Abstract
RNA viruses face dynamic environments and are masters at adaptation. During their short ‘lifespans’, they must surmount multiple physical, anatomical and immunological challenges. Central to their adaptative capacity is the enormous genetic diversity that characterizes RNA virus populations. Although genetic diversity increases the rate of adaptive evolution, low replication fidelity can present a risk because excess mutations can lead to population extinction. In this Review, we discuss the strategies used by RNA viruses to deal with the increased mutational load and consider how this mutational robustness might influence viral evolution and pathogenesis.
RNA viruses exhibit extremely high mutation rates, orders of magnitude greater than those of most DNA-based life forms1 (BOX 1). Although the measurement of viral mutation rates is a complex issue in itself, the studies carried out to date suggest that many RNA viruses generate 10−4 to 10−6 errors per nucleotide, which is equivalent to approximately one mutation per genome, per replication cycle2. Given the large population sizes observed in both experimental and natural infections with these viruses, every possible point mutation and many double-mutation combinations could theoretically be generated during each replication cycle within a population. Even a defined molecular clone quickly transforms into a collection of related sequences when introduced into cells1. This low replicative fidelity ensures that viral populations can generate and maintain mutations that allow them to quickly adapt to changes in the environment. The mutability and fleeting existence of each viral genome means that RNA virus populations exist as dynamic mutant networks in which sequences are continuously diversified and regenerated by mutation of related sequences (FIG. 1). The low replicative fidelity seems to be crucial for viral survival in the host ecosystem, as variants with abnormally low mutation rates are attenuated in vivo3–5.
Box 1. Mutation rates of RNA viruses and quasispecies theory.
A quasispecies refers to a population of genetically related viruses that are closely distributed around a consensus sequence1,84–87. Such a population is a flexible and diverse group of variants with different replicative capacities and fitnesses. For any given environment, the quasispecies hypothetically consists of a collection of genomes with many, or most, of the possible viable mutations represented. Many of these mutations are lost during multiple replication cycles, as they encode genomes of very low fitness (defined here as the relative capacity to produce infectious progeny) that cannot compete with fitter genomes during replication. However, within each round of replication, new mutations spontaneously emerge or re-emerge at a frequency that is directly determined by the error rate of the viral RNA polymerase.
The mutation rate measures the appearance of spontaneous mutations as a function of time. Observed mutation rates differ among species and also vary across the genome of a given species. This essential parameter is often measured as the number of nucleotide substitutions per base per generation. The mutation rate in unicellular eukaryotes and bacteria is roughly 0.003 mutations per genome per generation88. Because RNA viruses are replicated by RNA-dependent RNA polymerases that lack proofreading ability, they have the highest known per bp per generation mutation rates89. Double-stranded DNA viruses have mutation rates of between 10−6 and 10−8 mutations per bp per generation, whereas RNA viruses have mutation rates of between 10−4 and 10−6 mutations per bp per generation2. For comparison, recent genome sequencing studies estimate that the mutation rate of the human genome is ~1.1 × 10−8 mutations per bp per generation90,91. Importantly, the estimation of mutation rates described here suffers from a very limited number of experiments and non-standardized sampling of different species and genetic entities. With the use of new sequencing technologies, it is expected that this crucial evolutionary parameter will be better defined in the near future.
There has been considerable debate as to whether the high mutation rate of RNA viruses is adaptive or simply the by-product of selection for other traits such as replicative speed92. As detailed in the main text, many newly generated mutations are deleterious, and evolutionary theory suggests that high mutation rates drive selection for mutational robustness13.
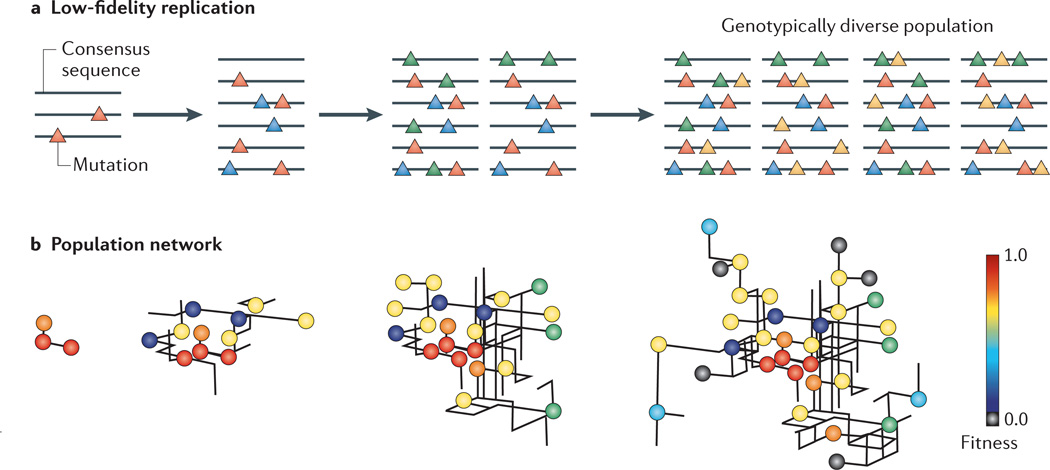
Figure 1. Viral populations as mutant networks.
a| The consensus sequence (grey line) is the average sequence of a population and might not be represented on any individual genome because of the extremely high genetic diversity of RNA virus populations. Low-fidelity replication, which is a characteristic feature of RNA viruses, results in a diverse population of unique genotypic variants while maintaining the same consensus genome sequence. Mutations acquired in each replication cycle are represented by differently coloured triangles. b| RNA virus populations can be depicted as networks in which the genetic variants (circles) of varying fitness are connected by mutational pathways (black lines).
The focus on mutation as a driving force in viral evolution has tended to overlook the tremendous cost of low replicative fidelity. Most mutations have deleterious effects on viral fitness. In vesicular stomatitis virus (VSV), more than 90% of random single-nucleotide mutations reduce replicative fitness, and 40% are lethal6. Similar trends have been found in tobacco etch virus and the phages ΦX174 and QΒ7. Furthermore, increasing error rates pharmacologically, with mutagenic nucleoside drugs8,9, or genetically, through the use of variant RNA-dependent RNA polymerases10–12, leads to viral extinction. These studies indicate that the mutation rate in RNA virus populations is perilously close to the maximum tolerable error rate. The mutational tolerance of a virus will determine the type (for example, variation in structural or non-structural proteins) and extent of genetic diversity that can be maintained in the population. Thus, viral population diversity results from both the generation of and the tolerance to mutations; these two factors together drive adaptation and viral evolution.
It has long been recognized that not all genotypic changes are expressed as alterations in phenotype, and in population genetics, this buffering of mutational effects is termed genetic robustness. Early work on genetic robustness was largely based on theory (reviewed in REF. 13), but a number of experimental studies over the past 10 years have established and extended the concept of genetic robustness and shown that this buffering allows a viral population to increase its genetic diversity without a dramatic alteration in phenotype. Importantly, these experimental systems have also begun to elucidate the molecular underpinnings of mutational tolerance and to identify the conditions in which genetic robustness is adaptive. Recent studies further suggest that the relationship between robustness and evolvability might be particularly important for viral pathogenesis14.
As a result of this recent work, we now have a clearer picture of how robustness influences the short- and long-term evolution of RNA viruses. In this Review, we begin by defining genetic robustness and how it can be measured, before considering how genetic robustness influences the composition of viral populations. We then discuss the mechanisms that contribute to genetic robustness and how they relate to viral pathogenesis, and finally consider the impact of genetic robustness on antiviral therapeutic strategies.
Robustness: defined and measured
de Visser and colleagues have provided a useful working definition of robustness: “Robustness is the invariance of phenotypes in the face of perturbation” (REF. 13). They further classify robustness on the basis of the source of perturbation, which can be either genetic or environmental. Environmental robustness refers to the stability of traits across different environments (for example, particular habitats or conditions, such as heat shock). Genetic robustness describes scenarios in which the perturbation (in other words, the mutation or mutations) is heritable. Conceptually, genetic robustness is a form of epistatic interaction; the degree to which genetic variation is expressed depends on the genetic background. Epistatic interactions among genes determine the phenotypic expression of mutant alleles, and if the collective effect of these interactions results in maintaining the phenotype, then this buffering manifests as robustness. Although the virology literature often describes mutational rather than genetic robustness, both terms refer to the same phenomenon. We use the term mutational robustness in our discussion below.
The simplest measure of mutational robustness is to quantify the mutational fitness effect of individual mutations. The mutational fitness effect has been determined in a number of viruses by introducing random point mutations into the viral genome and measuring their effects on replicative efficiency in competition assays6,15,16. Viruses that exhibit a small average mutational fitness effect are considered mutationally robust, whereas those exhibiting a large mutational fitness effect are considered fragile (or brittle). A less controlled, but equally valid, measure of robustness is to quantify the sensitivity of the virus to nucleoside analogues. Many of these nucleosides are misincorporated into viral genomes during replication and increase the observed mutation rate by templating mismatches14,17,18. These studies do not provide an absolute measure of robustness, given that each study focuses on a single virus or strain. Many discussions of the topic divide robustness into categories according to the evolutionary or mechanistic origins of the robustness13,19. For virologists, it is perhaps more useful to discuss robustness in the context of viral genome structure, replication and fitness. Below, we discuss the main factors that influence viral robustness.
Population size
RNA viruses have a tremendous reproductive capacity, generating thousands of progeny per genome. Despite frequent bottleneck events, a total population of millions of viruses in an infected host is not uncommon. Evolutionary theory suggests that a ‘safety in numbers’ phenomenon is at play, whereby mutational robustness exists at the population level despite the negative impact of mutation on each individual in the population (reviewed in REF. 19). The efficiency of negative selection is the product of the effective population size (Ne) and the average mutational fitness effect. Thus, in large populations, strong selection will quickly purge mutants with lower fitness, and the most fit sequence will dominate the mutant spectrum, accompanied by very low frequencies of numerous variants with intermediate fitness. This frequency distribution has been observed in host-derived populations of poliovirus14 and foot-and-mouth disease virus20. Mutational robustness can therefore be achieved by generating a large number of progeny, preserving the most fit sequence and ensuring its survival in a given environment. In this manner, large population sizes preserve the invariance of phenotype in the face of mutational perturbation.
Co-infection
A larger population size can also result in a higher multiplicity of infection such that a single host cell more often supports replication of at least two different viral genomes. In such cases, genetic complementation can increase the robustness of the viral population. This means that although a mutated viral protein can limit or even halt viral replication at a low multiplicity of infection, the defect is masked at a high multiplicity of infection because other genomes in the cell encode a functional version of the defective protein. This contribution to robustness is commonly observed in cell culture systems and explains why phenotypic differences among competing strains are harder to distinguish at high multiplicities of infection (for examples, see REFS 14,21). The importance of complementation to mutational robustness has been demonstrated by subjecting the RNA phage Φ6 to 300 passages at either a high or a low multiplicity of infection22. At a high multiplicity of infection, complementation was frequent and purifying selection against mutated genomes was reduced. However, because these defective genomes could be propagated, this strategy weakened long-term selection for mutational robustness. By contrast, at a low multiplicity of infection, viral populations evolved to become more mutationally robust.
Many RNA viruses undergo recombination or reassortment during replication. This exchange of genetic information can increase the genetic diversity of the population by combining previously unique mutations into the same genome. Although the impact of viral sex on mutational robustness and viral evolution remains somewhat obscure, it is clear that this process can also repair mutated genomes (reviewed in REF. 23). Recombination could also disrupt the genetic linkages between beneficial and detrimental mutations, thus reducing the impact of these detrimental mutations on the selection of the beneficial mutations. Given the complex relationship between complementation, recombination and robustness, the effective multiplicity of infection in infected individuals is likely to be a crucial factor influencing viral evolution.
Fitness landscapes and survival of the flattest
The phenotypic stability provided by mutational robustness confers a selective advantage, but is it necessarily adaptive? In principle, mutational robustness could evolve as a convenient side effect of selection for a different phenotype, such as increased inter-host survival or transmissability13. Perhaps the clearest argument in favour of an adaptive basis for mutational robustness is the resultant enhanced tolerance to the excess mutational load caused by a low replicative fidelity, as this tolerance suggests that high mutation rates probably select for robust genomes.
One way to illustrate robustness is using fitness landscapes, which represent the relationship between genotypes and fitness (FIG. 2). The ‘altitude’ at any given location is the fitness associated with that particular genotype. The ‘ground level’ is a representation of the range of genotypes in the sequence space. Two sequences that differ by a single mutation are neighbouring points at the ground level, whereas highly divergent sequences are well separated. The environment and its selective pressures determine the contours of the landscape. Fit but mutationally brittle populations occupy steep peaks, whereas robust populations reside on broader hills. When the mutation rate is high, the populations are pushed away from the fitness peaks and out into the surrounding sequence space lower down the slopes. Selection favours robust populations, as they are better able to accommodate mutations without a change in fitness (outward movement from the fitness peak corresponds to movement down a gentler slope). In experimental systems, it has been difficult to distinguish fitness from robustness (see below).
Figure 2. High mutation rates and survival of the flattest.
In a fitness landscape, the ‘ground level’ is a two-dimensional representation of genotypic sequence space, and the vertical axis gives the fitness value for each genotype or sequence.a| When the mutation rate is low, populations will be genotypically stable and cluster at the top of the fitness peak. The variant with the highest fitness (red) will easily outcompete all others. b| When the mutation rate is high, variants spread out over their corresponding peaks. The population on the flatter peak (blue) remains near its fitness optimum and has a higher mean fitness than the population located on the steeper peak (red). The flatter population will therefore prevail in competition with the population on the higher peak. Here, fitness and robustness are contrasted to show the importance of each in determining the dynamics of RNA virus populations. As described in the text, a population can theoretically be both fit and robust and thereby occupy a tall, broad peak. However, the experimental data currently available suggest that fitness and robustness are inversely correlated.
This process, whereby populations buffer the negative effects of mutation by migrating to regions of sequence space corresponding to flatter, selectively neutral fitness landscapes, is termed survival of the flattest. A pioneering in silico study used self-replicating digital organisms to show that selection does indeed favour slowly replicating, robust populations over their fitter, more fragile counterparts24. The first evidence for survival of the flattest in a biological system came from studies of RNA viroids in plants25. A viroid strain with a slightly larger neutral neighbourhood was able to outcompete another strain with a faster replication rate (a fitter strain) when these strains were propagated in a mutagenic environment (plants exposed to ultraviolet light). A similar study using two distinct populations of VSV found that selection favoured the slower replicating, mutationally robust population over a faster replicating population when the two populations were competed in the presence of mutagenic drugs26.
Both experimental studies clearly demonstrate survival of the flattest25,26, but the distinction between the fittest and flattest might be an artificial one. In theoretical discussions, fitness and robustness are often considered separately for the sake of argument and to illustrate that flatter populations can, in principle, outcompete fitter but more brittle ones (FIG. 2). This is not necessarily the whole story; a population can be both fit and robust, occupying a high, broad peak. Measurements from competition assays cannot distinguish between replicative fitness and mutational robustness, as both determine the number of successful progeny over multiple passages24. In a competition assay between two populations, a given population could dominate either by replicating faster (which is the most commonly used parameter for measuring fitness in the laboratory) or by producing progeny with preserved fitness. The latter outcome would be due to robustness, whereas the former scenario would indicate only that the population was dominant because it replicated faster. Perhaps this is the reason that selection for mutational robustness can be demonstrated in competition assays only at above-normal mutation rates, when the beneficial effect of increased mutational tolerance outweighs any reduction in replicative efficiency. This issue was addressed by comparing wild-type poliovirus to two poliovirus variants that contain a large number of synonymous mutations14,27,28. All three viruses have the same consensus amino acid sequence and exhibit similar replication kinetics. However, the three viral populations are genetically distinct and occupy unique fitness landscapes. One of the mutant populations was found to be less mutationally robust than the wild-type virus and also less fit in competition assays. The simplest interpretation of these data is that differences in the number of viable progeny generated during each round of replication drive the observed fitness differences. Furthermore, these data suggest that mutational robustness is an important component of viral fitness, buffering the negative effects of mutation even at basal RNA virus mutation rates.
Mechanisms of robustness at the genome level
The identification of VSV and poliovirus populations that differ in their mutational tolerance suggests that robustness can be defined at the level of the viral genome. In many RNA viruses, the RNA genome contains secondary and tertiary structures that are important for replication, packaging and other key functions. The robustness of such structures to mutation has been extensively studied in tRNA, ribozymes and other small RNA molecules for which the thermodynamics of folding is easily simulated. Early folding simulations of tRNA demonstrated that a large set of sequences (genotypes) could assemble into similar shapes (phenotypes)29,30. Because these diverse sequences are selectively neutral and connected through mutation, this type of genotype-to-phenotype map is often called a neutral network31. More recently, cycles of mutagenesis and selection have been used to create genotypically diverse ribozyme populations that retain their ability to cleave a phosphodiester bond in an RNA oligonucleotide32. The accumulated cryptic variation did not affect the phenotype and was selectively neutral in a given environment (mutationally robust), but because the mutations potentially provide a reservoir of beneficial mutations for future environmental changes, these ribozyme populations could also be environmentally robust. In RNA viruses, structures that regulate replication or translation are highly conserved. An in silico experiment with HIV, hepatitis C virus and dengue virus compared these conserved, functional elements to non-conserved RNA structures from the same species and found that the conserved functional elements were more mutationally robust than their non-conserved counterparts33. Similar phenomena have been documented in viroid structures34,35.
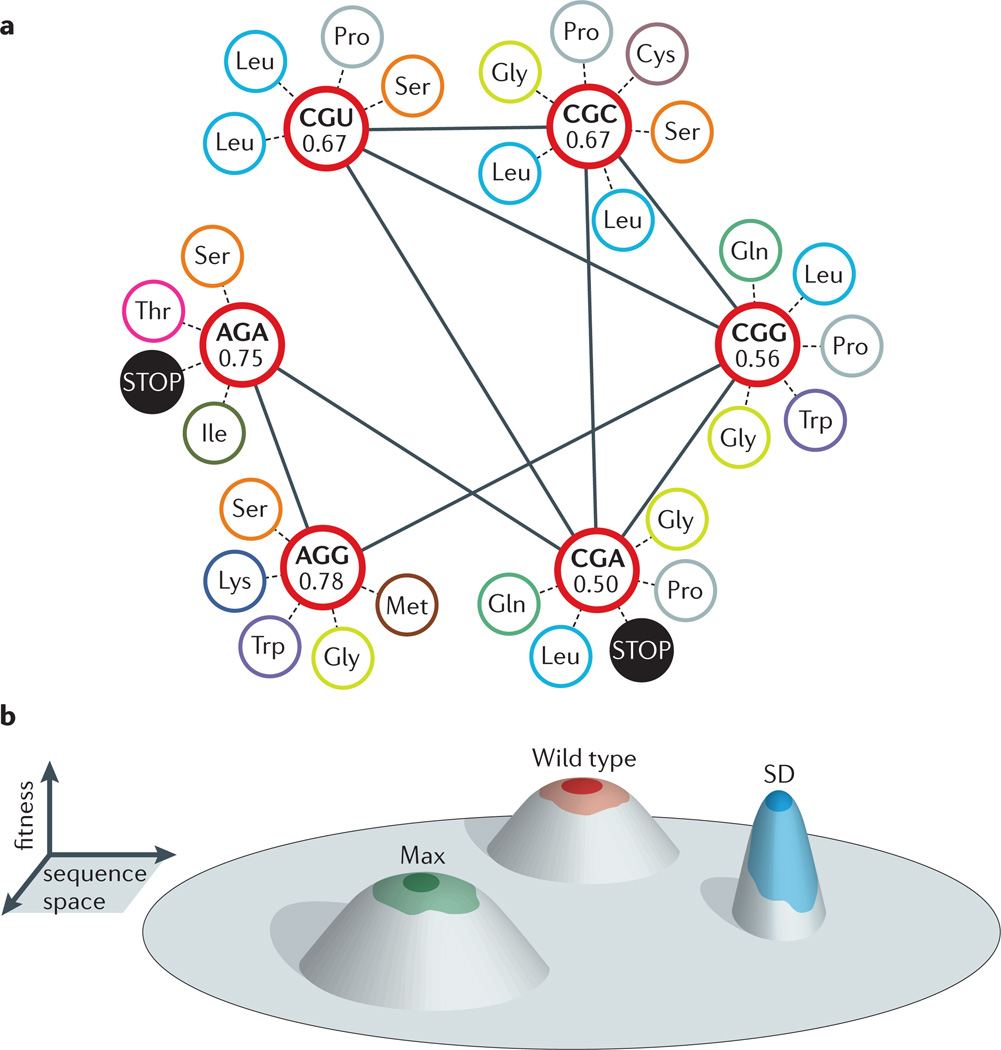
These data highlight the link between robustness and the degeneracy of RNA structural elements; multiple genotypes can give rise to the same phenotype. The genetic code is also degenerate, and nearly all amino acids are encoded by more than one codon. However, given the nature of the genetic code, synonymous codons will yield distinct amino acid changes when they are mutated (FIG. 3). For example, although both AGG and CGG code for arginine, the frequencies of mutations that give rise to non-synonymous codons are 78% and 56%, respectively, for these two synonymous codons. Furthermore, the amino acids introduced by single point mutations will also differ in their biochemical properties (such as hydrophobicity, polarity and charge). Codon usage can therefore determine how mutations are tolerated at the protein level. Bioinformatic studies of several RNA viruses indicate that the highly variable surface epitopes of viral proteins might be enriched for volatile codons that tend to mutate non-synonymously36,37. This is in contrast to more conserved domains, which contain codons with a lower predicted rate of non-synonymous substitutions. Theoretical studies have also highlighted the potential importance of codon volatility in rapidly evolving protein domains38.
Figure 3. Using synonymous mutation to place populations in distinct fitness landscapes.
a| A synonymous mutation alters the potential fitness impact of a subsequent mutation. Although two synonymous codons for arginine (such as AGG and CGG) are separated by a neutral A®C mutation, these codons differ in their propensity to mutate non-synonymously and non-conservatively. Shown are all six arginine codons (red circles) and, for each, the proportion of all potential mutations that would be non-synonymous. Synonymous mutations are indicated as solid lines, and non-synonymous mutations are indicated as dashed lines. b| Large-scale synonymous mutation preserves the consensus amino acid sequence, but relocates viral populations in sequence space. When these viruses replicate with a high mutation rate, the genetic architecture of the resultant populations differs, and the populations reside in distinct fitness landscapes. The figure shows the results of a poliovirus experiment in which the wild-type virus was compared with two variants, Max and SD, which contain 566 and 934 synonymous substitutions, respectively, in the 2,643-nucleotide sequence encoding the viral capsid protein. The SD variant was found to be the least mutationally robust (for example, it was hypersensitive to an RNA mutagen, relative to the Max variant and the wild type), suggesting that it resides on a steeper, less neutral landscape.
Codon choice has been proposed to be an important mechanism for robustness in RNA viruses14. In a recent study, wild-type poliovirus was compared to two variants with a large number of synonymous mutations14. These variants, Max and SD, were created by de novo gene synthesis and contain 566 and 934 substitutions, respectively, in the 2,643-nucleotide sequence encoding the viral capsid protein27,28. Because these variants were designed as negative controls for studies of codon bias and viral translation, their GC content, predicted free energy of RNA folding and codon usage frequency across all 64 codons were nearly identical to those of the wild-type virus. In the new study, the viral populations derived from replication of these three viruses were each found to contain a unique set of polymorphic amino acid substitutions in the capsid protein, and there was lower sequence similarity in the capsid sequence relative to the rest of the genome among the three viral strains14. The SD population was less mutationally robust than either the wild-type or Max populations, suggesting a role for codon choice in mutational tolerance. Interestingly, the synonymous substitutions in the Max variant were based on altering local codon pair bias, whereas the codons in the SD variant were randomly shuffled relative to the wild-type sequence. Perhaps the more drastic reorganization of the capsid sequence in the SD variant disrupted local patterns of codon volatility in the capsid proteins, thereby increasing the frequency of non-synonymous substitutions in structurally conserved domains.
Chaperones: cellular modulators of robustness
Another way to maintain the functionality of proteins carrying destabilizing mutations is through the activity of molecular chaperones39. Mutations that accumulate during viral replication can destabilize viral proteins and increase their tendency to misfold and aggregate. Because they face such a high mutation load, RNA viruses are likely to be highly dependent on the action of host chaperones and the quality control machinery to buffer mutational effects and to maintain the functionality of the viral proteome. Molecular chaperones are central regulators of the conformation and life cycle of proteins in the cell; these chaperones are fundamental for protein biogenesis, as they promote folding and assembly of newly translated polypeptides and assist their trafficking to organelles. Chaperones also monitor destabilized proteins, preventing their aggregation and promoting quality control through either refolding or facilitating the degradation of misfolded proteins. Accordingly, chaperones can preserve the functionality of proteins that have been destabilized by mutations, and thereby buffer detrimental mutations39. Most viruses use cellular chaperones during their life cycle both to solve their own protein-folding problems and to interfere with cellular processes, such as signal transduction40,41. Many examples of the interaction between cellular chaperones and viral factors at different stages of the infectious cycle have been reported in recent years42–52.
Although the role of chaperones as extrinsic modulators of viral diversity has not been systematically explored, some examples are beginning to emerge. For instance, the capsid precursor (P1) proteins of several picornaviruses, including poliovirus, rhinovirus and coxsackievirus, require heat shock protein 90 (HSP90) for folding and assembly41. Experiments in poliovirus showed that P1 uses HSP90 to fold into a conformation that is competent for processing by a viral protease; this processing is required for subsequent assembly of mature capsids41. Deep-sequencing analyses indicate that pharmacological inhibition of HSP90 results in a profound alteration of the mutation distribution in the viral population (R. Geller, A. Acevedo, J.F. and R.A., unpublished observations). Viral populations grown in HSP90-inhibited cells harboured substantially fewer amino acid substitutions (36% reduction) than viral populations grown in wild-type cells, suggesting that HSP90 enables expansion of the viable protein-coding sequence space in viral populations. Whether different chaperones preferentially buffer specific types of mutation remains to be determined, but if this were to be the case, it would imply that chaperone levels influence the evolutionary trajectory of the viral proteins in a given cell. In addition, the role of the quality control and degradation machineries, such as the ubiquitin–proteasome pathway, in purging dominant-negative or otherwise deleterious mutations from virus-infected cells is also relevant, but has not yet been studied. This function might be particularly important in the case of multisubunit complexes, such as the capsid, for which incorporation of misfolded subunits could create defective viral particles53.
Given the importance of chaperones to viral replication and robustness, it is not surprising that viral infection often elicits cellular stress responses that lead to enhanced expression of chaperones40,54. In principle, these responses could be triggered by the production of mutated, destabilized viral proteins during replication, as well as by the enhanced load put on the cellular biogenesis machinery by rapidly replicating viruses. It is intriguing to speculate that viral activation of cellular stress signalling pathways increases the availability of chaperones and other extrinsic modulators of robustness.
Robustness and evolvability
Our discussion thus far has focused largely on how mutational robustness buffers the negative impact of high viral mutation rates. However, the selective neutrality, or flatness, of the viral fitness landscape will also define the set of mutations that are viable and potentially adaptive. A key unresolved question, then, is whether mutational robustness increases or decreases evolvability, which we refer to as the capacity of a virus to increase in fitness through adaptation13. According to Fisher’s fundamental theorem, “The rate of increase in fitness of any organism at any time is equal to its genetic variance in fitness at that time.” (REF. 55.) Because robustness expands the neutral network of genotypes in a population, it should reduce phenotypic variation and the efficiency of natural selection. However, robustness will also increase the amount of cryptic genetic variation in a population, and these neutral mutations might have epistatic interactions with subsequent mutations, thus increasing the range of adaptive phenotypes that are available in the local sequence space. This pro-evolvability model is supported by thermodynamic studies of RNA folding29,30,56, and recent theoretical work suggests that robustness favours adaptive evolution when the number of phenotypes accessible to an individual through mutation is smaller than the total number of phenotypes in a given landscape57.
Experimental evidence from a number of systems also links robustness to increased evolvability. Mutagenesis studies with the cytochrome P450 system suggest that thermostability determines mutational robustness and the evolutionary capacity of proteins58. Perhaps the most intriguing results come from the recent studies of thermostable ribozymes32. As described above, in mutationally robust ribozyme populations that retained function, the cryptic variation was selectively neutral in the native environment. However, the accumulated diversity was nevertheless beneficial, as the robust populations adapted to a new substrate more rapidly than their less diverse ancestors. Therefore, mutational robustness allows a population to explore a range of genotypes that are neutral in one environment but potentially beneficial in another.
Work in the phage Φ6 model also suggests a positive correlation between robustness and evolvability. Using derived phage clones that were created by longterm passage22 and varied in mutational robustness, the authors of this study asked whether the clones differed in adaptation to a defined selective pressure59. Twelve clones from each lineage were subjected to intermittent heat shock for 50 additional generations, and the percentage of clones from each lineage that survived heat shock was used to quantify adaptation to this new environment. The mutationally robust strains demonstrated higher survival rates than the less robust strains, indicating that robustness accelerated the fitness gain over the preceding 50 generations. Similar to the ribozyme experiment, this work supports a model in which robustness allows for a greater exploration of sequence space, reducing the number of additional mutations required for fitness gain in a new environment.
This pattern of neutral network exploration and rapid adaptation has also been defined in a phylodynamic study of influenza A virus60. In this analysis, the linkage between genotypes and the phenotypes of the surface haemagglutinin (HA) was established using existing sequence databases and antigenic maps, which quantify cross-immunity to different epitopes. Influenza A virus H3N2 isolates were found to move throughout large neutral networks, accumulating genetic variation. This phenotypic stasis is punctuated by sudden changes in phenotype, in which the virus adopts a new antigenic structure. The viral dynamics are triggered by shifts in the host immune environment, aggregated across populations. Interestingly, these epochal shifts coincide with peaks in infections and are well described by epidemiological models of susceptible, infected and recovered individuals.
Evolvability and pathogenesis
Experimental measures of robustness and evolvability require controlled environments with defined selective pressures. By contrast, in nature, viral populations will encounter a range of environmental conditions over their lifespan (FIG. 4). Shifts in host tropism, immune pressure and cellular milieu create a highly dynamic fitness landscape to which the virus must adapt. Because optimal fitness in each environment is determined by a specific combination of mutations, it might be advantageous for the viral population to maintain an assortment of pre-adapted, or exapted, variants that are fewer mutational steps away from a fitness optimum32,61. In this model, mutational robustness would be a selective advantage, as it allows a greater exploration of the sequence space and a more rapid fitness gain.
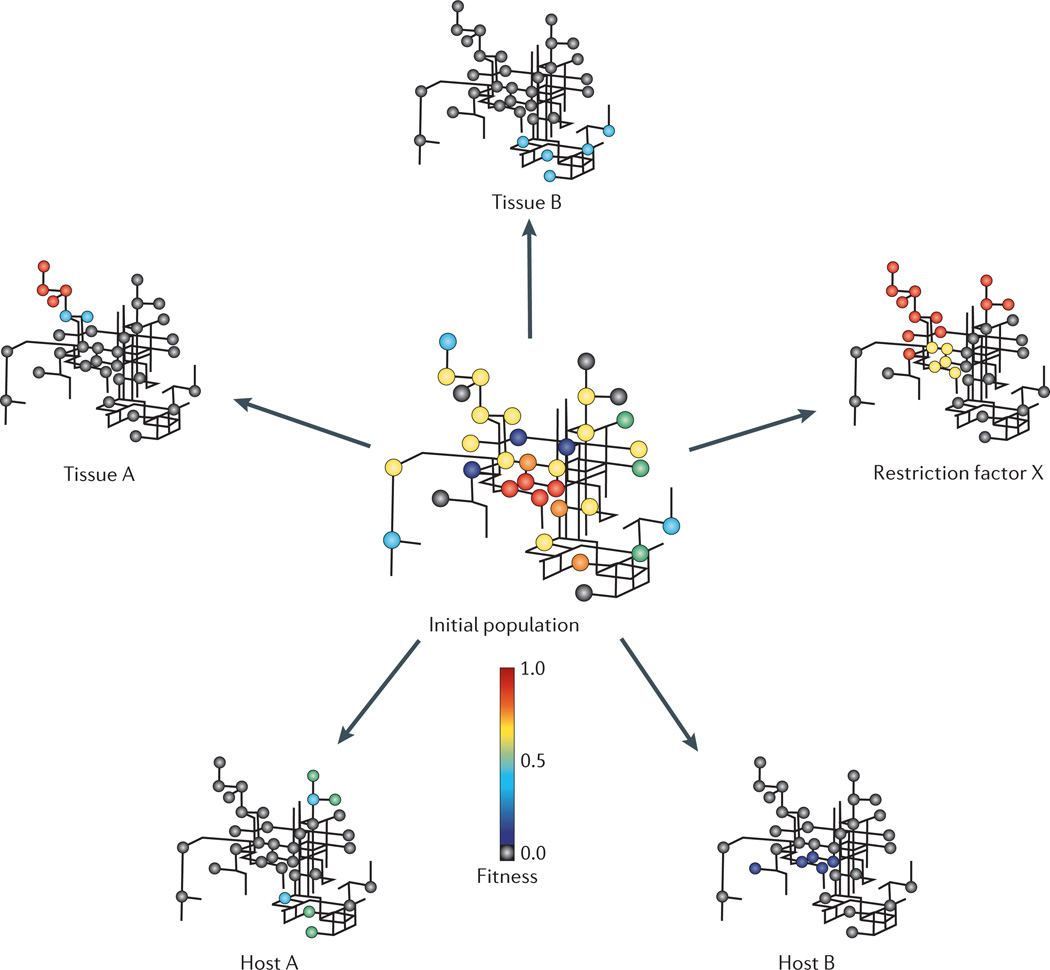
Figure 4. Dynamics of viral networks under selection in the host.
The initial viral population is represented by the pipe diagram at the centre of the figure. Each individual varient of the population is represented by a ball, coloured according to fitness, and these balls are connected to each other by a genetic network. Robustness allows the population to explore an extensive region of sequence space, resulting in a larger number of individuals that are able to adapt quickly to environmental challenges. As the viral population confronts changes in the environment, the mutant distribution changes to favour those better adapted to a particular condition, such as replication in a different tissue (tissue A versus tissue B), in a different host (naive versus immunologically primed) or in a different species (called zoonotic adaptation). Robustness and diversity might also allow the viral population to overcome immunological challenges such as that of antiviral restriction factors. With each of these challenges, the fitness landscape changes and so does the distribution of mutants in the population. The majority of the variants in the initial population are expected to be poorly adapted to a new environment and to have very low fitness (grey balls). However, given the high degree of phenotypic diversity in the initial founder population, there is a high chance that one or more variants (those with a fitness greater than 0 in the new environment) will quickly adapt and propogate in the new environment, whereas those of low fitness are expected to diminish over time. For example, a variant of intermediate fitness in host A (a green ball) could eventually predominate in this environment if the genotype of this variant is better adapted to this new environment than the other genotypes present in this host. However, if the new environment is host B, then an even less fit variant (dark blue) could theoretically reach fixation as the population is exposed to a different set of selective pressures. The nature of the selective pressure in each new environment is the ultimate factor that decides the distribution of mutants in the population.
This hypothesis was tested using the synonymous poliovirus variants described above14. Although the wild-type virus and both the SD and Max mutants all encode the same consensus amino acid sequence across the capsid, each virus was found to have a unique spectrum of low-frequency variants within the population. That is, the three populations had different cryptic genetic variation. Furthermore, the less mutationally robust SD population was attenuated relative to the wild type in a mouse pathogenesis model, whereas the more robust Max population was similar to the wild type in terms of virulence in vivo. These data support a model in which robust populations, which are located on the flatter fitness landscapes, are able to maintain sufficient cryptic variation for rapid adaptation to new selective pressures. Alternatively, the SD population might just have the ‘wrong’ set of variants compared to wild-type poliovirus, and these variants might reduce the ability of the SD population to adapt to the dynamic environment of the infected individual. In this alternative model, robustness would be a surrogate marker for the location of the population in the genetic sequence space. It is difficult to distinguish between these two models because the origin of a population in the sequence space defines the set of available mutations for that population and places it in a fitness landscape. This confounding effect of local sequence space might explain why another study found a negative correlation between robustness and evolvability62. In this study, the ability of two divergent VSV populations to gain fitness in a new cellular host was compared, and the less robust population proved to be more adaptable. Studies with larger numbers of viruses and distinct selective pressures will be needed to clarify which of these two models of robustness and evolvability is more generalizable.
Robustness and antiviral therapy
Given the importance of mutational robustness to the behaviour of RNA viruses, does robustness represent an opportunity for, or a barrier to, antiviral therapy? With their high mutation rates, RNA virus populations replicate near the limit of viability (the so-called error threshold). A population can remain at equilibrium despite a high mutation rate63,64; however, small increases in the mutation rate will disrupt this balance, and the population will lose meaningful genetic cohesion in an avalanche of errors. Although such an error catastrophe has not been observed experimentally, early studies of VSV and poliovirus showed that mutagenic nucleoside drugs are lethal to RNA virus populations8,65,66. Recent work on host cell restriction of retroviral infection suggests that some antiviral defence pathways rely on inducing lethal mutagenesis (reviewed in REF. 67). For example, members of the APOBEC family of RNA-editing enzymes deaminate C residues to U residues during reverse transcription of retroviral RNA. As a result, the progeny genomes exhibit G-to-A hypermutation and reduced infectivity68,69. It is important to note, however, that error catastrophe and lethal mutagenesis describe different processes. Bull et al. have explained the distinction most clearly: “an error catastrophe is an evolutionary shift in genotype space, whereas extinction is a demographic process, a drop in the absolute abundance of individuals in the population” (REF. 70). This distinction is more than semantic, as a population shift to a more mutationally robust region of genotypic space could delay or even prevent error catastrophe.
These observations have stimulated an interest in using lethal mutagenesis as a broadly effective therapeutic strategy for RNA virus infections (reviewed in REF. 71). Ribavirin and other nucleoside analogues have demonstrated efficacy against poliovirus, lymphocytic choriomeningitis virus, Hantaan virus, Lassa fever virus, foot-and-mouth disease virus and HIV66,72–76. Lethal mutagenesis was initially thought to be resistance-proof, as a newly arising drug resistance mutation would be shackled to a lethal mutation in the same genome. However, it is now clear that a viral polymerase can evolve biochemical resistance to mutagenic nucleosides by either excluding the drug from its active site or lowering its intrinsic error rate4,5,77,78.
Because high mutation rates will select for evolved robustness, RNA viruses could conceivably achieve resistance by moving to flatter, more neutral regions of the fitness landscape. In other words, viruses could adapt to the drug by becoming more tolerant to mutation. This possibility was examined using two closely related enteroviruses; human coxsackievirus B3 was found to be mutationally fragile, exhibiting substantially less genetic diversity and tolerance to non-synonymous mutation than poliovirus18. Human coxsackievirus B3 was also more susceptible to ribavirin-induced lethal mutagenesis. These results demonstrate the importance of relative robustness in determining the therapeutic efficacy of mutagenic drugs across viral species. In a separate study, the potential for the emergence of drug resistance through evolved robustness was examined directly by exposing lymphocytic choriomeningitis virus populations to 5-fluorouracil (5-FU) mutagenesis for nine passages. Both the evolved populations and their ancestral clones remained susceptible to lethal mutagenesis with higher concentrations of 5-FU, suggesting that robustness does not select for resistance73. These results are consistent with a recent theoretical treatment of the topic, in which the benefit of increased robustness was eclipsed by the detrimental effects of higher mutation rates on fitness79.
The manipulation of mutational robustness could be used to design strategies to restrict the evolution of live-attenuated viral vaccine strain candidates. The traditional process of attenuation through serial passage, forces a virus to adapt to a foreign host environment. The accumulated mutations move the population to a new region of sequence space that is unable to support efficient replication and spread in the native host. Studies have shown that small changes in the error rate of an RNA virus owing to mutations in the viral RNA-dependent RNA polymerase gene can substantially reduce population diversity, resulting in attenuation4,80. The study of synonymous poliovirus populations suggests that reorganizing the connectivity of viral mutant networks can achieve the same goal14. In this case, relocation of a population in sequence space rendered that population less mutationally robust. The ruggedness of the associated fitness landscape can reduce the capacity of the virus to generate fit progeny and adapt to host selective pressures. As others have shown, synonymous mutation has the additional advantages of preserving the antigenic composition of the wild-type virus and reducing the risk of reversion through recombination with homologous sequences in circulating strains27,28,81–83. A rational reduction in mutational robustness might allow for a finer control of evolvability, limiting the ability of a virus to escape immune surveillance and spread in a vaccinated host.
Conclusions and future directions
Mutational robustness is central to the evolution of living organisms. Although genetic variation is the fuel for natural selection, there are limits to the amount of variation that a population can tolerate without loss of fitness or viability. This is particularly true for RNA viruses, which have compact, tightly organized genomes and extremely high mutation rates. It is perhaps not surprising, then, to find that RNA viruses have developed mechanisms to cope with an increased mutational load. What it is more unexpected is the observation that robustness and cryptic genetic variation can have such crucial roles in the rapid and effective adaptation to dynamic environments. In view of the overall paucity of experimental work in this area, it is a particularly exciting time for the field, and there are currently many more questions than answers.
The precise nature of the robustness mechanisms is far from well defined at the moment. The relationship between the genetic structure of a given population and its phenotypic landscape is also unclear. Although many theoretical frameworks have been developed to delineate the relationship between robustness and adaptation, more experimental work is urgently needed to better define how viruses deal with their high mutation rates in the real world and to reveal the molecular mechanisms that underlie robustness at both the RNA and protein levels. Accordingly, it will be important to accurately define the mutation distribution of a viral population in order to establish the proportion of neutral mutations. This measurement would, in turn, serve as a baseline to better define whether robustness does indeed facilitate the increased diversity of viral populations, thereby providing a reservoir of mutations that could allow rapid adaptation to changes in the environment. It will also be of interest to understand how viral manipulation of the host protein and RNA homeostasis machineries, for instance through the induction of stress responses, promotes tolerance to mutations. Although the available data suggest that there are conditions which seem to favour robust populations, we still cannot say for certain whether viruses have evolved to become mutationally robust. We expect that the experimental dissection of robustness across RNA virus taxa will reveal important differences that are linked to viral phenotypes. For example, does robustness allow for the greater exploration of sequence space observed in chronic HIV and hepatitis C virus infection? Does the relative robustness determine which viruses are better targeted by lethal mutagenesis? Defining the evolutionary pay-off or disadvantage of mutational robustness is important because these factors could determine the evolutionary trajectory of viruses in their natural hosts and might hold the key to new antiviral strategies.
Glossary
- Fitness
The ability of an entity to survive and reproduce. In experimental virology, replicative efficiency is often used as a surrogate for fitness. In this Review, we define viral fitness as the capacity of a virus to generate infectious progeny
- Evolvability
The capacity of a virus or organism with a particular genotype to gain fitness over time after evolving in a given environment
- Epistatic interaction
An interaction between mutations such that their combined effect on fitness is different to that expected from their effects in isolation
- Mutational fitness effect
The effects of mutations on fitness; often described in a model that combines both the strength and distribution of these effects
- Bottleneck
In genetics a dramatic reduction in the number of individuals that can reproduce. Bottlenecks reduce genetic variation and are not necessarily selective events
- Negative selection
The removal of deleterious alleles from a population by natural selection. Also called purifying selection
- Effective population size
The size of an idealized population that would experience genetic drift in the same way as the actual population. The effective population size (Ne) is often smaller than the total population size
- Multiplicity of infection
In virology, the ratio of infectious particles to target cells
- Complementation
In the context of this Review the process by which a defective virus can take advantage of functional nucleic acid sequences or proteins from another virus that is infecting the same cell. As a result, the defective virus does not experience loss of fitness from its mutation (or mutations)
- Viral sex
The process by which genetic information is exchanged between two different strands of viral nucleic acid. In RNA viruses, this occurs most commonly through switching the replicative template (recombination) or through the exchange of genomic segments (reassortment)
- Fitness landscapes
Spatial models that link fitness values to individual sequences
- Sequence space
All possible mutations and combinations of mutations present in a given DNA or amino acid sequence
- Digital organisms
Self-replicating computer programs that mutate and evolve, often in competition with each other for CPU (central processing unit) cycles
- Synonymous mutations
Codon mutations that do not alter the amino acid specificity of the codons. By contrast, non-synonymous mutations do change the encoded amino acid
- Volatile codons
Codons with a propensity to mutate non-synonymously, as opposed to synonymously
- Codon bias
A difference in the observed frequencies of synonymous codons in a given set of sequences
- Codon pair bias
A difference in the observed frequencies of 6-nucleotide codon pairs in a given set of sequences
- Phylodynamic study
A study that develops a quantitative model, incorporating both a pathogen phylogeny and epidemiological or immunological data, to describe an infectious disease
- Error catastrophe
The loss of meaningful genetic information when a population is pushed beyond its maximum mutation rate. In theoretical models, the error catastrophe has been compared to a chemical phase transition, and a true error catastrophe has not been observed experimentally
- Lethal mutagenesis
The process whereby the number of viable individuals, or viruses, in a population is reduced through increases in the mutation rate
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Adam S. Lauring’s homepage:
http://www.med.umich.edu/microbio/bio/lauring.htm
Judith Frydman’s homepage:
http://www.stanford.edu/group/frydman/web
Raul Andino’s homepage:
http://andino.ucsf.edu/andino/index.html
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Holland J, et al. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 2. Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J. Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. A recent and comprehensive review of viral mutation rates.
- 3.Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffey LL, Beeharry Y, Bordería AV, Blanc H, Vignuzzi M. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc. Natl. Acad. Sci. USA. 2011;108:16038–16043. doi: 10.1073/pnas.1111650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanjuán R, Moya A, Elena SF. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc. Natl Acad. Sci. USA. 2004;101:8396–8401. doi: 10.1073/pnas.0400146101. The first study to rigorously examine mutational fitness effects in an RNA virus and demonstrate that the vast majority of mutations are detrimental to replicative fitness.
- 7.Sanjuán R. Mutational fitness effects in RNA and single-stranded DNA viruses: common patterns revealed by site-directed mutagenesis studies. Philos. Trans. R. Soc. 2010;B365:1975–1982. doi: 10.1098/rstb.2010.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CH, et al. Negative effects of chemical mutagenesis on the adaptive behavior of vesicular stomatitis virus. J. Virol. 1997;71:3636–3640. doi: 10.1128/jvi.71.5.3636-3640.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crotty S, et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nature Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 10.Eckerle LD, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham RL, et al. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nature Med. 2012;18:1820–1826. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnädig NF, et al. Coxsackievirus B3 mutator strains are attenuated in vivo . Proc. Natl Acad. Sci. USA. 2012;109:e2294–e2303. doi: 10.1073/pnas.1204022109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Visser JAGM, et al. Perspective: evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. A review by leaders in the field summarizing theoretical and experimental work on genetic robustness.
- 14.Lauring AS, Acevedo A, Cooper SB, Andino R. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe. 2012;12:623–632. doi: 10.1016/j.chom.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domingo-Calap P, Cuevas JM, Sanjuán R. The fitness effects of random mutations in single-stranded DNA and RNA bacteriophages. PLoS Genet. 2009;5:e1000742. doi: 10.1371/journal.pgen.1000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuevas JM, Domingo-Calap P, Sanjuán R. The fitness effects of synonymous mutations in DNA and RNA viruses. Mol. Biol. Evol. 2012;29:17–20. doi: 10.1093/molbev/msr179. [DOI] [PubMed] [Google Scholar]
- 17.Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J. Biol. Chem. 2005;280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graci JD, et al. Mutational robustness of an RNA virus influences sensitivity to lethal mutagenesis. J. Virol. 2012;86:2869–2873. doi: 10.1128/JVI.05712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elena SF. RNA virus genetic robustness: possible causes and some consequences. Curr. Opin. Virol. 2012;2:525–530. doi: 10.1016/j.coviro.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Wright CF, et al. Beyond the consensus: dissecting within-host viral population diversity of foot-and-mouth disease virus by using next-generation genome sequencing. J. Virol. 2011;85:2266–2275. doi: 10.1128/JVI.01396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Froissart R, et al. Co-infection weakens selection against epistatic mutations in RNA viruses. Genetics. 2004;168:9–19. doi: 10.1534/genetics.104.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montville R, Froissart R, Remold SK, Tenaillon O, Turner PE. Evolution of mutational robustness in an RNA virus. PLoS Biol. 2005;3:e381. doi: 10.1371/journal.pbio.0030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon-Loriere E, Holmes EC. Why do RNA viruses recombine? Nature Rev. Microbiol. 2011;9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilke CO, Wang JL, Ofria C, Lenski RE, Adami C. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature. 2001;412:331–333. doi: 10.1038/35085569. The initial demonstration of the ‘survival of the flattest’ effect using digital organisms.
- 25. Codoñer FM, Darós J-A, Solé RV, Elena SF. The fittest versus the flattest: experimental confirmation of the quasispecies effect with subviral pathogens. PLoS Pathog. 2006;2:e136. doi: 10.1371/journal.ppat.0020136. An examination of the survival of the fittest versus the survival of the flattest effects in plant viroids.
- 26. Sanjuán R, Cuevas JM, Furió V, Holmes EC, Moya A. Selection for robustness in mutagenized RNA viruses. PLoS Genet. 2007;3:e93. doi: 10.1371/journal.pgen.0030093. The demonstration of the survival of the flattest effect at elevated mutation rates in an RNA virus.
- 27.Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J. Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huynen MA, Stadler PF, Fontana W. Smoothness within ruggedness: the role of neutrality in adaptation. Proc. Natl Acad. Sci. USA. 1996;93:397–401. doi: 10.1073/pnas.93.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fontana W, Schuster P. Continuity in evolution: on the nature of transitions. Science. 1998;280:1451–1455. doi: 10.1126/science.280.5368.1451. [DOI] [PubMed] [Google Scholar]
- 31.van Nimwegen E. Influenza escapes immunity along neutral networks. Science. 2006;314:1884–1886. doi: 10.1126/science.1137300. [DOI] [PubMed] [Google Scholar]
- 32. Hayden EJ, Ferrada E, Wagner A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature. 2011;474:92–95. doi: 10.1038/nature10083. A landmark paper that shows how robust populations can accumulate cryptic genetic variation that is neutral in one environment but facilitates rapid adaptation in another.
- 33.Wagner A, Stadler PF. Viral RNA and evolved mutational robustness. J. Exp. Zool. 1999;285:119–127. [PubMed] [Google Scholar]
- 34.Sanjuán R, Forment J, Elena SF. In silico predicted robustness of viroid RNA secondary structures. II. Interaction between mutation pairs. Mol. Biol. Evol. 2006;23:2123–2130. doi: 10.1093/molbev/msl083. [DOI] [PubMed] [Google Scholar]
- 35.Sanjuán R, Forment J, Elena SF. In silico predicted robustness of viroids RNA secondary structures. I. The effect of single mutations. Mol. Biol. Evol. 2006;23:1427–1436. doi: 10.1093/molbev/msl005. [DOI] [PubMed] [Google Scholar]
- 36.Stephens CR, Waelbroeck H. Codon bias and mutability in HIV sequences. J. Mol. Evol. 1999;48:390–397. doi: 10.1007/pl00006483. [DOI] [PubMed] [Google Scholar]
- 37.Plotkin JB, Dushoff J. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc. Natl Acad. Sci. USA. 2003;100:7152–7157. doi: 10.1073/pnas.1132114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyers LA, Ancel FD, Lachmann M. Evolution of genetic potential. PLoS Comput. Biol. 2005;1:236–243. doi: 10.1371/journal.pcbi.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fares MA, Ruiz-González MX, Moya A, Elena SF, Barrio E. Endosymbiotic bacteria: GroEL buffers against deleterious mutations. Nature. 2002;417:398. doi: 10.1038/417398a. Pioneering work that explores how cellular chaperones could buffer mutational effects.
- 40.Mayer MP. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 2005;153:1–46. doi: 10.1007/s10254-004-0025-5. [DOI] [PubMed] [Google Scholar]
- 41.Geller R, Vignuzzi M, Andino R, Frydman J. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 2007;21:195–205. doi: 10.1101/gad.1505307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alfano C, McMacken R. Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of bacteriophage λ DNA replication. J. Biol. Chem. 1989;264:10709–10718. [PubMed] [Google Scholar]
- 43.Cripe TP, Delos SE, Estes PA, Garcea RL. In vivo and in vitro association of hsc70 with polyomavirus capsid proteins. J. Virol. 1995;69:7807–7813. doi: 10.1128/jvi.69.12.7807-7813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang D, et al. Heat shock cognate protein 70 is a cell fusion-enhancing factor but not an entry factor for human T-cell lymphotropic virus type I. Biochem. Biophys. Res. Commun. 1999;261:357–363. doi: 10.1006/bbrc.1999.1028. [DOI] [PubMed] [Google Scholar]
- 45.Gallie DR, Fortner D, Peng J, Puthoff D. ATP-dependent hexameric assembly of the heat shock protein Hsp101 involves multiple interaction domains and a functional C-proximal nucleotide-binding domain. J. Biol. Chem. 2002;277:39617–39626. doi: 10.1074/jbc.M204998200. [DOI] [PubMed] [Google Scholar]
- 46.Guerrero CA, et al. Heat shock cognate protein 70 is involved in rotavirus cell entry. J. Virol. 2002;76:4096–4102. doi: 10.1128/JVI.76.8.4096-4102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kampmueller KM, Miller DJ. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J. Virol. 2005;79:6827–6837. doi: 10.1128/JVI.79.11.6827-6837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu JS, et al. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J. Biol. Chem. 1998;273:30704–30712. doi: 10.1074/jbc.273.46.30704. [DOI] [PubMed] [Google Scholar]
- 49.Niewiarowska J, D’Halluin JC, Belin MT. Adenovirus capsid proteins interact with HSP70 proteins after penetration in human or rodent cells. Exp. Cell Res. 1992;201:408–416. doi: 10.1016/0014-4827(92)90290-o. [DOI] [PubMed] [Google Scholar]
- 50.Park SG, Lee SM, Jung G. Antisense oligodeoxynucleotides targeted against molecular chaperonin Hsp60 block human hepatitis B virus replication. J. Biol. Chem. 2003;278:39851–39857. doi: 10.1074/jbc.M301618200. [DOI] [PubMed] [Google Scholar]
- 51.Satyanarayana T, Gowda S, Ayllón MA, Dawson WO. Closterovirus bipolar virion: evidence for initiation of assembly by minor coat protein and its restriction to the genomic RNA 5′ region. Proc. Natl Acad. Sci. USA. 2004;101:799–804. doi: 10.1073/pnas.0307747100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agranovsky AA, et al. Beet yellows closterovirus HSP70-like protein mediates the cell-to-cell movement of a potexvirus transport-deficient mutant and a hordeivirus-based chimeric virus. J. Gen. Virol. 1998;79:889–895. doi: 10.1099/0022-1317-79-4-889. [DOI] [PubMed] [Google Scholar]
- 53.Crowder S, Kirkegaard K. Trans-dominant inhibition of RNA viral replication can slow growth of drug-resistant viruses. Nature Genet. 2005;37:701–709. doi: 10.1038/ng1583. [DOI] [PubMed] [Google Scholar]
- 54.Jockusch H, Wiegand C, Mersch B, Rajes D. Mutants of tobacco mosaic virus with temperature-sensitive coat proteins induce heat shock response in tobacco leaves. Mol. Plant Microbe Interact. 2001;14:914–917. doi: 10.1094/MPMI.2001.14.7.914. [DOI] [PubMed] [Google Scholar]
- 55.Fisher AR. The Genetical Theory Of Natural Selection. Claredon: 1930. [Google Scholar]
- 56. Wagner A. Robustness and evolvability: a paradox resolved. Proc. Biol. Sci. 2008;275:91–100. doi: 10.1098/rspb.2007.1137. An examination of the relationship between robustness and evolvability through analyses of RNA structures.
- 57. Draghi JA, Parsons TL, Wagner GP, Plotkin JB. Mutational robustness can facilitate adaptation. Nature. 2010;463:353–355. doi: 10.1038/nature08694. A theoretical study that defines situations in which robustness will favour evolvability.
- 58.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc. Natl Acad. Sci. USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McBride RC, Ogbunugafor CB, Turner PE. Robustness promotes evolvability of thermotolerance in an RNA virus. BMC Evol. Biol. 2008;8:231. doi: 10.1186/1471-2148-8-231. One of the first papers to explore the relationship between robustness and evolvability in an RNA phage.
- 60.Koelle K, Cobey S, Grenfell B, Pascual M. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science. 2006;314:1898–1903. doi: 10.1126/science.1132745. [DOI] [PubMed] [Google Scholar]
- 61.Gould SJ, Vrba ES. Exaptation—a missing term in the science of form. Paleobiology. 1982:4–15. [Google Scholar]
- 62.Cuevas JM, Moya A, Sanjuán R. A genetic background with low mutational robustness is associated with increased adaptability to a novel host in an RNA virus. J. Evol. Biol. 2009;22:2041–2048. doi: 10.1111/j.1420-9101.2009.01817.x. [DOI] [PubMed] [Google Scholar]
- 63.Biebricher CK, Eigen M. The error threshold. Virus Res. 2005;107:117–127. doi: 10.1016/j.virusres.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 64.Eigen M. Error catastrophe and antiviral strategy. Proc. Natl Acad. Sci. USA. 2002;99:13374–13376. doi: 10.1073/pnas.212514799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holland JJ, Domingo E, de la Torre JC, Steinhauer DA. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J. Virol. 1990;64:3960–3962. doi: 10.1128/jvi.64.8.3960-3962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl Acad. Sci. USA. 2001;98:6895–6900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malim MH. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos. Trans. R. Soc. 2009;B364:675–687. doi: 10.1098/rstb.2008.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mangeat B, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 69.Zhang H, et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bull JJ, Sanjuán R, Wilke CO. Theory of lethal mutagenesis for viruses. J. Virol. 2007;81:2930–2939. doi: 10.1128/JVI.01624-06. A study into the theory of error catastrophe, its relationship to lethal mutagenesis and the potential for evolved robustness during antiviral therapy.
- 71.Anderson JP, Daifuku R, Loeb LA. Viral error catastrophe by mutagenic nucleosides. Annu. Rev. Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- 72.McCormick JB, et al. Lassa fever. Effective therapy with ribavirin. N. Engl. J. Med. 1986;314:20–26. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- 73.Ruiz-Jarabo CM, Ly C, Domingo E, de la Torre JC. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) Virology. 2003;308:37–47. doi: 10.1016/s0042-6822(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 74.Severson WE, Schmaljohn CS, Javadian A, Jonsson CB. Ribavirin causes error catastrophe during Hantaan virus replication. J. Virol. 2003;77:481–488. doi: 10.1128/JVI.77.1.481-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Loeb LA, et al. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl Acad. Sci. USA. 1999;96:1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sierra S, Dávila M, Lowenstein PR, Domingo E. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J. Virol. 2000;74:8316–8323. doi: 10.1128/jvi.74.18.8316-8323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pfeiffer JK, Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl Acad. Sci. USA. 2003;100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.null) et al. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J. Virol. 2007;81:2012–2024. doi: 10.1128/JVI.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O’Dea EB, Keller TE, Wilke CO. Does mutational robustness inhibit extinction by lethal mutagenesis in viral populations? PLoS Comput. Biol. 2010;6:e1000811. doi: 10.1371/journal.pcbi.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vignuzzi M, Wendt E, Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nature Med. 2008;14:154–161. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- 81.Bull JJ, Molineux IJ, Wilke CO. Slow fitness recovery in a codon-modified viral genome. Mol. Biol. Evol. 2012;29:2997–3004. doi: 10.1093/molbev/mss119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Burns CC, et al. Genetic inactivation of poliovirus infectivity by increasing the frequencies of CpG and UpA dinucleotides within and across synonymous capsid region codons. J. Virol. 2009;83:9957–9969. doi: 10.1128/JVI.00508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burns CC, et al. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J. Virol. 2006;80:3259–3272. doi: 10.1128/JVI.80.7.3259-3272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 85.Eigen M. Viral quasispecies. Sci. Am. 1993;269:42–49. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- 86.Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 87.Holland JJ, la Torre, de JC, Steinhauer DA. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 88.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc. Natl Acad. Sci. USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elena SF, Sanjuán R. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 2005;79:11555–11558. doi: 10.1128/JVI.79.18.11555-11558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]