Abstract
Overgrowth disorders are a heterogeneous group of conditions characterised by increased growth parameters and variable other clinical features, such as intellectual disability and facial dysmorphism1. To identify novel causes of human overgrowth we performed exome sequencing in 10 proband-parent trios and detected two de novo DNMT3A mutations. We identified 11 additional de novo mutations through DNMT3A sequencing of a further 142 individuals with overgrowth. The mutations were all located in functional DNMT3A domains and protein modelling suggests they interfere with domain-domain interactions and histone binding. No similar mutations were present in 1000 UK population controls (13/152 vs 0/1000; P<0.0001). Mutation carriers had a distinctive facial appearance, intellectual disability and increased height. DNMT3A encodes a key methyltransferase essential for establishing the methylation imprint in embryogenesis and is commonly somatically mutated in acute myeloid leukaemia2-4. Thus DNMT3A joins an emerging group of epigenetic DNA and histone modifying genes associated with both developmental growth disorders and haematological malignancies5.
The control of human growth is highly complex and influenced by common and rare genetic variation6. The study of human overgrowth syndromes, which are characterised by increased prenatal and postnatal growth relative to age-related peers, has led to significant insights into fundamental biological processes involved in growth control1. As many growth disorders present as non-familial cases with a distinctive phenotype we hypothesised that de novo gene mutations may underlie some cases. To investigate this we are conducting trio-based exome sequencing in individuals with overgrowth and their parents using the Illumina TruSeq exome enrichment array. We are performing sequencing with an Illumina HiSeq 2000, aligning the data with Stampy, performing variant calling with Platypus, variant annotation with SAVANT and identifying potential de novo variants using a custom R script.
Review of the first 10 trios revealed two with apparent de novo mutations in DNMT3A (DNA cytosine 5 methyltransferase 3A), which was of immediate interest because of the functional relationship of this gene with EZH2, a known overgrowth predisposition gene we previously identified as the cause of Weaver syndrome7,8. By Sanger sequencing we confirmed the mutations, an inframe deletion p.Trp297del in COG0274 and a nonsynonymous mutation, p.Leu648Pro in COG0553, were present in the proband but not in the parents (Table 1, Supplementary Figure 1).
Table 1.
DNMT3A mutations and associated clinical features
| Case ID | Mutation | Protein alteration | Height /sd |
OFC /sd |
Age* /yrs |
Intellectual disability |
Other clinical features |
|---|---|---|---|---|---|---|---|
| COG0274 | c.889_891delTGG | p.Trp297del | 2.6 | 2.2 | 3.0 | moderate | Seizures |
| COG1770 | c.929T>A | p.Trp297del | 2.9 | 3.7 | 9.0 | moderate | Ventriculomegaly, umbilical hernia, scoliosis |
| COG1670 | c.934_937dupTCTT | p.Trp297del | 3.2 | 2.8 | 20.5 | moderate | |
| COG0141 | c.1594G>A | p.Trp297del | 2.3 | na | 5.9 | moderate | |
| COG0422 | c.1643T>A | p.Met548Lys | 1.9 | 1.2 | 4.1 | moderate | Atrio-septal defect |
| COG1288 | c.1645T>C | p.Cys549Arg | 1.8 | 3.8 | 11.3 | moderate | Atrio-septal defect, sagittal craniosynostosis |
| COG0553 | c.1943T>C | p.Leu648Pro | 3.4 | 5.1 | 19.0 | mild | Mild hemihypertrophy, umbilical hernia |
| COG1688 | c.2099C>T | p.Pro700Leu | 2.8 | 2.1 | 13.0 | moderate | Scoliosis |
| COG1695 | c.2245C>T | p.Arg749Cys | 4.0 | 3.8 | 12.0 | moderate | Vesico-ureteric reflux, patella subluxation |
| COG1512 | c.2297dupA | p.Arg767fs | 3.8 | 1.6 | 8.2 | moderate | |
| COG1771 | c.2512A>G | p.Asn838Asp | 4.2 | 1.7 | 13.4 | mild | Testicular atrophy, seizures, scoliosis |
| COG0109 | c.2705T>C | p.Phe902Ser | 2.7 | 1.3 | 9.8 | moderate | Mitral and tricuspid regurgitation, kyphoscoliosis |
| COG1677 | c.2711C>T | p.Pro904Leu | 3.7 | 1.2 | 11.0 | moderate |
This is the age at which growth parameters were measured.
Abbreviations: sd, standard deviations with reference to the mean (UK90 growth data); na, not available
To further evaluate the role of DNMT3A, we sequenced the full coding sequence and intron-exon boundaries of the gene by Sanger sequencing in a further 142 individuals with overgrowth in whom mutations in NSD1, EZH2 and PTEN and dysregulation of the 11p15 growth regulatory region had been excluded, and for whom parental DNA was also available (Supplementary Table 1). We identified an additional 11 de novo DNMT3A mutations. Thus in total, we found 13 different de novo DNMT3A mutations in 152 individuals with overgrowth phenotypes: 10 nonsynonymous mutations, two small frameshifting insertions and one inframe deletion (Figure 1, Table 1, Supplementary Figure 1). These data establish DNMT3A mutations as a cause of a novel human disorder, which we have termed ‘DNMT3A overgrowth syndrome’.
Figure 1.
DNMT3A structure and mutations. Schematic representation of the protein structure of DNMT3A with (a) de novo mutations identified in overgrowth cases placed above the protein and (b) nonsynonymous variants identified in controls placed below the protein.
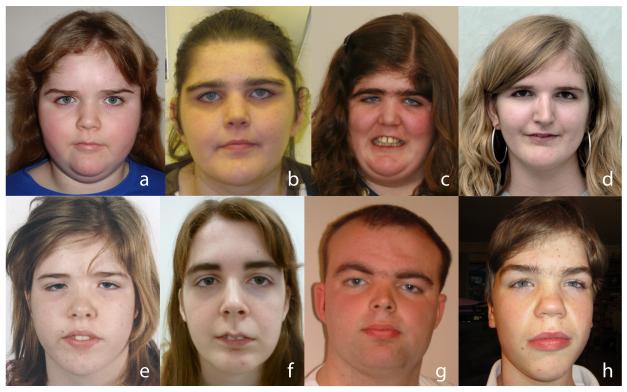
A consistent phenotype, characterised by a distinctive facial appearance, tall stature and intellectual disability, was evident amongst the thirteen individuals with de novo DNMT3A mutations (Figure 2, Table 1). The facial gestalt was characterised by a round face, heavy, horizontal eyebrows and narrow palpebral fissures (Figure 2). Height was increased in all individuals ranging from 1.8 to 4.2 (mean 3.0) standard deviations above the mean. Head circumference was also increased ranging from 1.2 to 5.1 (mean 2.5) standard deviations above the mean. Intellectual disability, which was described as moderate in 11 individuals and mild in the remaining two, is also a key feature of the condition. Other, less frequent clinical features were variably present (Table 1). More detailed phenotyping in larger series are required to evaluate if these are real associations of DNMT3A mutations and to better define the clinical spectrum of this new overgrowth syndrome.
Figure 2.
Characteristic facial appearance DNMT3A overgrowth syndrome.
The mutation, growth parameters and other clinical features are given in Table 1 under the appropriate COG ID: (a) COG1288; (b) COG1670; (c) COG0422; (d) COG1695; (e) COG1688; (f) COG0109; (g) COG0553; (h) COG1512. Specific consent to publish facial photographs was obtained for all individuals.
In eukaryotic DNA, methylation preferentially occurs at cytosine bases which are converted to 5-methylcytosine by four DNA methyltransferase enzymes: DNMT3A, DNMT3B, DNMT1 and DNMT3L2. DNMT3A and DNMT3B are essential for the establishment of new methylation marks following erasure of parental methylation patterns during early embryonic development and for the establishment of sex-dependent methylation marks of imprinted genes during gametogenesis9-11. By contrast, DNMT1 is essential for the maintenance and conservation of DNA methylation marks after DNA replication10. DNMT3L does not possess inherent enzymatic activity but appears to physically interact with DNMT3A and stimulate its enzymatic activity12-14.
DNMT3A contains three functional domains: a proline-tryptophan-tryptophan-proline (PWWP) domain; an ATRX-DNMT3A-DNMT3L-type zinc finger (ADD) domain and a C terminal DNA methyltransferase domain (Figure 1). It is noteworthy that all the mutations we identified were located in these domains. None were present in 1000 exomes from a UK population control series that were sequenced and analysed in a similar fashion. Moreover, no other frameshifting or nonsynonymous mutations in domains were present in the control exomes; four nonsynonymous variants were detected, but all were outside the functional domains (Figure 1). These data provide further evidence that the DNMT3A mutations in cases are pathogenic (13/152 vs 0/1000, P<0.0001).
DNMT3A is one of the most frequently mutated genes in acute myeloid leukemia (AML)3,4,15. Somatic DNMT3A mutations are detected in approximately one third of cytogenetically normal AML and have also been reported in other haematological neoplasms, such as myelodysplasia (MDS)16,17. Over half the somatic DNMT3A mutations target a single residue, Arg882, with the remainder being nonsynonymous and truncating mutations scattered through the gene 3,15. The somatic mutational spectra thus differs from the de novo mutations we identified in overgrowth cases; we did not detect any Arg882 mutations and only two of the mutations we report, Arg749Cys and Pro904Leu, are also in the 167 confirmed somatic DNMT3A mutations in haematological malignancies in the COSMIC database (Supplementary Table 2).
Protein structure modelling suggests the Arg882 somatic mutations affect DNA binding and functional analyses demonstrate mutations at this residue result in reduced methyltransferase activity, possibly through a dominant-negative mechanism4,15,18. To explore the potential impact of the DNMT3A overgrowth mutations we undertook protein structure modelling which revealed the residues targeted by nonsynonymous mutations in the MTase domain appear to be located at the interaction interface with the ADD domain, whereas those in the ADD domain are close to the histone H3 binding region (Figure 3). The Arg767 residue in the MTase domain that is mutated by a frameshifting insertion is situated at the interface between DNMT3A and DNMT3L. Thus, whilst none of the overgrowth mutations appear likely to affect DNA binding, their position and the role of the ADD domain in the recognition of unmethylated lysine 4 of histone H3 suggest they may interfere with domain-domain interactions and histone binding, thereby disrupting de novo methylation19.
Figure 3.
Mutations mapped onto a structural model of the DNMT3A-DNMT3L complex. Two orientations of a model of the DNMT3A monomer generated by superposition of structures of the DNMT3A ADD domain (cyan), the DNMT3A MTase domain (light orange) and full-length DNMTL (green) show mutations in the MTase domain in purple and mutations in the ADD domain in pink. The histone peptide bound to the ADD domain is shown in orange and the position of the DNA is inferred by superposition of the structurally homologous bacterial cytosine methyltransferase HhaI-DNA complex. The model shows that the mutations in the MTase domain appear to be located at the interaction interface with its ADD domain and at the interface with DNMT3L. Mutations in the ADD domain are close to the histone-binding region.
Taken together these data are intriguing as they show both clear differences and some overlap between the DNMT3A mutational spectra in malignancies and overgrowth. The mechanism of pathogenesis in DNMT3A overgrowth syndrome is currently unclear though a simple haploinsufficiency model appears unlikely given the small proportion of truncating mutations. Of note, parents of Dnmt3a knockout mice are grossly phenotypically normal, though it is unclear whether this is because loss of function of one Dnmt3a copy is not associated with overgrowth, or because the overgrowth phenotype is too subtle to detect in mice9. Further functional and mutational analyses will be of interest, to extend and illuminate these observations. Long-term follow-up of individuals with DNMT3A overgrowth syndrome, with particular focus on cancer incidence, will also be of interest. To date, none of the probands have developed malignancies, though the oldest is only 29 years of age. Haematological malignancies with somatic DNMT3A mutations typically occur in middle-aged individuals3,4, and it is thus possible that an increased cancer risk only manifests at older ages.
DNMT3A joins an emerging family of genes with dual roles in the pathogenesis of syndromic overgrowth and myeloid neoplasms. Other similar genes include EZH2 and NSD15. The overgrowth phenotypes associated with EZH2 and NSD1 constitutional mutations, which are called Weaver and Sotos syndrome respectively, are similar to each other and to DNMT3A overgrowth syndrome5,8,20. Somatic EZH2 mutations, both activating and inactivating, occur in AML and poor prognosis myeloproliferative neoplasms and myelodysplastic syndromes15,21. Somatic NSD1 point mutations are rare4 but the NUP90-NSD1 fusion protein, generated through the recurrent translocation, t(5;11)(q35.3;p15.5) is present in approximately 5% of childhood acute myeloid leukaemia22. Both EZH2 and NSD1 are histone methyltransferases that play key roles in regulation of transcription through histone modification and chromatin modelling. EZH2 catalyses the trimethylation of lysine residue 27 in histone H3 (H3K27me3) and is associated with transcriptional repression, whereas NSD1 preferentially catalyses methylation of lysine residue 36 of histone H3 (H3K36) and is primarily associated with transcriptional activation23,24. Of note, another gene involved in chromatin modification that is often mutated in AML and MDS, ASXL1, is also associated with a growth disorder15,25. Constitutional de novo truncating ASXL1 mutations cause Bohring-Opitz syndrome, a rare disorder characterised by severe undergrowth, severe intellectual disability, a characteristic facial appearance and flexion deformities25. This suggests other epigenetic regulatory genes somatically mutated in haematological malignancies, such as TET2, IDH1 and IDH2 would be worth evaluating in developmental growth disorders15.
METHODS
Patient samples
Individuals with overgrowth were recruited through the Childhood Overgrowth Study. The research has approval from the London Multicentre Ethics Committee (Reference: MREC/01/2/44) and informed consent was obtained from all participants and/or families. DNA was extracted from peripheral blood and was available from all probands and their parents. Detailed phenotypic information, through a standardised questionnaire and photographs was also obtained. Specific consent to publish facial photographs was obtained. A full list of collaborators is given in the Supplementary Note.
Control samples
We used lymphocyte DNA from 1000 population-based controls obtained from the 1958 Birth Cohort Collection, a continuing follow-up of persons born in the United Kingdom in one week in 1958. Biomedical assessment was undertaken during 2002-2004 at which blood samples and informed consent were obtained for creation of a genetic resource. http://www.cls.ioe.ac.uk/
Exome sequencing
We prepared DNA libraries from 1.5 μg blood-derived genomic DNA using the Paired-End DNA Sample Preparation Kit (Ilumina). DNA was fragmented using Covaris technology and the libraries were prepared without gel size selection. We performed target enrichment using the TruSeq Exome Enrichment Kit (Illumina) targeting 62 Mb of the human genome. The captured DNA libraries were PCR amplified using the supplied paired-end PCR primers. Sequencing was performed with an Illumina HiSeq2000 generating 2 × 101 bp reads.
Exome variant calling and de novo mutation detection
We mapped sequencing reads to the human reference genome (hg19) using Stampy version 1.0.1426. Duplicate reads were flagged using Picard version 1.60 (http://picard.sourceforge.net). Median coverage of the target at 15× was 91% across the 1030 individuals (1000 controls and 30 individuals from the 10 overgrowth trios), with a median of 47,215,315 reads mapping to the target. We used Platypus version 0.1.5 to perform variant calling27 and SAVANT, a custom strand-aware variant annotation tool written in Python which follows HGVS nomenclature and ensures consistent annotation of indels, shifting them to their most 3′ position in the transcript. This script is available on request. We used an R script to identify variants present in cases but not either parent which is available on request.
DNMT3A mutation analysis
We performed Sanger sequencing of PCR products from genomic DNA to confirm the mutations identified by exome sequencing, and to mutationally analyse the gene in the overgrowth series. We designed PCR primers to amplify the 22 coding exons and intron-exon boundaries of DNMT3A in 4 multiplex PCR reactions (Supplementary Table 1). The PCR was carried out using a Qiagen Multiplex PCR kit according to the manufacturer’s instructions. Products were sequenced with the original PCR primers or internal sequencing primers (exons 3, 6, 8, 10, 14 and 22) using the BigDye Terminator Cycle Sequencing Kit and an ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA,USA). Sequences were analyzed using Mutation Surveyor software v3.97 (SoftGenetics, State College, PA, USA), and verified by manual inspection. All mutations were confirmed by bidirectional sequencing of a second, independently amplified PCR product.
Statistical Analysis
The frequency of mutations in cases and controls was compared using a two-sided Fisher’s exact test.
Protein structure modelling of mutations
The model of the DNMT3A-DNMT3L complex was created using the crystal structures of the DNMT3A ADD domain (PDB 3A1B), the DNMT3A MTase domain (PDB 2QRV) and full length DNMT3L (PDB 2PVC). The bound DNA was positioned by superposing the crystal structure of bacterial cytosine methyltransferase HhaI in complex with DNA (PDB 1MHT). First, the structure of partial DNMT3L in complex with DNMT3A was superposed onto the structure of full-length DNMT3L to reveal the interactions of the full length DNMT3L with DNMT3A. Subsequently, the ADD domain of DNMT3A was positioned by superposing it onto the homologous region of the second molecule of full-length DNMT3L. Finally, the DNA was positioned by superposing the crystal structure of bacterial cytosine methyltransferase HhaI in complex with DNA onto the MTase domain of DNMT3A, placing the DNA at the SAM/DNA binding region. The image was depicted using PyMOL visualization software (The PyMOL Molecular Graphics System, Version 1.6.0.0 Schrödinger, LLC.).
Supplementary Material
Acknowledgements
We thank the families for their participation in our research and the physicians and nurses that recruited them. Samples were collected through the Childhood Overgrowth Collaboration; a full list of collaborators in on the Supplementary Note. We are grateful to Margaret Warren-Perry, Darshna Dudakia and Jess Bull for assistance in recruitment, and to Ellen Moran (Department of Genetics, NYU Hospital for Joint Diseases, New York, USA) and Alexandra Murray (Institute of Medical Genetics, University Hospital of Wales, Cardiff, UK) for their clinical input for COG1770 and COG0109 respectively. We thank Ann Strydom for assistance in preparing the manuscript. We are grateful to Dr Gerton Lunter and Dr Márton Münz (Wellcome Trust Centre for Human Genetics) Oxford University) for their contribution to the development of the custom annotation tool SAVANT. We acknowledge use of services provided by the Institute of Cancer Research Genetics Core Facility, which is managed by Sandra Hanks and Nazneen Rahman. We acknowledge NHS funding to the Royal Marsden/ICR NIHR Biomedical Research Centre. We also thank Mariani Foundation Milan for supporting the clinical activity of UOS Genetica Clinica Pediatrica, Fondazione MBBM, AO S Gerardo Monza, Italy. This research and was supported by the Wellcome Trust (100210/Z/12/Z) and the Institute of Cancer Research, London.
Footnotes
Accession numbers
DNMT3A mutation nomenclature corresponds to Ensembl Transcript ID ENST00000264709.
Competing Financial Interests
None
References
- 1.Tatton-Brown K, Weksberg R. Molecular mechanisms of childhood overgrowth. Am J Med Genet C Semin Med Genet. 2013;163:71–5. doi: 10.1002/ajmg.c.31362. [DOI] [PubMed] [Google Scholar]
- 2.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–22. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 3.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–15. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 5.Tatton-Brown K, Rahman N. The NSD1 and EZH2 Overgrowth Genes, Similarities and Differences. Am J Med Genet C Semin Med Genet. 2013;163:86–91. doi: 10.1002/ajmg.c.31359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durand C, Rappold GA. Height matters-from monogenic disorders to normal variation. Nat Rev Endocrinol. 2013;9:171–7. doi: 10.1038/nrendo.2012.251. [DOI] [PubMed] [Google Scholar]
- 7.Rush M, et al. Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation. Epigenetics. 2009;4:404–14. doi: 10.4161/epi.4.6.9392. [DOI] [PubMed] [Google Scholar]
- 8.Tatton-Brown K, et al. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2:1127–33. doi: 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 10.Smallwood SA, Kelsey G. De novo DNA methylation: a germ cell perspective. Trends Genet. 2012;28:33–42. doi: 10.1016/j.tig.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Kaneda M, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–3. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 12.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci U S A. 2002;99:16916–21. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–93. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 14.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–23. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Wahab O, Levine RL. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood. 2013;121:3563–72. doi: 10.1182/blood-2013-01-451781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcucci G, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2012;30:742–50. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikoloski G, van der Reijden BA, Jansen JH. Mutations in epigenetic regulators in myelodysplastic syndromes. Int J Hematol. 2012;95:8–16. doi: 10.1007/s12185-011-0996-3. [DOI] [PubMed] [Google Scholar]
- 18.Kim SJ, et al. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013;122:4086–89. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otani J, et al. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–41. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatton-Brown K, et al. Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am J Hum Genet. 2005;77:193–204. doi: 10.1086/432082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund K, Adams PD, Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia. 2013;28:44–49. doi: 10.1038/leu.2013.288. [DOI] [PubMed] [Google Scholar]
- 22.Cerveira N, et al. Frequency of NUP98-NSD1 fusion transcript in childhood acute myeloid leukaemia. Leukemia. 2003;17:2244–7. doi: 10.1038/sj.leu.2403104. [DOI] [PubMed] [Google Scholar]
- 23.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–64. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Morishita M, di Luccio E. Structural insights into the regulation and the recognition of histone marks by the SET domain of NSD1. Biochem Biophys Res Commun. 2011;412:214–9. doi: 10.1016/j.bbrc.2011.07.061. [DOI] [PubMed] [Google Scholar]
- 25.Hoischen A, et al. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat Genet. 2011;43:729–31. doi: 10.1038/ng.868. [DOI] [PubMed] [Google Scholar]
- 26.Lunter G, Goodson Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–9. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rimmer A, Lunter G, McVean G. Platypus: an integrated variant caller. 2012 www.well.ox.ac.uk/platypus.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.