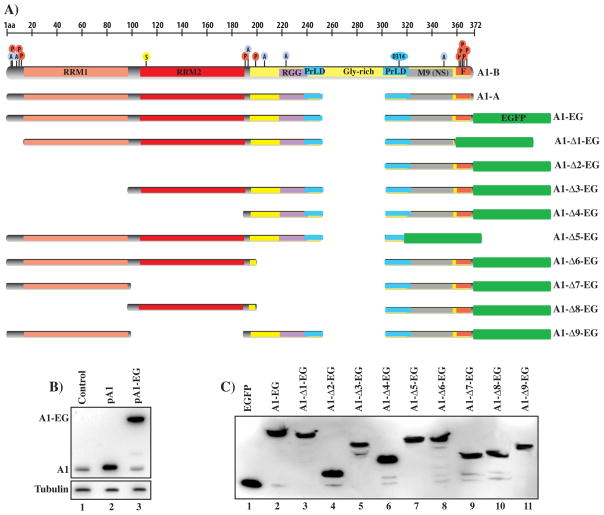
Fig. 1.
(A) Schematic representation of hnRNP A1 isoforms (A1-A, A1B) and deletion clones map. The protein RBDs and other key structural and functional features are indicated, M9 (nuclear localization sequence), F (F-peptide), PrLD (prion like domain). The location of residues modified by acetylation (A), phosphorylation (P) and SUMOylation (S) is indicated. The sequences present in the major deletion clones are indicated. (B) Expression of hnRNP A1 in control HEK-293 cells or transfected with the hnRNP A1-A expression clone (pA1) or the plasmid expressing the hnRNP A1-EGFP fusion protein. Proteins are detected with the anti-hnRNP A1 antibody. (C) Expression of the hnRNP A1–EGFP fusion proteins in HEK-29 cells. Proteins are detected with the anti-GFP antibody.
Much of hnRNP A1 functions are dependent on its ability to recognize specific nucleic acid sequences. The two RRMs, despite their sequences and structural similarities, are neither redundant nor functionally equivalent, [9]. The precise contribution of the RGG-box to the overall RNA binding activity of hnRNP A1 is not well understood, although in-vitro experimental data indicates that it is responsible for the cooperative binding of the protein to its target RNA [9, 10].
hnRNP A1 plays a prominent role in viral replication and pathogenesis. Biochemical and functional studies aimed at the characterization of the complex mechanisms regulating the expression of the HIV-1 genome, have shown that hnRNP A1 is part of a complex regulatory network that regulates viral mRNA export, stability and splicing [1]. The mechanisms that regulate HIV-1 gene expression utilize both, cellular and viral factors, which interact with several cis-acting sequences located within the viral transcripts. Transcription of the integrated viral genome is mediated by several cellular transcription factors and the viral protein Tat, which binds a short sequence, named TAR, at the 5′ end of the viral transcript [11]. The integrated viral genome is transcribed into a single primary transcript, which undergoes a complex series of splicing events to generate several mRNAs coding for the nine viral genes [12]. HIV-1 splicing regulation relies on the presence of intronic and exonic sequences as well as cellular splicing factors that interact with these elements [13].
We have previously identified a complex cis-acting element located within the first tat coding exon that is required for the splicing regulation of tat coding mRNAs [14, 15]. hnRNP A1 binds a short regulatory sequence, named exonic splicing silencer 2 (ESS2), to repress splicing of the tat messenger. A similar regulatory mechanism has been also characterized within the second tat coding exon [16, 17]. Over-expression of hnRNP A1 in an ex-vivo cellular system down-regulates expression of the transcriptional transactivator Tat, which results in a sharp reduction in the transcription of the viral genome and a 100 fold drop in the production of new virions [18].
The understanding of how each structural domain and sequence motif of hnRNP A1 contribute to the functions of the proteins is key in understanding a number of molecular mechanisms and contribute to the development of novel therapies for genetic and viral diseases. We set up to study the role that the different domains and functional motifs of this protein play in cellular localization, splicing and viral replication.
Furthermore, we wanted to determine if a minimal truncated isoform of this protein could recapitulate the effect of the full-length protein in HIV-1 splicing and replication. Here we report that additional sequence motifs other than the M9 domain are required for the proper cellular localization of the protein and that the RGG-box is not essential for the protein functions in alternative splicing. An hnRNP A1 clone lacking the RGG-box and a portion of the PrLD is properly localized and can functionally substitute for the ubiquitously expressed A1-A isoform. Furthermore, analysis of protein and gene sequence databases indicates that an isoform of hnRNP A1 lacking the RGG-box, similar to the functional deletion clone we characterized, may be expressed in specific human tissues.
2. Materials and methods
2.1. Plasmids and cell transfections
The hnRNP A1 deletion clones library was obtained by PCR amplification and cloning into the pEGFP-N1 vector (Clontech) of hnRNP A1-A fragments from the pGT7-A1 expression vector (donation from Dr. A. Krainer, CSHL). Single hnRNP A1-A domains and subdomains were amplified utilizing primers F1 (agctccgcggtcttcaccctgccgtcatgtctaag), F2 (agctccgcggatgcccgaacagctgaggaagctcttc), F3 (atgcccgcggatgggaagcaattttggaggtggtg), F4 (agctccgcggatggtctccagagaagattctcaaagac), F5 (atgcccgcggatggctagtgcttcatccagccaaag), R1 (atgcggatccccaaatcttctgccactgccatagctac), R2 (atgcggatccccaccgccatagccaccttggtttcg), R3 (atgcggatccccgtaattcccaaaatcattgtagc), R4 (atgcccgcggcccacttcaatttttccatactgttc), R5 (atgcccgcggggcacctggtctttgagaatcttc), R6 (atgcccgcggtccagaaccacttcgacctctttg), R7 (atgcccgcggcctttctgaatgacaatcttatccac) and cloned following the diagram shown in figure S1. HEK-293 cells were maintained at below 80% confluence in D-MEM (Gibco BRL) supplemented with 8% fetal calf serum and gentamicin. Cells were transfected in 24 well plates with Lipofectamine 2000 (Invitrogen) with 0.1 μg of the hnRNP A1 constructs and 0.4 μg of the reporter plasmid pLTR-S1Xm-R (REF) or the HIV-1 molecular clone pNL4-3 (NIH AIDS Research & Reference Reagent Program). Western blot analysis was carried out with the following antibodies: hnRNP A1 (mAb 9H10) (provided by Dr. G. Dreyfuss, University of Pennsylvania), GFP (B-2, Santa Cruz Biotech), β-Tubulin (Sigma).
2.2. RNA extraction and PCR analysis
Total RNA was extracted 48 after transfection with the Total RNA Isolation Kit (Agilent) and DNase treated with Turbo DNase (Ambion). RNA was reverse transcribed utilizing a random pd(N)6 primer and Superscript II RT (LifeTechnologies). Quantitative PCR analysis of the viral transcripts was obtained as previously described [18] utilizing primers pGagF (ttcttcagagcagaccagagc) and PGagR (gctgccaaagagtgatctga) for gag/pol specific mRNA, Pex8F (ttgctcaatgccacagccat), Pex8R (tttgaccacttgccacccat) for total viral mRNA, PTatF (aggggcggcgactgaattgggt), PTatR (gattgggaggtgggttgctttg) for tat specific mRNA. Each sample was normalized for the relative content in the housekeeping gene GAPDH. qPCR was performed utilizing a Stratagene Mx3005P real time PCR system, SYBR green dye and analyzed with MxPro V3.0 software. Each assay was carried out with a minimum of three independent transfections while qPCR assays were carried out in duplicates. Semi-quantitative RT-PCR analysis of the pLTR-mS1X-R transcripts was performed with the primer pair PFL1 (aaagcttgccttgagtgcttca), PRL1 (gatgagctcttcgtcgctgt) and 29 cycles of amplification.
2.3. Viral replication assays
TZM-bl cells (NIH AIDS Research & Reference Reagent Program) were seeded 24 hours before infection in 96 well plates at 20% confluence in 200 μL of D-MEM supplemented with 8% fetal calf serum and gentamicin. Supernatant collected from the HEK-293 cells 72 hours after the transfection carried out with the proviral constructs pNL4-3 and each hnRNP A1 coding construct was utilized to infect the TZM-bl cells. At 48 hours post infection cells were lysed and luciferase expression was assayed and quantified utilizing a BMG PolarStar Omega reader and MARS data analysis software.
2.4. Fluorescence Microscopy
HEK-293 cells were transfected with the EGFP tagged clones and cultured on poly-Lysine coated glass coverslips. At 24 hours post-transfection, cells were fixed with 4% p-formaldehyde in PBS for 30 min. For cytosol labeling the cells were permeabilized with 0.2% Triton X-100 for 10 min and washed three times with PBS. The cells were incubated with Alexa Fluor-594 Phalloidin (Life-Technologies) at room temperature for 30 min and washed three times with PBS. Cells were mounted onto slides using Vectastain (Invitrogen). For immunofluorescence assays cells were permeabilized with 0.2% Triton X-100 for 10 minutes and washed three times with PBS. Cells were blocked in blocking buffer (10% normal goat serum and 1% BSA in PBS) for 1 hour at 37 °C and incubated with anti-SC35 antibody (a gift from Dr. J. Stevenin, INSERM, Strasbourg, France) or anti-hnRNP A1 antibody 4B10 (SC Biotech). Cells were washed three times with PBS, and then incubated with Alexa Fluor 647 secondary IgG (Invitrogen) 1:2000 in PBS containing 1% normal goat serum and 1% BSA) for 30 minutes at room temperature. Cells were washed three times with PBS, and mounted onto slides using Vectastain (Invitrogen). Samples were visualized and analyzed using a laser confocal microscope (Zeiss LSM700) and image analysis was performed using the ImageJ software.
3. Results
3.1. hnRNP A1 deletion library
We have previously shown that the hnRNP A1-A isoform, which is prevalently expressed in most tissues, modulates viral splicing and replication. To study the roles played by the functional domains of hnRNP A1 in viral replication we constructed a library of EGFP tagged hnRNP A1 expression clones carrying single or multiple deletions of complete or partial functional domains. In building the library we included protein-coding isoforms predicted by the ENSEMBL database but lacking a functional characterization. The expression level of the tagged clones was comparable to the one obtained with the clone expressing the wild type A1-A isoform sequence (Fig. 1B and 1C), which we have previously utilized to inhibit viral splicing and replication [18].
All clones were analyzed for their ability to properly localize within the nuclear compartment, regulate a viral splicing substrate and inhibit HIV-1 replication. The 9 clones shown in figure 1 are representative of the key findings obtained by analyzing the 31 clones library (Fig. S1 and Tab. 1). Clones with deletions of partial domains that failed to distinguish functionally from the deletion of the complete domain are not shown in figure 1.
3.2. The RBDs contribute to the nuclear localization of hnRNP A1
hnRNP A1 is predominantly localized in the nucleus at steady-state, but can shuttle between the nucleus and the cytoplasm in response to specific signals [6, 7]. The nuclear import of hnRNP A1 is mediated by the direct interaction of the M9 sequence with two transport receptors of the karyopherin-β family, Transportin 1 and 2 (Trn1, Trn2) [19–21]. This sequence also acts as a nuclear export signal, allowing export and cytoplasmic accumulation of hnRNP A1 in response to physiological signals [22]. Comparison of the cellular localization of the endogenous hnRNP A1 with the transiently expressed A1-EG clone showed that addition of the EGFP tag did not alter the predominant nuclear localization of the protein (Fig. 2, A1 vs A1-EG).
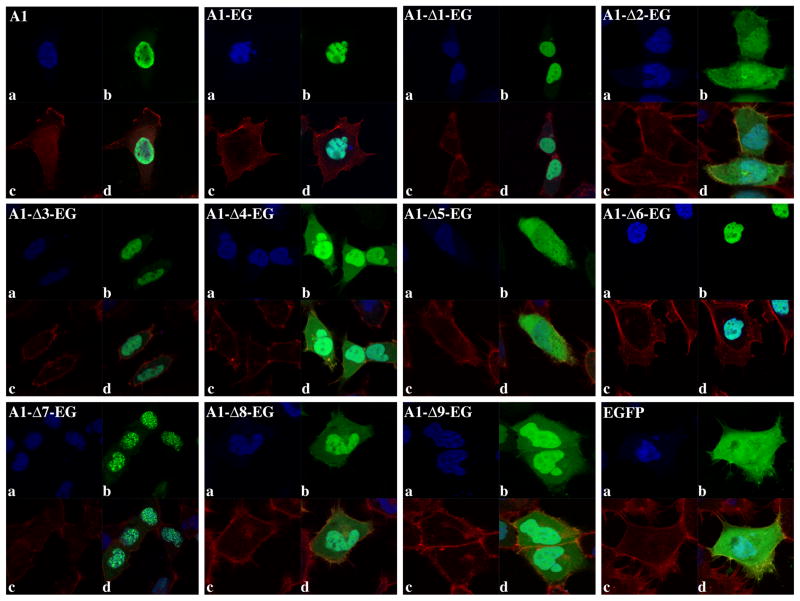
Fig. 2.
Localization of endogenous hnRNP A1 and EGFP tagged wild type and deletion clones. (a) Nuclear DAPI staining. (b) Endogenous hnRNP A1 (A1) was detected by immunofluorescence while tagged hnRNP A1-EGFP clones were detected by fluorescence microscopy (green). (c) Cytosol is visualized with the Phalloidin actin stain (red). (d) Overlay and localization of the endogenous and tagged proteins within the nuclei and cytosol.
In agreement with previous results [5] deletion of a fragment containing the M9 sequence caused a loss of nuclear localization (Fig. 2, A1-Δ5-EG), nevertheless clones containing M9 but lacking other sequences failed to localize primarily within the nucleus (clone A1-Δ2-EG). Thus, the sole presence of the M9 sequence was not sufficient to establish the prevalent nuclear localization of the protein. Although previous reports indicated that the M9 sequence was necessary and sufficient for the prevalent nuclear localization of hnRNP A1 [5], sequences other than the M9 have also been shown to contribute to the cellular localization of hnRNP A1.
The F-peptide (Fig. 1A) is a short sequence located at the C-terminus of the protein that contains 7 serine residues, which are hypophosphorylated under normal physiological conditions and became phosphorylated in response to osmotic shock. Phosphorylation of the F-peptide weakens the interaction between hnRNP A1 and Trn1, thus increasing the cytoplasmic localization of the protein [7, 23]. It is plausible that the presence of the F-peptide in the A1-Δ2-EG clone could prevent nuclear import of hnRNP A1and cause its accumulation in the cytoplasm. Nevertheless, deletion of the F-peptide from the A1-Δ1-EG or other clones (data not shown) did not alter the cellular distribution of the protein, indicating that F-peptide exerted no specific inhibition of nuclear import. Further analysis of the deletion clones showed that addition of one or more RBDs to the M9 sequence could partially restore nuclear localization. While addition of both RRMs or RRM2 and the RGG box to the M9 induced the predominant nuclear localization of the protein (Fig. 2 clones A1-Δ3-EG, A1-Δ6-EG), addition of the RRM1 in combination with the RGG box only partially restored it (Fig. 2 A1–Δ9-EG). Addition of the sole RRM1 to M9 (A1-Δ7-EG) induced localization of the protein into discrete sub-nuclear structures. Several RNA binding proteins regulating mRNA processing have been shown to congregate in nuclear speckles, which are nuclear domains enriched in pre-mRNA splicing factors. Nevertheless, the gene product expressed by the A1-Δ7-EG clone does not colocalize with the nuclear speckles marker SC35 (Fig. S2). Localization of the protein generated by the clone A1-Δ7-EG in nuclear aggregates with perinuclear distribution suggests an association with specific subsets of RNAs or RNA processing and export factors. Taken together, these data indicate that, although the M9 sequence is essential for the prevalent nuclear localization of this protein, one or multiple RBDs are also required.
3.3 The RGG box is not required for the alternative splicing activity of hnRNP A1
hnRNP A1 is one of the main cellular factors regulating the alternative splicing of cellular and viral genes. Biochemical and cellular studies have uncovered several mechanisms utilized by hnRNP A1 to modulate splicing [1]. In previous work, we have identified a complex splicing regulatory sequence within the first coding exon of the HIV-1 tat coding mRNA, which is required for proper splicing of the viral transcript [15]. This regulatory element is composed of overlapping exonic splicing enhancer (ESE) and silencer (ESS) sequences (Fig 3A). A detailed biochemical analysis showed that hnRNP A1 binds the ESS to repress splicing of the tat coding mRNAs while binding of SC35, a member of the SR proteins family of splicing regulators, promotes it. hnRNP A1 binding to the ESS inhibits recognition by the splicing machinery of the upstream, tat specific, 3′ splice site by masking the SC35 binding site [15].
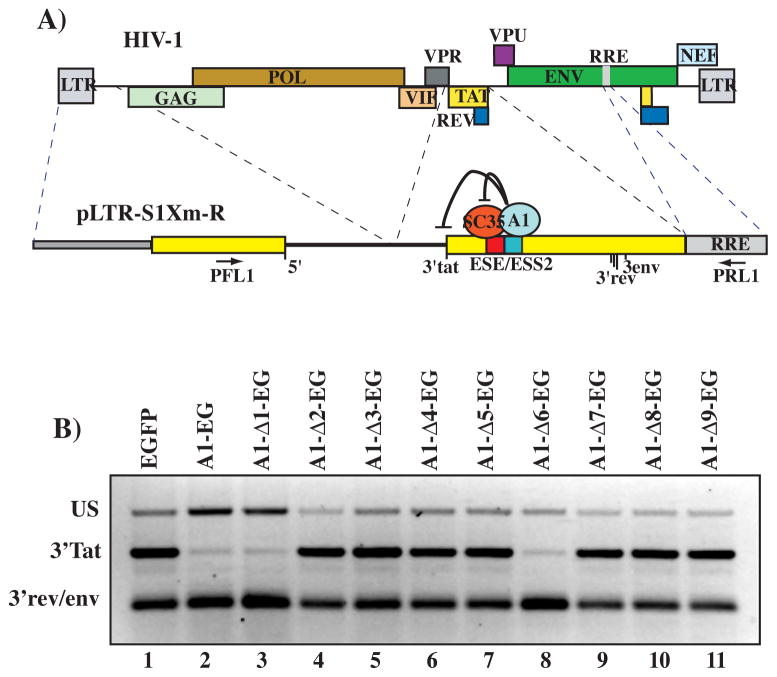
Fig. 3.
(A) Schematic map of the pLTR-S1Xm-R reporter minigene. Dashed lines indicate the viral sequences utilized to construct the minigenes in relation to the HIV-1 genome (top). The mechanism regulating splicing at the tat specific splice site (3′tat) via the binding of competing splicing factors SC35 and hnRNP A1 to a complex regulatory sequence (ESE/ESS2) is shown. Location of the cluster of 3′ splice sites utilized to generate viral rev and env coding mRNAs is also indicated. Location of the primers utilized in the semi-quantitative RT-PCR assay is shown. (B) Alternative splicing of the pLTR-S1Xm-R reporter. HEK 293 cells were co-transfected with the pLTR-S1Xm-R reporter and the indicated hnRNP A1-EGFP fusion clone or the control EGFP expression vector. RT-PCR analysis shows the unspliced (US) and mRNAs splicing to the splice sites 3′tat and 3′rev/env cluster.
To study the effects that deletion of the multiple hnRNP A1 domains have on the protein splicing regulatory functions, we have utilized a minigene derived from HIV-1 sequences that mimics the splicing regulation of the viral mRNA. Co-transfection of the minigene pLTR-S1Xm-R with the A1-EG clone in the absence of the viral transcriptional trans-activator Tat resulted in a marked decrease of splicing to the tat specific splice site (Fig 3B). A similar result was obtained with the clone A1-Δ1-EG indicating that the F-peptide was not required for proper splicing activity.
A clone containing the RRM1 and RRM2 domains but lacking the RGG-box (A1-Δ6-EG) induced a splicing pattern similar to the one obtained with the wild type sequence, thus suggesting that the contribution of the RGG-box to splicing regulation is minimal. This is in agreement with work showing that both RRM1 and RRM2 are required for hnRNP A1 splicing activity [9] and although previous reports indicated that the RGG-box conveys the RNA affinity specificity to the Gly-rich domain [24, 25] the present data suggest a lesser role for the Gly-rich domain and the RGG-box in splicing. Nevertheless, analysis of the relative ratios of unspliced and spliced mRNAs showed that expression of the wild type and A1–D1-EG mutant correlates with an increase in the amount of unspliced mRNA detected, while expression of the A1-Δ6-EG does not. Thus, the A1-Δ6-EG clone might have distinct functions in splicing than the full-length protein suggesting a role for the RGG-box in splicing regulation.
3.4 hnRNP A1 deletion mutants can inhibit HIV-1 replication
hnRNP A1 has been shown to regulate viral transcription, mRNA processing and translation through a number of independent mechanisms [1, 26–28]. Transcription of the HIV-1 genome is regulated by the viral protein Tat [29, 30] and over-expression of hnRNP A1 results in a decrease of the tat coding mRNAs, thus reducing the transcription rate of the viral genome and the production of functional virions [18]. To study the role of the different hnRNP A1 domains in HIV-1 replication we co-transfected human HEK-293 cells with the proviral vector pNL4-3, which codes for a replication competent copy of the viral genome, and the tagged expression library constructs. The transfection efficiency of HEK-293 cells was estimated to be over 90% as monitored by fluorescence, thus ensuring co-expression of the proviral and the tagged hnRNP A1 expression constructs. Viral replication was analyzed by collecting the supernatant from the transfected HEK-293 cells and infecting TZM-bl cells [31], which contain a copy of the luciferase gene under control of the HIV-1 LTR promoter. Once HIV-1 is integrated into the cellular genome it expresses the viral protein Tat, which in turn activates the viral LTR promoter. Thus, luciferase expression is utilized as a reliable reporter for viral replication. Expression of the wild type hnRNP A1 sequence decreased viral replication to 1–2% of the control (Fig 4B), this was consistent with our previous results achieved by over-expressing the untagged protein [18]. A strong inhibition of viral replication was also obtained with expression of the A1-Δ1-EG clone, which lacks the F-peptide phosphorylation sites. Consistently with the data obtained in the splicing assay, expression of the clone lacking the RGG-box (A1-Δ6-EG) resulted in a 10-fold decrease in viral replication while only marginal changes were observed upon expression of the other clones.
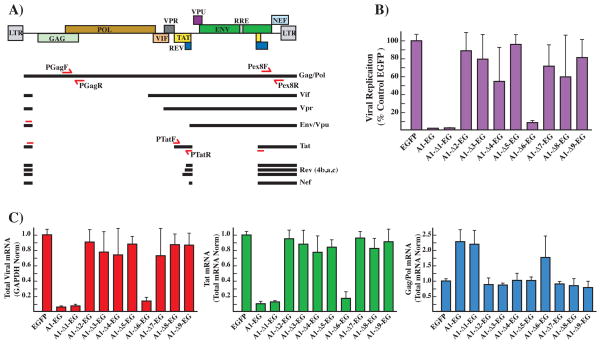
Fig. 4.
(A) Schematic representation of the pNL4-3 proviral clone. The relative position of the viral genes is indicated on the map on top. The main mRNAs generated from the single viral transcript are indicated. Locations of the primers utilized in the qPCR assays to amplify the total viral mRNA (Pex8F, Pex8R) and specific for the gag/pol (PGagF, PGagR) and tat (PTatF, PTatR) mRNAs are indicated. (B) Viral replication assay. HEK-293 cells were co-transfected with the proviral clone pNL4-3, the control pEGFP or the indicated hnRNP A1-EGFP fusion clone. TZM-bl reporter cells were infected with the supernatant from HEK-293 cells 72 hours after transfection. Luciferase activity was measured 48 hours after infection and it is directly proportional to the amount of virion contained in the infection supernatant. 100% replication was defined as luciferase activity obtained by incubating the indicator cell line with the supernatant from the HEK-293 cells transfected with the viral clone pNL4-3 and the pEGFP control plasmid. (C) qPCR analysis of viral transcripts. Each graph summarizes the quantification by qPCR analysis of the indicated viral mRNA species extracted from 293T cells transfected with the proviral clone pNL4-3 and the indicated expression clone. The amount of each mRNA species generated in the transfection containing the control pEGFP was assigned the value 1. Total viral mRNA expression was normalized for expression of the GAPDH housekeeping gene. Expression of the gag/pol and tat mRNAs relative to the total amount of viral transcript was calculated by normalizing the amount of specific transcripts for the total amount of viral mRNA in each assay.
To determine the effect that the hnRNP A1 deletion clones have on the levels of total viral mRNA and on the splicing of specific viral mRNA isoforms we performed RT-qPCR with primers sets designed to anneal either to a region common to all viral mRNAs or to specific spliced mRNAs species as previously described [18].
Expression of the wild type hnRNP A1 protein decreased total viral RNA production to 5% of the control; a similar result was achieved by expressing the A1-Δ1-EG clone while the A1-Δ6-EG clone reduced it to 15%, other clones did not induce significant changes. These results are consistent with the role of hnRNP A1 in the repression of splicing of the tat coding mRNAs and the resulting decrease in the transcription rate of the viral genome and impaired production of functional virions [18].
To confirm that the decrease in the amount of viral transcripts was caused by the relative decline in the splicing of tat coding mRNAs, we quantified their relative abundance and compared it with the unspliced gag/pol mRNAs. Since the total amounts of viral transcripts varies upon over-expression of the hnRNP expression clones, the relative amount of the single mRNA species (tat and gag/pol) was normalized for the total amount of viral mRNA present in each reaction. Expression of the wild type A1 sequence, the A1-Δ1-EG and A1-Δ6-EG clones caused a decrease of the spliced tat coding mRNAs and a corresponding increase in the unspliced gag/pol mRNAs (Fig. 4C). The effects on viral replication and splicing caused by expression of the clone lacking the RGG-box (A1-Δ6-EG) are similar but less severe to the ones triggered by the wild type protein (Fig 4B and 4C). Since hnRNP A1 has been shown to play multiple roles in viral replication and to regulate viral splicing in multiple regions of the viral transcript it is plausible that these additional functions might require the RNA binding specificity or protein-protein interactions provided by the deleted RGG-box or neighboring sequences.
3.5. The A1-Δ6-EG clone encodes for a predicted hnRNP A1 isoform
14 hnRNP A1 splicing variants, 9 of which coding, are predicted by the ENSEMBL database. Two of the coding variants, isoforms A1-A and A1-B, have been experimentally confirmed. A third isoform named isoform-2 (ENSEMBL ENST00000547276) is also predicted by the Uniprot (Uniprot identifier P09651-3) and UCSC Genes databases (Fig. 5). hnRNP A1 isoform-2 carries a central deletion of residues 203 to 307, which contains the RGG-box. Our A1-Δ6-EG clone, which carries the deletion of residues 201 to 303, is highly similar to the isoform-2 sequence. Our data indicate that a clone carrying sequences similar to the isoform-2 of hnRNP A1 can partially substitute for the more abundant A1-A isoform. Nevertheless, the isoform lacking the RGG-box is unlikely to exhibit the full range of functions displayed by the main A1-A isoform. Since a cDNA clone corresponding to isoform-2 has been isolated and sequenced from a human retinoblastoma (GenBank: BC103707) it is plausible that this isoform might be present in a number of human tissues, or under specific physiological conditions, possibly at a lower level and with different RNA specificity and functionality, than the more abundant A1-A and A1-B.
Fig. 5.
(A) Genomic map of the major hnRNP A1 isoforms (A1-A: ENSEMBL ENST00000546500; A1-B: ENSEMBL ENST00000340913) and the predicted A1-isoform 2 (ENSEMBL: ENST00000547276). (B) Schematic map of the main hnRNP A1 protein isoforms (A1-A: Uniprot P09651-2; A1-B: Uniprot P09651) and the minor A1-isoform 2 (Uniprot P09651-3).
4. Discussion
hnRNP A1 is a widely expressed, multifunctional protein with roles in several mechanisms regulating cellular and viral gene expression. Studies aimed at the functional and biochemical characterization of this protein indicated that multiple domains participate to confer its RNA binding specificity and functionality. The structural and functional domains of this protein have been mostly studied utilizing biochemical and biophysical approaches, therefore less is known about the role that such sequences might play in a cell-based system. We aimed at identifying the functions of single and partial domains in a cellular system and isolate a minimal hnRNP A1 derived sequence that retained the splicing regulatory and antiviral properties of the full-length protein.
Here we have shown that a minimal deletion mutant, lacking one of the protein RBDs, the RGG-box, and a portion of the prion-like domain can functionally substitute for the wild type protein. Furthermore, the truncated clone we isolated is similar to an isoform, which is predicted by several databases and detected in human retinoblastoma. The RNA binding properties of hnRNP A1 have been widely studied with focus on the N-terminal fragment, composed of the RRM1 and RRM2 RBDs, which are required to confer proper RNA binding specificity to the protein [9, 32]. Less is known about the third RBD, the RGG-box, and although its contribution to the overall binding specificity of the protein was considered modest by some [33], other reports indicate that its role is significant [4, 10, 34]. Furthermore, since the RGG-box selectively interacts in vitro with itself and other hnRNPs [35], it seems to be required for the cooperative binding profile of hnRNP A1 with itself and hnRNP H [34, 36]. Our data indicate that splicing regulation of a HIV-1 derived minigene is not impacted by the deletion of the RGG-box.
It is plausible that hnRNP A1 might regulate splicing through multiple mechanisms, as previously summarized [1], therefore deletion of the RGG-box might impact this protein splicing functions differently in different systems. Previous work indicated that the predominant nuclear localization of hnRNP A1 was solely dependent on the short M9 sequence [5], and, although recent lines of evidence showed that proper nuclear trafficking and cytoplasmic localization is also dependent on the phosphorylation of the F-peptide and the binding to cognate RNAs [37], little is known about the role played by other sequences on the cellular localization of the protein. Our results show that, although the M9 sequence is essential for the nuclear localization of hnRNP A1, a key function is also played by the RBDs. Clones lacking all three RBDs lack a prevalent nuclear localization similarly to clones lacking the M9 sequence (Fig. 2 cfr. A1-Δ2-EG with A1-Δ5-EG), while addition of one or more RBDs can either partially (RGG-box or RRM2) or completely (RRM1 alone or RRM2 in combination with either RGG-box or RRM1) restore nuclear localization. It is plausible that the discrepancies between our findings and those of others [5] could be due to the different linkers or tag sequences utilized, which might have functionally substituted for the protein RBDs.
The characterization of truncated, functionally active, hnRNP A1 isoforms could aid our understanding of the molecular mechanisms underlying this protein functions in disease. hnRNP A1 diverse activities in cellular metabolism and proliferation correlate with its functions in human disease and pathogenesis. Variations in the relative abundance of hnRNP A1 have been shown to alter the splicing pattern of key genes involved in apoptosis and tumorogenesis [38–41]. Similarly, deregulation of hnRNP A1 expression relates to changes in the expression of cellular factors regulating neural development and contribute to neurodegenerative diseases, such as Alzheimer’s disease (AD), spinal muscular atrophy (SMA) and multiple sclerosis (MS) [42]. hnRNP A1 can also act as a prion-like protein [8]. Families with inherited degeneration affecting muscle, brain, motor neuron and bone carry distinctive mutations within the hnRNP A1 PrLD located within the Gly-rich region. The mutated protein can oligomerize and enhances the ability of the wild-type hnRNP A1 to polymerize into fibrils and, in an animal model, the formation of cytoplasmic inclusion. The hnRNP A1 deletion clone A1-Δ6-EG we characterized in this work retains the splicing functions of the protein but lacks a portion of the PrLD domain. The isolation of hnRNP A1 isoforms with a different range of functions and lacking portions of the Gly-rich domain that contributes to its self-assembling properties and decreased solubility when synthesized in a prokaryotic system suggests possible therapeutic implications in the treatment of both genetic and viral disease.
5. Conclusions
In conclusion we isolated an hnRNP A1 clone lacking the RGG-box RBD that can functionally substitute for the ubiquitously expressed A1-A isoform and regulate HIV-1 splicing and replication. Our study also highlights a role for the hnRNP A1 RBDs in the intracellular localization of the protein.
Supplementary Material
Tab. S1. Summary of the results obtained with the hnRNP A1 clone library. Nuclear localization of the individual clones is qualified as strong (+++), middle (++) or weak (+). The splicing inhibition exerted by the individual clones on the pLTR-S1Xm-R substrate is qualified as strong (+++), middle (++) or weak (+). Inhibition of viral replication in 293T cells transfected with the pNL4-3 clone and the individual clones of the library is qualified as strong (+++), middle (++) or weak (+).
Fig. S1. Schematic representation of hnRNP A1 isoforms and complete deletion clones library. The protein structural and functional features are indicated. The sequences present in each deletion clone are indicated together with the forward (F) and reverse (R) primers utilized to clone the hnRNP A1 fragments amplified from the pGT7-A1 clone (coding for the major A1-A isoform) into the pEGFP-N1 expression vector.
Fig. S2. A1-Δ7-EG deletion clone localization. DAPI staining shows the prevalent nuclear localization of the A1-Δ7-EG deletion clone (EGFP). Nuclear speckles are visualized utilizing monoclonal antibody for the marker SC35. The merge of the images shows the lack of co-localization between the nuclear speckles and the A1-Δ7-EG clone.
Highlights.
hnRNP A1 RGG-box is not required for HIV-1 splicing regulation.
hnRNP A1 RRMs are required for nuclear localization.
A novel hnRNP A1 isoform can functional substitute for the major A1-A isoform.
Acknowledgments
We thank Dr. Jianning Wei for helpful comments and technical advice. This work was supported by NIH/NIAID grant R15 AI093229 to M.C and by the purchase of major equipment by the Schmidt Foundation.
Footnotes
Conflict of interest
Authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jean-Philippe J, Paz S, Caputi M. hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci. 2013;14:18999–19024. doi: 10.3390/ijms140918999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buvoli M, Cobianchi F, Bestagno MG, Mangiarotti A, Bassi MT, Biamonti G, Riva S. Alternative splicing in the human gene for the core protein A1 generates another hnRNP protein. EMBO J. 1990;9:1229–1235. doi: 10.1002/j.1460-2075.1990.tb08230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci. 2009;66:1239–1256. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nadler SG, Merrill BM, Roberts WJ, Keating KM, Lisbin MJ, Barnett SF, Wilson SH, Williams KR. Interactions of the A1 heterogeneous nuclear ribonucleoprotein and its proteolytic derivative, UP1, with RNA and DNA: evidence for multiple RNA binding domains and salt-dependent binding mode transitions. Biochemistry. 1991;30:2968–2976. doi: 10.1021/bi00225a034. [DOI] [PubMed] [Google Scholar]
- 5.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 7.Allemand E, Guil S, Myers M, Moscat J, Caceres JF, Krainer AR. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc Natl Acad Sci U S A. 2005;102:3605–3610. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayeda A, Munroe SH, Cáceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A, Wilson SH. Studies of the strand-annealing activity of mammalian hnRNP complex protein A1. Biochemistry. 1990;29:10717–10722. doi: 10.1021/bi00500a001. [DOI] [PubMed] [Google Scholar]
- 11.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 12.Purcell DFJ, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoltzfus CM, Madsen JM. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr HIV Res. 2006;4:43–55. doi: 10.2174/157016206775197655. [DOI] [PubMed] [Google Scholar]
- 14.Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zahler AM, Damgaard CK, Kjems J, Caputi M. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J Biol Chem. 2004;279:10077–10084. doi: 10.1074/jbc.M312743200. [DOI] [PubMed] [Google Scholar]
- 16.Tange TO, Damgaard CK, Guth S, Valcarcel J, Kjems J. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. Embo J. 2001;20:5748–5758. doi: 10.1093/emboj/20.20.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 18.Jablonski JA, Caputi M. Role of cellular RNA processing factors in human immunodeficiency virus type 1 mRNA metabolism, replication, and infectivity. J Virol. 2009;83:981–992. doi: 10.1128/JVI.01801-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siomi MC, Eder PS, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 21.Rebane A, Aab A, Steitz JA. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA. 2004;10:590–599. doi: 10.1261/rna.5224304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 23.van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Caceres JF. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. Embo J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mears WE, Rice SA. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J Virol. 1996;70:7445–7453. doi: 10.1128/jvi.70.11.7445-7453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karn J, Stoltzfus CM. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med. 2012;2:a006916. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monette A, Ajamian L, Lopez-Lastra M, Mouland AJ. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: implications for HIV-1 gene expression. J Biol Chem. 2009;284:31350–31362. doi: 10.1074/jbc.M109.048736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Najera I, Krieg M, Karn J. Synergistic stimulation of HIV-1 Rev-dependent export of unspliced mRNA to the cytoplasm by hnRNP A1. J Mol Biol. 1999;285:1951–1964. doi: 10.1006/jmbi.1998.2473. [DOI] [PubMed] [Google Scholar]
- 29.Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brady J, Kashanchi F. Tat gets the “green” light on transcription initiation. Retrovirology. 2005;2:69. doi: 10.1186/1742-4690-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayeda A, Munroe SH, Xu RM, Krainer AR. Distinct functions of the closely related tandem RNA-recognition motifs of hnRNP A1. RNA. 1998;4:1111–1123. doi: 10.1017/s135583829898089x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weighardt F, Biamonti G, Riva S. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 34.Damgaard CK, Tange TO, Kjems J. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA. 2002;8:1401–1415. doi: 10.1017/s1355838202023075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. hnRNP A1 selectively interacts through its gly-rich domain with different RNA-binding proteins. J Mol Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 36.Fisette JF, Toutant J, Dugre-Brisson S, Desgroseillers L, Chabot B. hnRNP A1 and hnRNP H can collaborate to modulate 5′ splice site selection. Rna. 2010;16:228–238. doi: 10.1261/rna.1890310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lichtenstein M, Guo W, Tartakoff AM. Control of nuclear export of hnRNP A1. Traffic. 2001;2:261–267. doi: 10.1034/j.1600-0854.2001.1o002.x. [DOI] [PubMed] [Google Scholar]
- 38.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boukakis G, Patrinou-Georgoula M, Lekarakou M, Valavanis C, Guialis A. Deregulated expression of hnRNP A/B proteins in human non-small cell lung cancer: parallel assessment of protein and mRNA levels in paired tumour/non-tumour tissues. BMC Cancer. 2012;10:434. doi: 10.1186/1471-2407-10-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pino I, Pio R, Toledo G, Zabalegui N, Vicent S, Rey N, Lozano MD, Torre W, Garcia-Foncillas J, Montuenga LM. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer. 2003;41:131–143. doi: 10.1016/s0169-5002(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 41.Venables JP. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28:378–386. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 42.Bekenstein U, Soreq H. Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: From structural insights to post-transcriptional regulatory roles. Mol Cell Neurosci. 2013;56:436–446. doi: 10.1016/j.mcn.2012.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tab. S1. Summary of the results obtained with the hnRNP A1 clone library. Nuclear localization of the individual clones is qualified as strong (+++), middle (++) or weak (+). The splicing inhibition exerted by the individual clones on the pLTR-S1Xm-R substrate is qualified as strong (+++), middle (++) or weak (+). Inhibition of viral replication in 293T cells transfected with the pNL4-3 clone and the individual clones of the library is qualified as strong (+++), middle (++) or weak (+).
Fig. S1. Schematic representation of hnRNP A1 isoforms and complete deletion clones library. The protein structural and functional features are indicated. The sequences present in each deletion clone are indicated together with the forward (F) and reverse (R) primers utilized to clone the hnRNP A1 fragments amplified from the pGT7-A1 clone (coding for the major A1-A isoform) into the pEGFP-N1 expression vector.
Fig. S2. A1-Δ7-EG deletion clone localization. DAPI staining shows the prevalent nuclear localization of the A1-Δ7-EG deletion clone (EGFP). Nuclear speckles are visualized utilizing monoclonal antibody for the marker SC35. The merge of the images shows the lack of co-localization between the nuclear speckles and the A1-Δ7-EG clone.