Abstract
Microscopical and molecular analyses were used to investigate the diversity and spatial community structure of spring phytoplankton all along the estuarine gradient in a macrotidal ecosystem, the Baie des Veys (eastern English Channel). Taxa distribution at high tide in the water column appeared to be mainly driven by the tidal force which superimposed on the natural salinity gradient, resulting in a two-layer flow within the channel. Lowest taxa richness and abundance were found in the bay where Teleaulax-like cryptophytes dominated. A shift in species composition occurred towards the mouth of the river, with the diatom Asterionellopsis glacialis dramatically accumulating in the bottom waters of the upstream brackish reach. Small thalassiosiroid diatoms dominated the upper layer river community, where taxa richness was higher. Through the construction of partial 18S rDNA clone libraries, the microeukaryotic diversity was further explored for three samples selected along the surface salinity gradient (freshwater - brackish - marine). Clone libraries revealed a high diversity among heterotrophic and/or small-sized protists which were undetected by microscopy. Among them, a rich variety of Chrysophyceae and other lineages (e.g. novel marine stramenopiles) are reported here for the first time in this transition area. However, conventional microscopy remains more efficient in revealing the high diversity of phototrophic taxa, low in abundances but morphologically distinct, that is overlooked by the molecular approach. The differences between microscopical and molecular analyses and their limitations are discussed here, pointing out the complementarities of both approaches, for a thorough phytoplankton community description.
Introduction
Estuaries form transition zones linking freshwater and marine biomes. Due to mixing of both distinct water bodies, they are characterized by pronounced gradients of physical and chemical components [1]. These factors strongly influence the phytoplankton community structure and other microbial eukaryotes along the resulting continuum. Major estuaries are usually classified into three types based on their longitudinal salinity distribution and flow characteristics: i) highly stratified or salt wedge, ii) partially mixed, or iii) well mixed [2]. However, for many systems wherein physical forces are highly variable, such as the shallow macrotidal estuaries [3], assignation to one estuary type considering temporal (seasonality, tidal cycle variation) and spatial (lower, intermediate, upper estuary) variations is difficult.
Protists are key components of aquatic food webs, both as major primary producers and as important consumers of bacteria in the “microbial loop” [4]. In recent years, the rise of molecular microbial ecology has opened the possibility of studying protist diversity independently of morphological considerations. Such molecular environmental surveys revealed a high diversity of eukaryotic lineages and contributed to our current understanding of microbial food web structure and biogeochemical processes in aquatic systems [5], [6]. This approach has been applied in a wide variety of ecosystems, including oceanic/coastal waters, freshwater ecosystems, and many extreme environments such as anoxic systems or deep-sea vents [7]–[10]. Most studies have focused on small size protists (<3–5 μm) which usually escape detection with traditional microscopy and are difficult to isolate. Insight of many novel eukaryotic lineages divergent from known protist sequences suggests that a large fraction of these communities still remains to be discovered [11].
Microbial communities inhabiting aquatic transition systems received relatively little attention in the past. Most investigations were based on morphological approaches and carried out in well-known and vast estuaries of the world, focusing generally on the large phytoplanktonic fractions [12], or restricted to a specific part of the estuary [13]. Very few diversity surveys using molecular techniques have been conducted so far on phytoplankton and other protists inhabiting rivers or marine–freshwater transition zones, except for some recent studies restricted to a single point and following temporal dynamics [14], [15].
The Vire River flows into the Baie des Veys which is located on the French coast, facing the English Channel. While much effort has been made to study temporal dynamics of phytoplankton and primary production in this macrotidal estuarine ecosystem [16]–[19], detailed studies of protistan diversity and their spatial patterns in such transitional waters are still lacking. Planktonic and benthic microalgae within the Baie des Veys have been mainly characterized using microscopy techniques [16], [17], and only one study has been conducted in the Vire River estuary, limited to a single station located in the lower zone [19].
In this context, we investigated here the diversity and spatial distribution of the spring phytoplankton community along the entire freshwater-to-marine continuum of the Vire River (Baie des Veys). This spatial investigation involved exhaustive taxonomic identifications and cell counting using the traditional microscopical method. Through the construction of 18S rDNA gene clone libraries for three selected samples along the surface salinity gradient, the genetic diversity of microeukaryotes was explored, providing a first insight into the protistan diversity that may occur in such a transition zone. The data offered an opportunity to compare the parallel use of morphological and molecular approaches for identifying taxa and measuring diversity in transitional waters.
Materials and Methods
1. Ethics Statement
The present study was not carried out in a protected area or on private land. Therefore, no specific permission was required. We confirmed that the field study did not involve endangered or protected species. Only water samples were collected (no animals), therefore, not subject to regulation.
2. Study area and sampling strategy
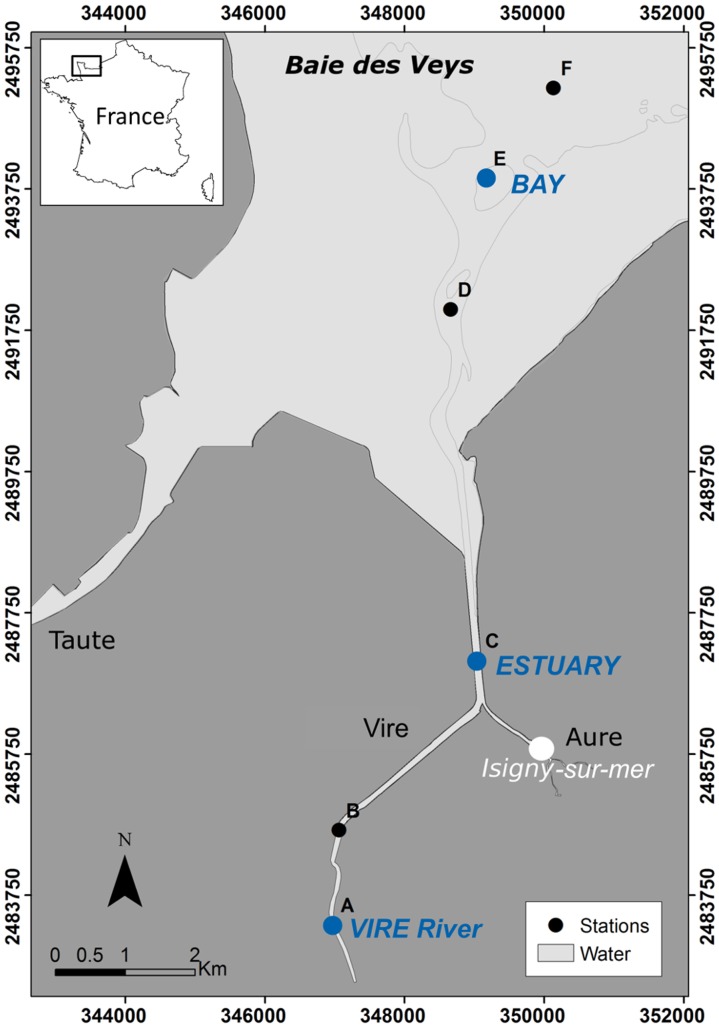
The Baie des Veys is an intertidal estuarine ecosystem of the eastern English Channel, located in Normandy, north-western France (Figure 1). With a maximum tidal range of 8 m and a small intertidal area (37 km2, [20]), this macrotidal estuary is highly tide-influenced. Freshwater that enters the southern part of the bay derives from the discharge of four rivers, notably the main river, the Vire (length = 128 km), which has an annual mean discharge of 15 m3 s−1, with important variations throughout the year. The freshwater inputs from this channel are the main source of nutrients in the bay and induce each year a relatively high primary production during the spring diatom bloom [18].
Figure 1. Map of the Vire River estuary (Baie des Veys) with location of the sampling stations (A–F).
Blue dots indicate sampling points analyzed by both microscopy and clone library approaches (referred to as VIRE River, ESTUARY and BAY). Lambert II coordinates system.
Sampling was undertaken on 27th April 2010, at the end of the high river discharge period (4.85 m−3 s−1) and during the slack high tide, so that no tidal variation influenced our measurements. Six sampling sites (stations A–F, Figure 1) were established covering the whole salinity gradient of the Vire River estuary. Water was collected at the surface of all stations (samples As→Fs) and at 1 m above the bottom for stations B to E (samples Bb→Eb) with a 5-L Niskin bottle (stations A and F were only sampled at the surface due to technical constraints). All samples were analyzed by microscopy and three of the surface samples (As, Cs and Es) belonging respectively to the freshwater, brackish water and marine part of the estuarine gradient, were also used for clone library analysis (hereafter referred to as VIRE River, ESTUARY and BAY, Figure 1).
3. Physicochemical and biological measurements
Physicochemical profiles (depth, temperature, salinity) of the water column were obtained with a multi-parameter probe (Hydrolab Data sonde 5 Options, USA), and vertical profiles of irradiance were measured with an underwater quantum sensor (LICOR LI-1400, Nebraska, USA). Inorganic nutrients analysis was performed in the laboratory with a Bran+Luebbe Autoanalyzer AA3 according to Aminot & Kérouel [21]. Chlorophyll a biomass (Chl a) was estimated by fluorometry (Trilogy 7200-000 - Turner Designs, California, USA) according to the method of Welschmeyer [22] as described in Bazin et al. [23].
4. Morphological identification of the phytoplankton community
First observations with light microscopy (LM) on living material were carried out to establish a preliminary floristic list. For further identification and counting, sub-samples were rapidly fixed after collection with glutaraldehyde (final concentration 1%) and stored in darkness at 4°C until analysis. Phytoplankton was identified to the lowest possible taxonomic level using appropriate literature and keys for marine and freshwater environments (e.g. [24]–[26]). Further examination was made by scanning electron microscopy (SEM) using a JEOL JSM-6400. Heterotrophic dinoflagellates were included in the overall analysis.
Cells were enumerated using the Utermöhl settling method [27]. Because phytoplankton density can vary considerably along the estuarine gradient, volume of samples and settling time were adjusted (3 mL–10 mL for at least 48 hours) to ensure the complete sedimentation of the organisms [28]. Taxa were quantified at 400× in randomly-selected microscopic fields, with a Leica DMI3000B inverted microscope. A minimum of 500 individual units were counted, leading to a counting error not exceeding 10% [29].
5. DNA collection, PCR and cloning
To examine the overall eukaryotic community in the three selected surface samples (VIRE River, ESTUARY and BAY), 500 mL to 2 L of water was filtered onto 0.7-μm pore size glass fiber filters (Whatman) with no initial prefiltration.
As described in detail in Bazin et al. [23], total DNA was extracted using the Invisorb Spin Plant mini Kit (Invitek, Berlin, Germany) with modification of the first steps of the manufacturer's protocol, including cutting filters into pieces and cell disruption by thermal shocks (three freeze-thaw cycles: liquid nitrogen/+65°C).
Eukaryotic 18S rRNA genes (≈1800 bp) were amplified using the eukaryotic-specific primer set Euk A/Euk B [30] according to Bazin et al. [23]. Reactions were performed at two different annealing temperatures: 55 and 50°C, and pooled. The conditions were as follows: an initial hot-start at 95°C for 10 min, followed by 30 cycles (95°C for 1 min, 1.5 min at 50/55°C, 72°C for 2 min) and a final extension at 72°C for 10 min. Several replicates of PCR products were pooled and cleaned with the Wizard PCR clean-up system kit (Promega).
Clone libraries were constructed using the pCR2.1 TOPO-TA cloning kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The presence of inserts in the putative positive colonies was checked using flanking vector primers (M13). PCR products containing amplicons of the target size were purified and sequenced with an ABI Prism 3100 (Applied Biosystems, Foster City, CA, USA). The 18S rDNA was partially sequenced using the internal standard primer 895R (5′-AAATCCAAGAATTTCACCTC-3′) which covers conserved and rapidly evolving regions, and resulted in reads of 500–800 bp.
6. Taxonomic affiliation and phylogenetic analyses
All the sequences were manually checked, trimmed and edited using the SeqAssem software [31], and then compared to those available in public databases (GenBank) using the NCBI BLASTn web application [32]. Potential chimeras were detected with the online softwares Bellerophon [33] and KeyDNAtools [34]. After removal of low-quality sequences, metazoan sequences, and suspected chimeras, the remaining sequences were aligned using the slow and iterative refinement method FFT-NS-I with MAFFT 6.9 software [35]. The resulting alignment was checked and corrected manually. Based on this alignment, the sequences were clustered into distinct operational taxonomic units (OTUs) with MOTHUR 1.13 [36] using a similarity threshold of 98% that roughly corresponds to the genus/species level [37].
Phylogenetic trees including additional selected sequences from both GenBank and ARB databases were reconstructed using both neighbor joining (NJ, Jukes-Cantor distance) and maximum likelihood method (ML) with MEGA 5. Bootstrap support values (BP) were calculated from 1000 replicates for NJ tree and were reported on the ML tree, for which BP were from 100 replicates. The sequences reported in this paper have been deposited in the GenBank database under accession numbers JX645081- JX645156.
7. Diversity and statistical analyses
For the morphological approach, taxa richness and abundance were determined from floristic lists and cell counts. Taxa richness for the molecular data was estimated with MOTHUR, based on OTUs defined at 98% sequence similarity level. Rarefaction curves (Sobs) were generated for the three clone libraries and OTU richness was estimated by the non-parametric Chao 1 diversity estimator [38]. The comparison of taxa diversity between the samples was carried out through the Jaccard similarity index, by considering taxa (and OTUs) composition only. Principal Component Analysis (PCA) was performed to group samples according to the environmental variables (standardized and ln(x+1) transformed), and Correspondence Analysis (CA) was applied to a matrix of the relative contribution of phytoplankton in order to establish relationships between taxonomic composition and sampling stations. All statistics were computed with the PAST software [39].
Results
1. Environmental context
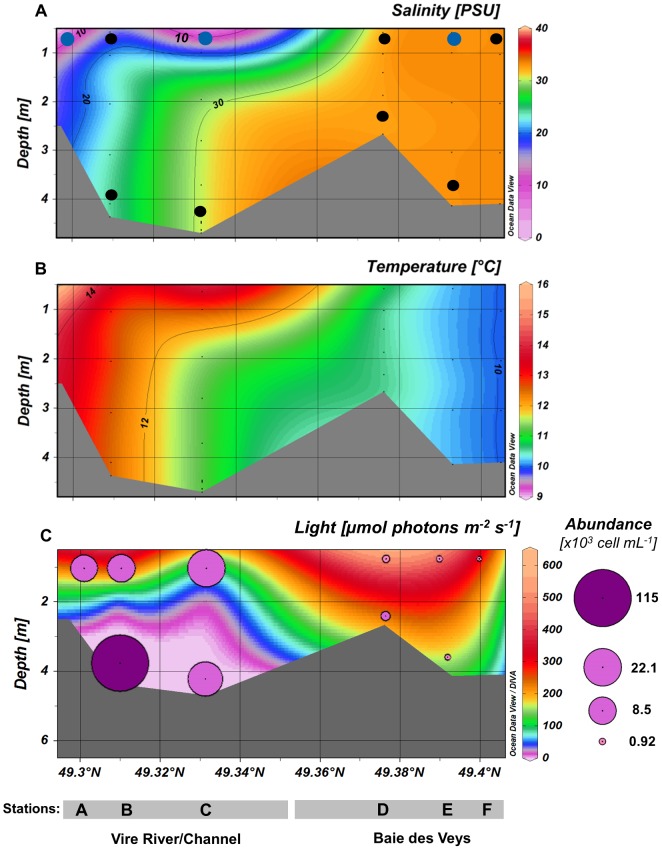
The Vire River estuary is characterized by shallow waters (depth = 2.7–4.7 m along the whole transect at the time of sampling) (Figure 2). Clear horizontal and vertical gradients characterized the distribution of salinity, temperature and nutrients (Figure 2A–B and Figure S1) in the first part of the transect (stations A–D), emphasizing the water column stratification (i.e. a two-layer system) within the Vire channel at high tide. As confirmed by the PCA analysis including nine environmental variables (temperature, salinity, nitrate, nitrite, phosphate, silicate, Chl a concentrations and total phytoplankton abundance, Figure S2), three main area can be discriminated along the estuarine continuum. An “upper surface layer” in the river channel [samples As( = VIRE River), Bs, Cs( = ESTUARY)], defined by oligohaline (0.5–5) to mesohaline (5–18) waters, high nutrients from terrestrial runoff, warmer temperatures (13.8–16°C) and high phytoplankton abundance (8.5–22×103 cells mL−1), was distinguished from the bottom brackish layer (samples Bb-Cb), which was characteristic of the estuarine “silt plug” (or estuarine turbidity maximum, ETM). The latter was characterized by polyhaline water (18–30) and total darkness due to very high light attenuation in the water column (attenuation coefficient Kd = 2.4–1.8 for B–C, knowing that Kd = 0.035 for pure water) associated with highest phytoplankton abundance (>115×103 cells mL−1 in Bb) (Figure 2C). No vertical gradient was noted for stations D–F (including sample Es = BAY) due to well-mixed water column within the bay (Figures 2, S1 and S2). This “coastal” area was defined by higher salinities (euhaline >30) but lower temperatures (<11°C), as well as lower nutrients, Chl a, and phytoplankton abundances (≤1.2×103 cells mL−1).
Figure 2. Profiles of salinity (A), temperature (B), irradiance and phytoplankton abundance (C) along the estuarine gradient.
(Stations A→F). Black dots on the profile (A) represent sampling points collected for the global phytoplankton analysis by microscopy, and the blue dots indicate those analyzed by both microscopy and clone library approaches. A logarithmic scale was used for the representation of phytoplankton cell abundance on the profile of irradiance(C).
2. Spatial pattern in phytoplankton community structure
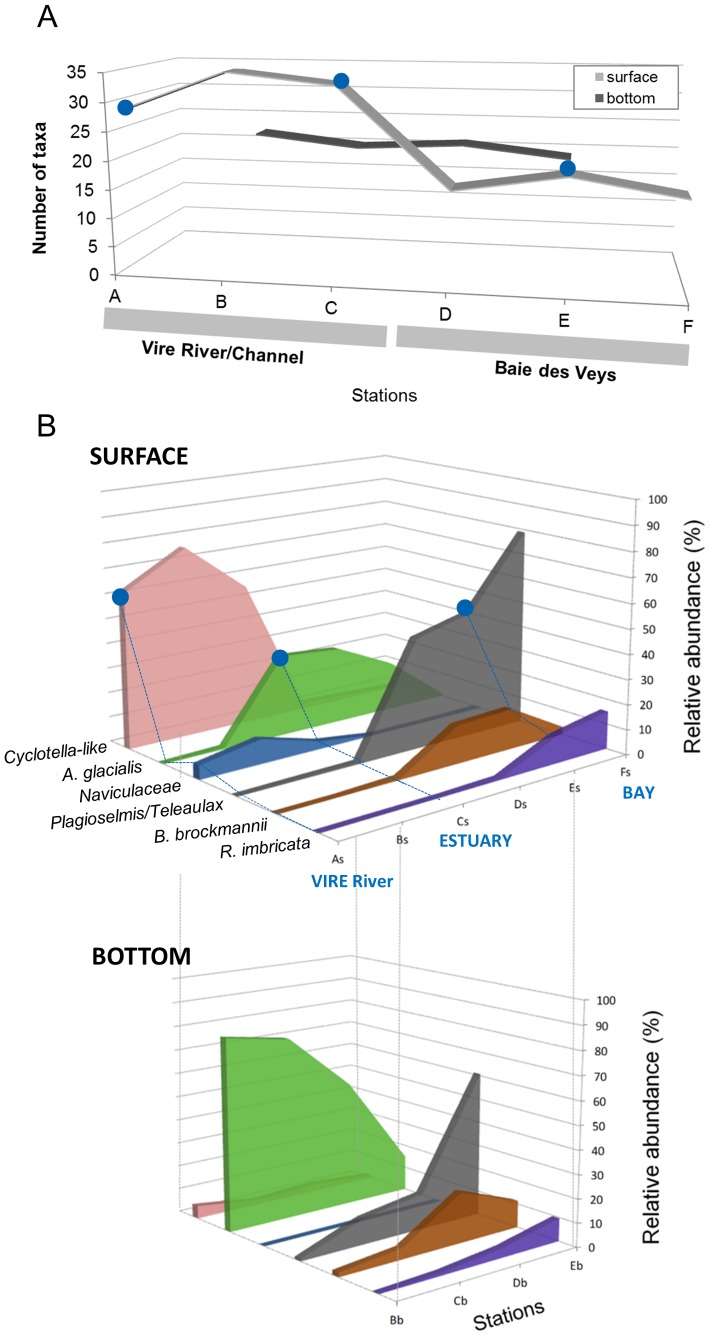
A distinct horizontal gradient in algal cell abundance (Figure 2C), taxa richness (Figure 3A) and taxa composition (Figure 3B) was observed along the whole transect, with vertical variations in the water-stratified zone in the Vire channel (B–C). A total of 86 different species were observed (floristic list, Table S1) corresponding to 63 genera, and six taxa were regarded as dominant species (>10% in at least one sampling point, Figure 3B).
Figure 3. Taxa richness and distribution of dominant species along the estuarine gradient.
(A) Total phytoplankton richness. (B) Relative abundances of dominant taxa (accounting for >10% of total phytoplankton in at least one sample) at the surface (top) and depth (down). A. glacialis = Asterionellopsis glacialis, B.brockmanni = Brockmanniella brockmannii and R.imbricata = Rhizosolenia imbricata. Blue dots indicate sampling points analyzed by both microscopy and clone library approaches (referred to as VIRE River, ESTUARY and BAY).
Major taxa in the marine section
Lowest taxa richness and abundances (<1.2×103 cells mL−1) were found in the Baie des Veys (stations D–F, including BAY) (Figures 2C, 3A). The Cryptophyceae, exclusively represented by the Teleaulax/Plagioselmis group, dominated the phytoplankton community in the northern part of the bay reaching 79% of total species abundance in Fs (Figure 3B). Predominance of these flagellates was reduced toward the mouth of the estuary, where a shift towards dominance of diatoms (Bacillariophyceae) was observed with a relative contribution reaching 84% at the bottom of station D. The most abundant diatoms in the bay were the pennates Asterionellopsis glacialis (max. = 53% of total species in Db), Brockmanniella brockmannii (21% in Db) and the centric Rhizosolenia imbricata (15% at Fs) (Figure 3B).
Due to similarity in species composition, samples from the north of the bay (E–F), regardless of the depth, were grouped in the same cluster on the CA ordination diagram (Figure S3). This highlights the well-mixed water column in the coastal part of the transect unlike the stratified mixing zone in the channel.
Major taxa in the brackish stratified section
Bacillariophyceae were the major class in the mixing zone (stations B–C), but clear differences in taxa composition were detected between surface and depth (Figures 3 and S3).
In the brackish bottom water (Bb-Cb) derived from the bay and moving landward in the Vire River channel, cell abundances increased sharply (Figure 2C). This formed a dramatic phytoplankton accumulation in the bottom water of station B (silt plug) predominated by Asterionellopsis glacialis (76%–81% of the total phytoplankton, Figure 3B) with more than 115×103 cells mL−1, i.e. 100 to 200-fold higher than in the bay.
The surface brackish layer Bs-Cs (ESTUARY), influenced by the upstream river, showed the highest phytoplankton diversity observed along the transect (Figure 3A), reaching twice as many species than in the north of the bay (Bs vs. Fs). A large part of this diversity (up to 69%) comprised taxa occurring at low cell concentrations ( = rare taxa, <1% of total species abundance). The genus Cyclotella and associated small centric diatoms (designated here as Cyclotella-like) were predominant (57–76%, Figure 3B). Further SEM observations of this group led to the identification of a great diversity of morphologically similar small taxa (<10 μm for the most part), not identifiable under the light microscope, e.g.: Cyclotella meneghiniana, Discostella sp., Cyclostephanos dubius, Stephanodiscus minutulus, S. hantzschii, Thalassiosira guillardii. Presence of green algae from the class Chlorophyceae was significant (3–7.5% of total abundance), including the genera Monoraphidium, Scenedesmus and Chlamydomonas.
According to the CA ordination (Figure S3), Cs (ESTUARY) had an intermediate place on the diagram with no typically occurring species, while Bs plotted close to the freshwater sample As, with chlorophyte species and freshwater diatoms as most typical taxa.
Major taxa in the freshwater section
The Cyclotella-like group was predominant in the freshwater station A = VIRE River (63% of total abundance). Other noteworthy diatoms were the freshwater species Nitzschia acicularis (7%) and some unidentified Naviculaceae (6%). Compared to the other stations, chlorophytes were most abundant (20.8% of total abundance), including members of the Chlorophyceae (12%) and Trebouxiophyceae (9%).
Other algal groups
The other taxa identified, including dinoflagellates (4 taxa), haptophytes (2), chrysophytes (2), and euglenophytes (1), were much less abundant along the transect (<1% of total abundance).
3. Clone libraries analysis
The following three eukaryotic rDNA clone libraries were constructed: the “VIRE River” library from the upstream freshwater sample As (1 psu), “ESTUARY” from the brackish sample Cs (8.5 psu), and “BAY” from the marine sample Es (33 psu). After removal of low-quality results, a total of 287 partial microeukaryotic sequences were obtained. Clustering of sequences based on a 98% similarity level revealed a total of 67 different operational taxonomic units (OTUs, Table 1), with 37, 13 and 26 OTUs for VIRE River, ESTUARY and BAY respectively. Rarefaction curves computed for each library (to obtain an estimate of phylotype diversity relative to sampling effort, i.e. number of clones sequenced) never reached saturation (Figure S4). However, a comparison between the curves showed that higher microeukaryotic diversity was found in the freshwater and marine samples rather than in the brackish one. The non-parametric richness estimator Chao1 yielded values of 60, 27 and 61 OTUs for VIRE River, ESTUARY and BAY respectively. Therefore, the recovered OTUs represented only 43 to 62% of the estimated OTU richness (Chao1), meaning that an important part of the genetic protistan diversity remains unsampled. The ESTUARY library was more substantially undersampled because of the strong predominance of an OTU affiliated to Thalassiosira guillardii, which contained 68 sequences out of the 101 obtained.
Table 1. OTUs recovered in this study (clustering at a 98% similarity threshold).
| Distribution of clones | ||||||||||||
| OTU | Division | Class/or other group | Closest match | Accession | Identity (%) | Origin (for uncultured clones) | Closest cultured match | Identity (%) | Total | VIRE | ESTUARY | BAY |
| V99 | Stramenopiles | Chrysophyceae | Mallomonas portae-ferreae | GU935618 | 99.6 | 1 | 1 | 0 | 0 | |||
| B83 | Uncult. eukaryote clone LG47-07 | AY919816 | 99.8 | Lake George, oligotrophic (USA) | Paraphysomonas foraminifera | 99.4 | 4 | 1 | 0 | 3 | ||
| V56 | Uncult. eukaryote clone AMT15_15_10m | GQ863816 | 98.8 | Upwelling region, Atlantic (Canaries) | Paraphysomonas butcheri | 98.7 | 1 | 1 | 0 | 0 | ||
| V09 | Uncult. stramenopile clone MLB53.161 | EU143922 | 97.4 | Lake Taihu, subtropical (China) | Uroglena sp. | 94.4 | 1 | 1 | 0 | 0 | ||
| V92 | Uncult. eukaryote clone RW2_2011 | AB721074 | 96.5 | Freshwater, Kiryu (Japan) | Uroglena sp. | 96.1 | 2 | 2 | 0 | 0 | ||
| V69 | Spumella-like flagellate (isolate JBAS36) | AY651079 | 99.6 | Freshwater, Nag Pokhari (Nepal) | Pedospumella encystans | 99 | 2 | 2 | 0 | 0 | ||
| V78 | Spumella-like flagellate (isolate JBC27) | AY651093 | 93.3 | Small pond, Huqiu (China) | Chrysosphaera parvula | 90.9 | 2 | 2 | 0 | 0 | ||
| V97 | Uncult. eukaryote clone PG5.22 | AY642735 | 97.5 | Lake Godivelle, oligotrophic (France) | Paraphysomonas imperforata | 95.1 | 1 | 1 | 0 | 0 | ||
| V145 | Uncult. eukaryote clone Ch8A2mF4 | JF730784 | 99.2 | Char Lake (Arctic) | Ochromonas tuberculata | 95.5 | 13 | 13 | 0 | 0 | ||
| V81 | Uncult. eukaryote clone WH8eA1 | JF730838 | 99.8 | Ward hunt Lake (Arctic) | Ochromonas tuberculata | 96.4 | 1 | 1 | 0 | 0 | ||
| B18 | Uncult eukaryote clone CYSGM-7 | AB275090 | 98 | Marine sediment, methane seep (Japan) | Kephyrion sp. | 96.7 | 21 | 0 | 1 | 20 | ||
| B59 | Uncult. eukaryote clone Q2B03N10 | EF172974 | 97.6 | Sargasso Sea (25m depth) | Synura curtispina | 91 | 1 | 0 | 0 | 1 | ||
| Es64 | Bacillariophyceae | Thalassiosira guillardii | AF374478 | 99.7 | 68 | 0 | 68 | 0 | ||||
| Es37 | Thalassiosira pseudonana | DQ093367 | 99.8 | 4 | 0 | 4 | 0 | |||||
| V141 | Cyclotella meneghinia | AY496207 | 100 | 6 | 6 | 0 | 0 | |||||
| V105 | Stephanodiscus hantzschii | DQ514914 | 99.8 | 26 | 10 | 16 | 0 | |||||
| V38 | Discostella pseudostelligera | DQ514905 | 99.8 | 10 | 7 | 3 | 0 | |||||
| Es86 | Thalassiosira nordenskioeldii | DQ514886 | 99.9 | 1 | 0 | 1 | 0 | |||||
| Es109 | Diatoma tenuis var. elongatus | EF423403 | 99.9 | 1 | 0 | 1 | 0 | |||||
| Es123 | Asterionellopsis glacialis | X77701 | 99.8 | 1 | 0 | 1 | 0 | |||||
| Es110 | Navicula phyllepta | EU938308 | 100 | 1 | 0 | 1 | 0 | |||||
| V154 | Navicula gregaria | HM805037 | 99.9 | 1 | 1 | 0 | 0 | |||||
| V163 | Surirella brebissoni | AJ867029 | 99.7 | 1 | 1 | 0 | 0 | |||||
| B128 | Rhizosolenia imbricata | AY485510 | 100 | 7 | 0 | 0 | 7 | |||||
| B140 | Bolidophyceae | Uncult. eukaryote clone BLACKSEA_48 | HM749950 | 99.5 | Southern Black Sea | Bolidomonas pacifica | 95.7 | 1 | 0 | 0 | 1 | |
| V16 | Bicosoecids | Uncult. eukaryote clone P34.6 | AY642710 | 98.1 | Lake Pavin, oligomesotrophic (France) | Adriamonas peritocrescens | 93.4 | 1 | 1 | 0 | 0 | |
| V44 | MAST-2 | Uncult. eukaryote clone W8eD9 | JF730854 | 99.9 | Ward hunt Lake (Arctic) | Pirsonia verrucosa | 88.5 | 1 | 1 | 0 | 0 | |
| B110 | MAST-4 | Uncult. eukaryote clone H3S8Ae5 | JQ781882 | 100 | North pacific ocean | Pirsonia verrucosa | 88.9 | 1 | 0 | 0 | 1 | |
| B75 | MAST-12 | Uncult. eukaryote clone BAQA21 | AF372755 | 96 | Marine anoxic sediments, Berkeley (USA) | Oblongichytrium sp. | 84.1 | 1 | 0 | 0 | 1 | |
| V122 | Cryptophyta | (nucleus) | Cryptomonas curvata | AM051189 | 99.9 | 21 | 21 | 0 | 0 | |||
| V83 | Cryptomonas pyrenoidifera/C. marssonii | EU163587/AM051192 | 99.7 | 1 | 1 | 0 | 0 | |||||
| V42 | Cryptomonas tetrapyrenoidosa/C. ovata | AB240954/AF508270 | 100 | 3 | 3 | 0 | 0 | |||||
| V151 | Cryptomonas borealis/C. ovata | AM051188/AB240952 | 93.9 | 1 | 1 | 0 | 0 | |||||
| V23 | Uncult. Cryptophyte, clone STFeb_146 | HM135081 | 99.9 | Lake Stechlin, oligotrophic (Germany) | Teleaulax amphioxeia | 98.7 | 1 | 1 | 0 | 0 | ||
| V113 | Uncult. eukaryote, clone D7 | FN263278 | 100 | Southern Baltic sea | Geminigera cryophila | 99.6 | 6 | 1 | 1 | 4 | ||
| B67 | Teleaulax acuta | AB471786 | 100 | 3 | 0 | 0 | 3 | |||||
| B147 | Cryptophyceae sp. | GQ375265 | 99.7 | Northern Baffin Bay (Arctic ocean) | Falcomonas daucoides | 98.6 | 3 | 0 | 0 | 3 | ||
| B96 | (nucleomorph) | Falcomonas daucoides | AJ420689 | 86.2 | 2 | 0 | 0 | 2 | ||||
| B23 | Haptophyta | Prymnesiophyceae | Haptolina hirta | AJ246272 | 98.7 | 1 | 0 | 0 | 1 | |||
| B121 | Phaeocystis globosa | AF182112 | 99.6 | 1 | 0 | 0 | 1 | |||||
| B01 | Picobiliphytes | Uncult. eukaryote clone RA000907 | DQ222877 | 99.2 | English Channel | 1 | 0 | 0 | 1 | |||
| B107 | Centrohelids | Pterocystis sp. unidentified helio 5 | AY749610 | 95.6 | Okareka Lake (New Zealand) | Heterophrys myriopoda | 94.9 | 1 | 0 | 0 | 1 | |
| V33 | Alveolata | Dinophyceae | Gymnodinium eucyaneum | JQ639760 | 99.8 | 1 | 1 | 0 | 0 | |||
| B131 | Gyrodinium spirale | AB120001 | 99.7 | 8 | 0 | 0 | 8 | |||||
| B49 | Uncult. eukaryote clone M1_18B06 | DQ103837 | 99.5 | The anoxic Mariager Fjord (Denmark) | Peridinium umbonatum | 93.4 | 5 | 0 | 0 | 5 | ||
| B115 | Uncult eukaryote clone SA1_4B9 | EF527151 | 99.4 | The anoxic Framvaren Fjord (Norway) | Euduboscquella sp. | 96.2 | 1 | 0 | 0 | 1 | ||
| V98 | Ciliophora | Uncult. eukaryote clone VNP11 | DQ409125 | 99.4 | Lacustrine reservoir, hyper-eutrophic (France) | Strobilidium caudatum | 90.1 | 2 | 2 | 0 | 0 | |
| V84 | Uncult. alveolate clone 1-D5 | FN689891 | 99.8 | Gulf of Finland (Finland) | Tintinnidium sp. 1 | 90.5 | 1 | 1 | 0 | 0 | ||
| V85 | Uncult. ciliate clone AY2009D10 | HQ219435 | 100 | Lake Aydat, eutrophic (France) | Strobilidium caudatum | 90 | 1 | 1 | 0 | 0 | ||
| V61 | Uncult. eukaryote clone VNP38 | DQ409135 | 100 | Lacustrine reservoir, hyper-eutrophic (France) | Pseudochilodonopsis fluviatilis | 93.2 | 2 | 2 | 0 | 0 | ||
| B116 | Tintinnopsis rapa (isolate 241) | JN831834 | 99.4 | 2 | 0 | 0 | 2 | |||||
| V127 | Chlorophyta | Chlorophyceae | Chlamydomonas noctigama | JN903979 | 99.7 | 2 | 2 | 0 | 0 | |||
| V102 | Uncult. eukaryote clone KRL01E11 | JN090871 | 100 | Lake Karla (Greece) | Chlamydomonas kuwadae | 99.4 | 7 | 5 | 1 | 1 | ||
| V140 | Neochlorosarcina negevensis | AB218715 | 99.1 | 1 | 1 | 0 | 0 | |||||
| V146 | Mychonastes huancayensi/M. jurisii | GQ477050/GQ477038 | 99.9 | 2 | 2 | 0 | 0 | |||||
| V131 | Atractomorpha echinata | AF302772 | 100 | 1 | 1 | 0 | 0 | |||||
| Es74 | Prasinophyceae | Nephroselmis olivacea | X74754 | 98,1 | 1 | 0 | 1 | 0 | ||||
| B125 | Uncult Crustomastix, clone PROSOPE.CM-5m. | EU143398 | 96.9 | Mediterranean Sea | Crustomastix stigmatica (pras1) | 94.6 | 1 | 0 | 0 | 1 | ||
| B129 | Micromonas pusilla strain | AY955010 | 99.7 | 1 | 0 | 0 | 1 | |||||
| V05 | Fungi | Chytridiomycota | Uncult. eukaryote clone B86-172 | EF196796 | 98.2 | Freshwater, Alpen (France) | Spizellomyces pseudodichotomus | 94.4 | 1 | 1 | 0 | 0 |
| Es32 | Uncult. Chytridiomycota clone PFD5SP2005 | EU162640 | 99.6 | Lake Pavin, oligomesotrophic (France) | Kappamyces laurelensi | 94.8 | 9 | 7 | 2 | 0 | ||
| V31 | Environmental clade LKM | Uncult. eukaryote clone E-C4_1 | HM628660 | 99.7 | Slow sand filter biofilm | Mortierella microzygospora | 87.1 | 2 | 2 | 0 | 0 | |
| V136 | Unknown lineage | Uncult. fungus clone C10 | JN054676 | 100 | Wastewater treatment plant | Rhizophlyctis rosea | 87.5 | 1 | 1 | 0 | 0 | |
| B64 | Cercozoa | Phytomyxea | Uncult. eukaryote clone TAGIRI-5 | AB191413 | 96.3 | Anoxic marine sediment (Japan) | Phagomyxa bellerocheae | 87.8 | 1 | 0 | 0 | 1 |
| B65 | Filosoa | Uncult. eukaryote clone 9_149 | EU545751 | 99.8 | Marine sediment, East Sea | Botuliforma benthica | 95,4 | 1 | 0 | 0 | 1 | |
| B87 | Uncult. eukaryote clone 9_25 | EU087251 | 100 | Marine sediment, East Sea | Protaspis oviformis | 98,8 | 3 | 0 | 0 | 3 | ||
| B144 | Thaumatomastix sp. | GQ144681 | 100 | Boundary Bay (Canada) | Thaumatomonas coloniensi | 94.4 | 1 | 0 | 0 | 1 | ||
A representative clone for each OTU and its phylogenetic affiliation are provided. Highest BLAST match and closest cultured organisms from GenBank are given with accession numbers and percentage similarity. The remaining columns indicate the distribution and the number of clones found in each clone library (VIRE River, ESTUARY, and BAY).
Among the 287 sequences identified, up to 94% (falling in 56 OTUs) had at least 98% similarity with known sequences based on BLAST analysis, and were widely distributed across the major eukaryotic lineages. The following first-rank taxa were represented: stramenopiles, Cryptophyceae, Haptophyta, Alveolata, Chlorophyta, Fungi, Cercozoa and two subgroups of eukaryote incertae sedis (centrohelids and picobiliphytes) (Table 1). However, 30 out of the 68 total OTUs had less than 98% similarity with named and cultured organisms, meaning that sizeable fractions of protistan groups in the environment still remain to be sequenced and characterized. Most of these clones belonged to heterotrophic representatives (e.g. Fungi, Ciliates, Cercozoa, some Dinophyceae and stramenopiles). Overall, the heterotrophic/mixotrophic OTUs accounted for 44% of all OTUs.
Total sequences were dominated by stramenopiles [accounting for 182 clones (64%) falling in 29 OTUs (43%)], followed by alveolates (9% of all clones in 9 OTUs), cryptophytes (14% of all clones in 8 OTUs) and chlorophytes (<6% of all clones in 8 OTUs). The other taxonomic groups were less abundant in the libraries, each accounting for less than 5 OTUs.
Stramenopiles. The 29 stramenopiles OTUs retrieved from our study were distributed across four major lineages (Table 1). The photosynthetic groups included the diatoms (Bacillariophyceae, 12 OTUs) detected in all libraries but predominant in the estuarine sample (74% of all diatom sequences, 8 OTUs), and the picoplanktonic class Bolidophyceae as a singleton (i.e. an OTU containing only one sequence) in the bay. All the diatom sequences showed high similarity with cultivated strains (99.7–100%) and were for the most part affiliated with genera also identified by microscopy in this study. Half of the diatom OTUs (6) belonged to thalassiosiroid lineages found in the Vire channel (3 OTUs in ESTUARY, 1 OTU in VIRE River, and two present in both libraries), including several Stephanodiscaceae (Cyclotella meneghiniana, Discostella pseudostelligera, Stephanodiscus hantzschii) and small representatives of Thalassiosiraceae (e.g. T. pseudonana, T. guillardii). Thalassiosira guillardii was the predominant OTU in ESTUARY (67% of total clones) but not detected in the other libraries. Only one diatom OTU was found in BAY and belonged to Rhizosolenia imbricata, which was the most abundant diatom species detected by microscopy in this part of the bay.
Non-photosynthetic groups of stramenopiles were represented by singletons belonging to Bicosoecida and three lineages of uncultured novel Marine Stramenopiles (MASTs). MAST-2 is a clade hitherto known from exclusively marine waters, found in diverse oceanic and coastal areas [40], [41]. However, our analysis showed that the clone V44 from VIRE River, together with two other freshwater clones (99.9% similarity) recovered from an Arctic lake (W8eD9, [42]) and an oligotrophic temperate lake (STFeb_251, [43]), clustered with high bootstrap values with marine MAST-2 sequences (Figure S5). Clone B75 from the bay was positioned within the radiation of MAST-12, which is mainly composed of sequences from oxygen-depleted habitats [44], and the clone B110 belonged to the MAST-4 lineage known to be widely distributed in temperate samples [45].
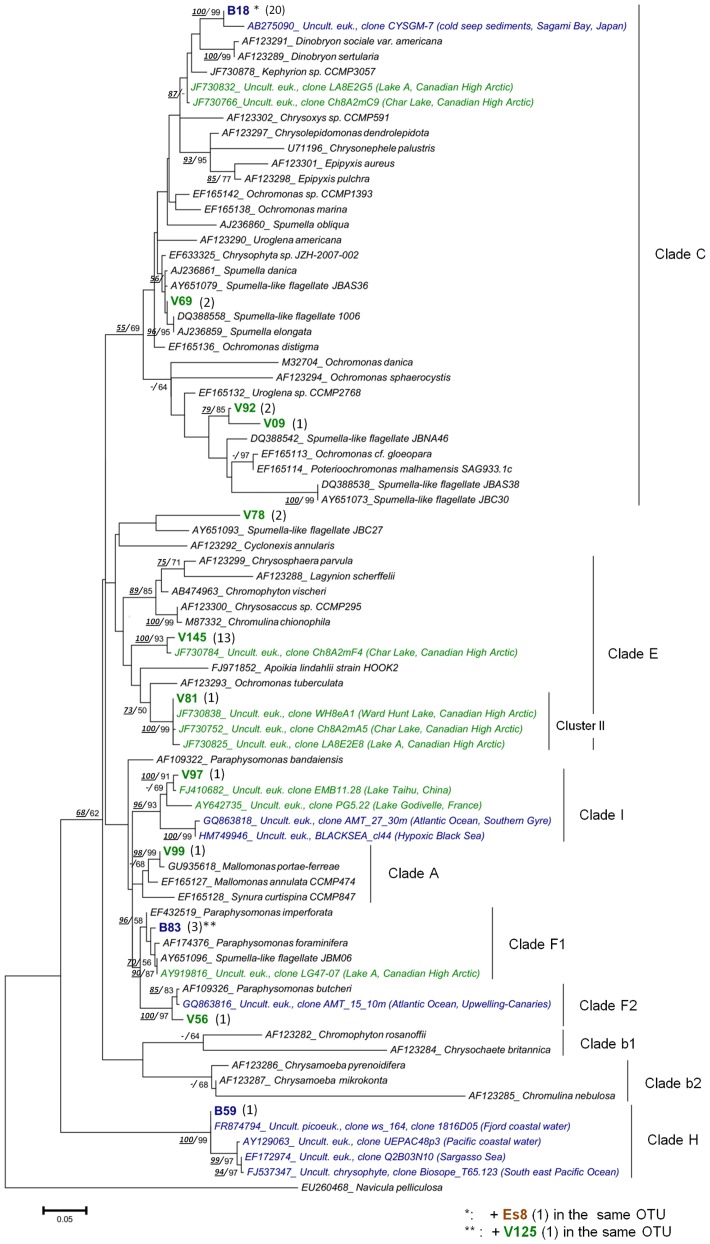
The chrysophytes sensu lato (i.e. including Synurophyceae) were represented by 50 clones falling in 12 OTUs, and were therefore, with the diatoms, the most diverse lineage detected in this molecular survey (Table 1). Most of these OTUs (10) were recovered from VIRE River, while 3 OTUs were detected in BAY, and only one in ESTUARY. Phylogenetic analyses placed these sequences in seven clades among the twelve recently proposed by del Campo & Massana [46] (Figure 4), with a tree topology in agreement with those previously described [46], [47]. An OTU from the freshwater library (V99) belonged to the clade A ( = Synurophyceae) containing photosynthetic organisms, and was closely related to Mallomonas portae-ferrae (99.6% similarity). Two OTUs (V56, B83) were closely related to cultured organisms of the heterotrophic genus Paraphysomonas (clades F1, F2), while four others OTUs were affiliated to clade C, but their internal position within the clade remains uncertain. Among the latter, clone B18 was representative of the predominant OTU (20 clones) in BAY library, which was moderately related to loricate organisms such as Kephyrion sp. [42] (96.7% similarity), and the riverine clone V69 was highly similar to non-green ‘Spumella-like’ strains, all isolated from soil (99.6% similarity with JBAS36 [48]). Three OTUs (V81, V97, and B59) belonged with high bootstrap support (96–100%) to three lineages (cluster II, clade I, clade H) containing only environmental sequences to which they were closely related (closest matches: 97.5–99.8%). Among them, the freshwater OTU V81 fell into the cluster II containing sequences solely recovered from high Arctic lakes [42]. The clone V97 together with other sequences from freshwater systems (subtropical or oligotrophic lakes) clustered with high support with marine sequences (Black Sea, Atlantic Ocean) previously described as members of the clade I [46] or ‘Marine B’ [49]. This clade was until now defined as exclusively marine; however, the present phylogenetic analysis showed that it actually contains both freshwater and marine representatives.
Figure 4. Maximum likelihood (ML) tree showing the position of the Chrysophyte OTUs.
OTUs were obtained from the VIRE River (V), the ESTUARY (Es) and the BAY (B) clone libraries. Tree construction was based on an alignment of 79 partial sequences (ca 500 align positions). The diatom Navicula pelliculosa was used as outgroup. The number of clones per OTU is indicated in brackets. Sequences from cultured taxa appear in black and environmental sequences in green (freshwater), blue (marine) or brown (brackish/estuary). Bootstrap values (>50%) obtained from the neighbor-joining tree and those from ML tree are indicated (NJ /ML).
Alveolates
Nine OTUs were affiliated to alveolates but no sequences were found in the ESTUARY library (Table 1). Dinophyceae were represented by 4 OTUs, among which two were closely related to the cultivated and known species Gymnodinium eucyaneum, Gyrodinium spirale. The two other dinoflagellate OTUs (B49, B115, Table 1) showed rather high similarities with uncultured clones from anoxic fjord waters [10], [50]. Among them, B115 was related to the syndinean Euduboscquella sp known to be a parasite of tintinnids [51].
Cryptophyceae
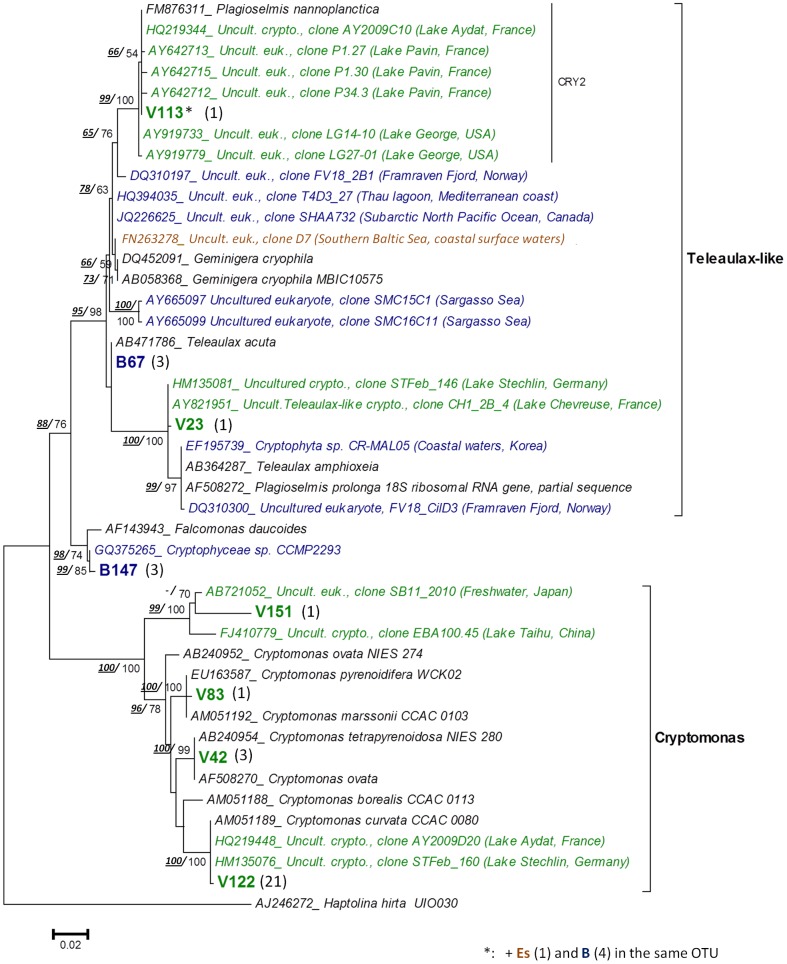
A total of 8 OTUs were affiliated with nuclear or nucleomorph cryptophyte sequences, and distributed in three major lineages within the order Cryptomonadales (Figure 5, Table 1). Four OTUs recovered from the Vire River were closely related to Cryptomonas species, e.g. C. ovata and C. curvata; (99.7–100% similarity) and rather moderately C. borealis (93.9% similarity). An OTU from the river (V23) along with one unique to BAY library (B147) and another present in all clone libraries (representative: V113), belonged to the Teleaulax-like cluster (bootstrap support 98%, Figure 5) that includes Geminigera and Plagioselmis genera lineages [52]. Another OTU from the bay (B147) was closely related to the species Falcomonas daucoides (98.6%).
Figure 5. Maximum likelihood (ML) tree showing the position of the Cryptophyte OTUs (order Cryptomonadales).
Tree construction was based on an alignment of 44 partial sequences (ca 650 align positions). The haptophyte Haptolina hirta was used as out-group. (See legend of Figure 4 for details).
Chlorophyta
Most of the chlorophyte sequences (Table 1) were retrieved from the VIRE River library (11 clones) and were affiliated to the class Chlorophyceae, falling in 5 OTUs with high similarity to known genera (from 99 to 100%). They corresponded either to small sized coccoid species such as Mychonastes sp. and Neochlorosarcina negevensis, or to the flagellated Chlamydomonas species. The class Prasinophyceae was detected as singletons in the brackish and marine libraries, including clones highly similar to Micromonas pusilla, Nephroselmis oliveacea, and to an environmental clone of the Crustomastix lineage [34].
Other groups
Among the remaining OTUs, two singletons from the BAY library were affiliated to the haptophytes (Table 1), both related to members of the Prymnesiophyceae (Phaeocystis globosa, Haptolina sp.). Finally, the marine picoplanktonic group Picobiliphytes was represented by an OTU in the bay (B01), highly similar to an environmental clone (99.2% similarity with RA000907.54) recovered from the English Channel [53].
4. Molecular versus Morphological diversity
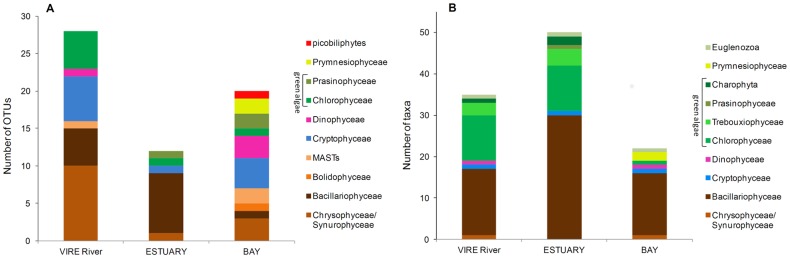
To compare diversity estimates by microscopical and molecular methods, sequences from exclusively heterotrophic lineages were removed from analysis (e.g. Fungi, Cercozoa, and Ciliates). Taxa richness estimated with both approaches showed substantially different trends between the three samples (Figure 6). Microscopic analyses (LM+SEM) revealed a much higher taxonomic richness in the ESTUARY sample, whereas, conversely, number of OTUs found in ESTUARY was lower than in both VIRE River and BAY libraries.
Figure 6. Phytoplankton taxonomic composition (taxa richness) in three surface samples: Molecular vs. Morphological approaches.
VIRE River (station A), ESTUARY (station C) and BAY (station E). (A) Number of OTUs according to the taxonomic affiliation of the 18S rRNA gene sequences. (B) Number of taxa identified by microscopy (LM + SEM).
Taxa composition of the different communities at a high taxonomy level (class/division, Figure 6) showed differences between the two approaches. The clone library analysis identified a greater diversity within stramenopiles, including lineages of nano- and/or pico-size flagellates (Chrysophyceae, MASTs, Bolidophyceae). Moreover, in spite of being identified by microscopy, a greater diversity within Cryptophyceae and Dinophyceae was also recovered with the molecular survey. On the other hand, diatoms and green algae were better detected with the morphological approach.
Only three OTUs were shared between the clone libraries VIRE River and ESTUARY (Table 1), and one between ESTUARY and BAY, reflecting a low similarity between communities (Jaccard index: 11% and 10% respectively). The microscopic survey indicated that 21 taxa were shared between VIRE River and ESTUARY (floristic list, Table S1) with a Jaccard's similarity of 39%, and 8 taxa were found in both samples ESTUARY/BAY resulting in a 18% similarity. The morphological survey therefore revealed higher similarity between both sample combinations than the 18S rDNA clone library approach did.
Discussion
Our taxonomic investigation along the Vire River estuary and in the Baie des Veys took place just at the beginning of the diatom spring bloom, which usually occurs every year in the bay [18]. Physicochemical parameters and chl a concentrations estimated within the bay were consistent with previous studies carried out in the same season (for more details: [17]–[19]). Although we studied the estuarine continuum at a single date, by using two contrasting but complementing methods we enhanced our assessment of the global microeukaryote diversity in this site hitherto considered a relatively well-known ecosystem.
1. Community structure and diversity along the macrotidal Vire estuarine gradient in early spring bloom
Examination of species assemblages (see Figures 3 and S3) suggested a gradual change of the phytoplankton composition along the estuarine continuum influenced by salinity and tidal force. The latter involves a stratification of the water column at high tide, with significant vertical differences both in hydrographic and biological data. This two-layer circulation system characteristic of this macrotidal estuary has been previously introduced byJouenne et al. [18]. At the time of sampling, water stratification within the mixing zone of the channel (stations B–C) was particularly pronounced because the Vire River discharge was low (<½ the mean annual discharge) compared to the tidal current (spring tide). Salinity thus increased rapidly in the freshwater surface layer, while seawater intrusion into the bottom layer extended upstream over 7 km from the mouth section (Figure 2). A spectacular phytoplankton bloom (>100×103 cells mL−1) occurred in the most upstream brackish reach of the bottom seawater (station B bottom – 24 psu), with cell abundances among the highest reported in estuaries [54], [55]. This bottom zone of the estuarine gradient corresponds to the estuarine turbidity maximum (ETM) or “silt plug”, characterized by high organic matter accumulation resulting in strong light-limitation [15]. Presence of an ETM is reported here for the first time in the Vire River estuary.
During our study, three main communities and their associated water mass overlapped each other, longitudinally and vertically along the estuarine continuum.
Within the “coastal” community in the well-mixed water column of the Baie des Veys, diatoms were one of the dominant groups, as commonly found throughout the year [18]. However, we report here for the first time that the nanoplanktonic group Teleaulax/Plagioselmis-like (Cryptophyceae) was preponderant in this area. According to the clone library content, two different OTUs belonging to the Teleaulax-like cluster probably correspond to the taxa enumerated, including one closely related to Teleaulax acuta. Other sequences however corresponded to Falcomonas daucoides, a species with cells superficially resembling Plagioselmis and Telelaulax when observed by light microscopy, due to a similar acute posterior end [52]. Given this similarity, individuals of F. daucoides were probably included in the cell count of the Teleaulax-like group, although morphological differences could be revealed with electron microscopy [56]. This illustrates the limits of the quantitative approach with light microscopy that may overlook an important part of the overall diversity [43], [57]. It also reflects the need to further investigate this cryptophyte group, whose importance and recurrence as part of marine/estuarine communities has been recently emphasized (reports of the dominance of these flagellates is increasing[58]), but remains largely ignored due to difficulties in taxonomic identifications and suspicions of several phylogenetic misclassifications [59].
Shift in species composition occurred toward the river mouth and the channel, where the two-layer system in the mixing zone led to the presence of two different phytoplankton communities.
A “bottom-brackish” community was mainly composed of coastal euryhaline species transported at high tide by the tidal current into the bottom of the Vire channel.
The unexpected phytoplankton bloom restricted to the polyhaline bottom layer (24 psu) in the ETM zone was mainly composed of the euryhaline diatom Asterionellopsis glacialis (81% of the total phytoplankton), while typical marine species disappeared as they were advected upstream. Low irradiance caused by high turbidity in the silt plug can constrain development of phytoplankton and prevent it from using available nutrients [60], however, our microscopical observations certified that the A. glacialis bloom was composed of long chain-forming, physiologically healthy cells. It is likely that, given the transience of the slack high-tide (30 min-1h), the A.glacialis bloom will spend only a short time in darkness, thus providing a fresh stock of cells that may enrich the bay communities at ebb tide.
An “upper-layer” community occurred in the oligohaline (1psu) freshwater tidal reach, and spread to the downstream mesohaline part of the continuum (5–18 psu) depending on species distribution ranges. Taxa diversity was higher in this part of the estuary than in the silt plug and the bay, with freshwater green algae as important contributors in terms of species number and abundance. Small centric Cyclotella-like diatoms predominated this “upper-layer” community. More detailed examination of this group with SEM was consistent with clone library results, confirming the existence of a great number of small thalassiosiroid taxa (mostly <10 μm), with 8 different species at least that would have escaped detection without such a thorough analysis.
Presence of some lineages (e.g. the poorly known groups of MASTs and picobiliphytes) and diversity within groups of morphologically similar taxa (e.g. cryptophytes, small centric diatoms) were estimated for the first time in the Vire estuary. Among them, the Chrysophyceae were the most noticeable. Most of these flagellates have been recorded in freshwater systems, but in lakes especially [42], [46]; here, our molecular approach revealed a large diversity of these golden algae in the Vire River sample (10 OTUs), suggesting their ecological importance in dynamic and transitional freshwater ecosystems as well. Only three OTUs were found in the Baie des Veys, but one could be an abundant member of the bay's community (most represented OTU in the BAY library).
Taxa identified by our molecular approach were widely distributed throughout the chrysophyte clades [46], including autotrophic members (e.g. Mallomonas) but most OTUs were affiliated to colourless bacteriovorous taxa Paraphysomonas spp., Spumella sp., and unidentified ‘Spumella-like’ flagellates [48]. Based on traditional microscopy, only two chrysophytes were observed along the estuarine continuum: Mallomonas sp. and the mixotrophic Ollicola vangorii (the latter was identified using SEM). The discovery of sequences in the Vire estuary (river and bay) affiliated only with environmental sequences originating from geographically distant sites (e.g. Black Sea, Arctic lakes – [61], [42]) and developing in a variety of ecosystems (fresh waters, soils, marine, or extreme environments), confirm ubiquity of the Chrysophyceae and the need for further investigation.
2. Morphological and molecular approaches of in situ phytoplankton diversity: application to estuarine waters
The traditional microscopy approach has been used as a rule for ecological studies of phytoplankton assemblages in dynamic estuaries ecosystems [62], [63], being suitable to study morphologically distinctive species and certain higher-taxon rank protistan groups [64]. Morphological identifications are unfortunately often limited to taxa previously described by cultivation-based techniques, and assaying the whole protistan community using a single microscope-based method becomes very difficult. Due to their small size, many protists may remain undetected while others are not recognized due to their cryptic nature [48], [65]. This has been clearly shown in our study, with several taxa/lineages (e.g. within chrysophytes, cryptophytes, chlorophytes) absent from our floristic list, but revealed by our molecular approach. The whole community-targeting molecular strategy, applied to three representative surface samples of the estuarine gradient, was carried out to broaden our assessment of the overall protistan diversity. In the past decade, phylogenetic analyses of the eukaryotic 18S rRNA genes have successfully described protists in a variety of aquatic environments, although such methods also include limitations, being prone to multiple sources of biases and therefore not completely reliable [10], [66]–[68]. To our knowledge, this approach has rarely been used in transitional waters such as estuarine systems [14], [15].
While providing a deeper insight into diversity of microeukaryotic communities, combination of both morphological and molecular strategies also allowed us to determine the limitations and advantages associated with each method. Our results highlight several characteristics: i) presence of chlorophytes and diatoms was lower in clone libraries than estimated by microscopy, irrespective of the location along the estuarine gradient, ii) pico- and nano-size protists were a major component in the libraries, and iii) heterotrophic lineages were clearly enhanced in the molecular approach.
The chlorophytes and diatoms lacking in our clone libraries (e.g. Scenedesmus spp., Cylindrotheca closterium) correspond to “large” species with more distinguishable features than heterotrophic flagellates or picoplankton, which explains why they are well represented in morphological approach [57]. However, most of these “easily identifiable” taxa had low densities within communities, probably leading to dilution of their sequences among the total pool of template DNA and explaining their non-detection with PCR amplification. The molecular approach was rather helpful to improve diversity estimate of the above photosynthetic lineages by recovering their smallest members that escaped our microscopical observations. Consequently, sequences related to the pico-sized Micromonas pusilla and Crustomastix (Prasinophyceae) in the bay, and others belonging to small thalassiosiroid diatoms (<10 μm) in the Vire River, were identified for the first time in this area.
The plentifulness of heterotrophic/mixotrophic small eukaryotes in the Vire River and the Baie des Veys, as revealed by the clone libraries (44% of the total OTUs retrieved), probably indicates that these communities are a major component of the microbial food web with important ecological implications in this estuarine ecosystem. The extent of their diversity and trophic interactions with phototrophic estuarine components are still poorly known and require further investigations [69]. Nevertheless, our results also corroborate similar studies highlighting that approaches based on 18S rRNA gene clone libraries using universal primers are heavily biased toward heterotrophs (e.g. alveolates and stramenopiles), to the detriment of autotrophic organisms [49]. Here, the use of microscopy partially addressed this bias and revealed the high diversity among phototrophic taxa.
The lack of congruity between morphological and molecular analyses was especially evident in the estuarine brackish surface sample (sample Cs = ESTUARY library). While microscopical observations led to the identification of a broad phytoplanktonic diversity in the sample including many less abundant recognizable taxa (floristic list, Table S1), the OTUs richness recovered was low because of over-representation of Thalassiosira guillardi related clones. The predominance of this diatom in the sample associated with the restricted sampling effort (i.e. the limited number of clones sequenced) might explain the inefficiency of our PCR-based clone library strategy. As commonly reported in previous studies (including those based on larger data sets [10], [37]), our rarefaction analyses showed that none of the clone libraries was sufficiently large to reach saturation, and therefore part of the complete protist diversity was inevitably missed. In addition to this undersampling and as discussed earlier, the non-recovery (or underrepresentation) of some taxa in clone libraries can be explained by many underlying biases, such as PCR-primer incompatibilities [66], [67], competition for primers [70] because of large variations in 18S rRNA gene copy number among taxa [68], and variable cloning efficiency. Thereby, in contrast to the microscopy-based approach, quantification of taxa based on the number of OTUs obtained with the clone library strategy is not possible. Moreover, assigning OTUs to a specific taxon rank is difficult because sequence identities vary widely with taxa considered. As suggested by other authors [10], [37], [71], we chose a 98% similarity threshold to discriminate at the genus/species level. This cut-off level associated with only partial 18S rDNA sequences provided an image of the phytoplankton diversity obviously somewhat underestimated, but for several groups, taxa discrimination was finer than with conventional microscopy, e.g. among cryptophytes (at least 4 different Cryptomonas species and 3 Teleaulax/Plagioselmis-like genera detected in clone libraries were missed by microscopy). Measures of protistan diversity at genotype levels are therefore difficult to compare with traditional studies relying on morphology-based taxonomic ranks [10].
Despite limitations above-mentioned, both approaches tend to be rather congruent when considering the most abundant taxa in the studied samples, as most of them, identified and counted by microscopy, were also detected by our molecular approach ( = five of the six dominant taxa, e.g. A. glacialis, Teleaulax-like, many small thalassiosiroids, R. imbricata).
Finally, the combination of morphological and molecular approaches, both implying advantages but also limitations, were clearly complementary, providing access to greater protistan diversity than with a single method.
3. Conclusion
In the present study, the traditional microscopical approach allowed us to detail the changes in phytoplankton diversity occurring at a specific time along the entire estuarine gradient.
Additional 18S rDNA analysis as well as electron microscopy for three samples along the surface of the salinity gradient revealed significant additional diversity overlooked using only light microscopy and unnoticed during previous studies in the area. Several lineages such as the Chrysophyceae and Teleaulax-like taxa in the cryptophytes would need further characterization in the future, in terms of diversity extent (accurate identifications) and ecological implications along the estuarine continuum.
As in previous studies [23], [43], [57], our results confirm that both approaches – whether morphological or molecular – are complementary and that whatever the method, only a fraction of the whole phytoplankton community will be captured. Such combination of methods appears to be fully relevant to investigate more thoroughly the diversity of phytoplankton communities that can occur in complex ecosystems such as the transitional waters. This provides an overview of the ‘hidden’ diversity that would have escaped detection without such in-depth analysis. Our study also suggests that a significant part of the eukaryotic microbial diversity still remains to be uncovered, even in temperate estuarine waters yet considered as relatively well-known. For a molecular approach on typical estuarine samples with high diversity of low abundant taxa or predominance of few species, we suggest the use of a multi-primer PCR strategy and high-throughput sequencing, to increase the probability of detecting a broad variety of taxa and to avoid undersampling due to overrepresentation of some OTUs.
Supporting Information
Profiles of nutrient concentrations along the estuarine continuum. Silicate, phosphate and nitrate/nitrite were analyzed in the samples collected in the water column. (A logarithmic scale was used for the data representation).
(PDF)
Principal component analysis (PCA) of environmental parameters. Samples (dots) and variables (green lines) are displayed for the first two axes. Blue dots: surface samples and red dots: samples of the near-bottom water layer.
(TIFF)
Correspondence analysis (CA) based on phytoplankton composition (relative-abundance matrix). Taxa considered in the analysis accounted for >1% of total phytoplankton in at least one sample. Blue dots: surface samples and red dots: bottom samples. *Freshwater diatoms included the following pennates: Asterionella formosa, Nitzschia acicularis, and undetermined Naviculaceae. *Chlorophytes included Chlorophyceae (Scenedesmus spp., Monoraphidium contortum, Chlamydomonas sp.) and Trebouxiophyceae (Micractinium sp., Dictyosphaerium pulchellum).
(DOC)
Rarefaction curves determined for the three 18S rRNA gene libraries (VIRE River, ESTUARY and BAY). Curves were constructed at 98% sequence similarity cut-off value.
(TIFF)
Maximum likelihood (ML) tree showing the position of the non-photosynthetic stramenopiles OTUs (Bicosoecida, MASTs). OTUs were obtained from the the VIRE River (V), the ESTUARY (Es) and the BAY (B) clone libraries. Tree construction was based on an alignment of 96 partial sequences (ca 490 align positions). Number of clones per OTU is indicated in brackets. Sequences from cultured taxa appear in black, and environmental sequences in green (freshwater), blue (marine) or in brown (brackish/estuary). Bootstrap values (>50%) are indicated. The Dinophyceae Gyrodinium fusiforme and Peridinium umbonatum were used as outgroup.
(TIF)
Phytoplankton taxa identified by microscsopy. Samples corresponded to surface (As → Fs) and bottom (Bb → Eb) waters. “(SEM)” refers to additional taxa identified in the surface samples As, Cs and Es (also selected for clone library analysis, referred to as VIRE River, ESTUARY and BAY) using scanning electron microscopy.
(DOC)
Acknowledgments
The authors wish to thank Frédéric Guyot (Centre de Recherche en Environnement Côtier, UCBN, Luc-sur-mer) for logistical support during the cruise; Antoine Mabire, Sophie Picot, Florien Mauduit, and Hugo Lebredonchel for their help in the field. We are very grateful to IFREMER Port-en-Bessin (particularly Olivier Pierre-Duplessix) for the loan of equipment and the nutrient measurements, Pierre Bazin for his contribution to the construction of clone libraries, Gabriele Curdt-Hollmann (EPSAG, Georg-August-Universität Göttingen, Germany) for DNA sequencing, and Didier Goux (CMABio, UCBN) for the high-quality analyses using SEM. We also thank Drs. Christophe Lelong and Guillaume Rivière for their help and advice on the molecular approach; and Pr. Chantal Billard for reviewing the English language of the manuscript.
Funding Statement
This work was funded by the University of Caen Basse-Normandie. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. McLusky DS (1993) Marine and estuarine gradients — An overview. Netherlands Journal of Aquatic Ecology 27: 489–493. [Google Scholar]
- 2.Pritchard DW (1989) Estuarine classification - a help or a hindrance. In: Neilson BJ, Kuo A, Brubaker J, editors. Estuarine Circulation. Clifton, NJ: Humana Press. pp. 1–38.
- 3. Trigueros JM, Orive E (2000) Tidally driven distribution of phytoplankton blooms in a shallow, macrotidal estuary. J Plankton Res 22: 969–986. [Google Scholar]
- 4. Azam F, Fenchel T, Field JG, Gray JS, Meyer-Reil LA, et al. (1983) The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 10: 257–263. [Google Scholar]
- 5. Díez B, Pedrós-Alió C, Massana R (2001) Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol 67: 2932–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moon-van der Staay SY, De Wachter R, Vaulot D (2001) Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409: 607–610. [DOI] [PubMed] [Google Scholar]
- 7. Edgcomb VP, Kysela DT, Teske A, De Vera Gomez A, Sogin ML (2002) Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc Natl Acad Sci U S A 99: 7658–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Massana R, Balagué V, Guillou L, Pedrós-Alió C (2004) Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiol Ecol 50: 231–243. [DOI] [PubMed] [Google Scholar]
- 9. Šlapeta J, Moreira D, López-García P (2005) The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc Biol Sci/The Royal Society 272: 2073–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Behnke A, Barger KJ, Bunge J, Stoeck T (2010) Spatio-temporal variations in protistan communities along an O2/H2S gradient in the anoxic Framvaren Fjord (Norway). FEMS Microbiol Ecol 72: 89–102. [DOI] [PubMed] [Google Scholar]
- 11. Moreira D, López-García P (2002) The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol 10: 31–38. [DOI] [PubMed] [Google Scholar]
- 12. Muylaert K, Sabbe K, Vyverman W (2009) Changes in phytoplankton diversity and community composition along the salinity gradient of the Schelde estuary (Belgium/The Netherlands). Estuar Coast Shelf Sci 82: 335–340. [Google Scholar]
- 13. Lionard M, Muylaert K, Hanoutti A, Maris T, Tackx M, et al. (2008) Inter-annual variability in phytoplankton summer blooms in the freshwater tidal reaches of the Schelde estuary (Belgium). Estuar Coast Shelf Sci 79: 694–700. [Google Scholar]
- 14. Vigil P, Countway P, Rose J, Lonsdale D, Gobler C, et al. (2009) Rapid shifts in dominant taxa among microbial eukaryotes in estuarine ecosystems. Aquat Microb Ecol 54: 83–100. [Google Scholar]
- 15. Herfort L, Peterson TD, McCue L, Zuber P (2011) Protist 18S rRNA gene sequence analysis reveals multiple sources of organic matter contributing to turbidity maxima of the Columbia River estuary. Mar Ecol Prog Ser 438: 19–31. [Google Scholar]
- 16. Klein C, Claquin P, Bouchart V, Le Roy B, Véron B (2010) Dynamics of Pseudo-nitzschia spp. and domoic acid production in a macrotidal ecosystem of the Eastern English Channel (Normandy, France). Harmful Algae 9: 218–226. [Google Scholar]
- 17. Ubertini M, Lefebvre S, Gangnery A, Grangeré K, Le Gendre R, et al. (2012) Spatial variability of benthic-pelagic coupling in an estuary ecosystem: consequences for microphytobenthos resuspension phenomenon. PloS ONE 7(8): e44155 doi:10.1371/journal.pone.0044155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jouenne F, Lefebvre S, Véron B, Lagadeuc Y (2007) Phytoplankton community structure and primary production in small intertidal estuarine-bay ecosystem (eastern English Channel, France). Marine Biology 151: 805–825. [Google Scholar]
- 19. Jouenne F, Lefebvre S, Véron B, Lagadeuc Y (2005) Biological and physicochemical factors controlling short-term variability in phytoplankton primary production and photosynthetic parameters in a macrotidal ecosystem (eastern English Channel). Estuar Coast Shelf Sci 65: 421–439. [Google Scholar]
- 20. Desprez M, Ducrotoy J-P, Sylvand B (1986) Fluctuations naturelles et évolution artificielle des biocenoses macrozoobenthiques intertidales de trois estuaires des côtes françaises de la Manche. Hydrobiologia 142: 249–270. [Google Scholar]
- 21.Aminot A, Kérouel R (2007) Dosage automatique des nutriments dans les eaux marines: méthodes en flux continu. Ifremer. Ed Quae. 187 p. [Google Scholar]
- 22. Welschmeyer N (1994) Fluorometric analysis of chlorophyll chlorophyll b and pheopigments. Limnol Oceanogr 39: 1985–1992. [Google Scholar]
- 23. Bazin P, Jouenne F, Deton-Cabanillas AF, Pérez-Ruzafa Á, Véron B (2014) Complex patterns in phytoplankton and microeukaryote diversity along the estuarine continuum. Hydrobiologia 726: 155–178. [Google Scholar]
- 24.Bourrelly P (1981) Les algues d'eau douce - Tome 1,2 et 3. Paris: Ed Boubée. 572, 577 and 606 p. [Google Scholar]
- 25.Tomas CR (1997) Identifying Marine Phytoplankton. San Diego: Academic Press. 858 p. [Google Scholar]
- 26.John DM, Whitton BA, Brook AJ (2002) The Freshwater Algal Flora of the British Isles: An Identification Guide to Freshwater and Terrestrial Algae. Cambridge: Cambridge University Press. 714 p. [Google Scholar]
- 27. Utermöhl von H (1931) Neue Wege in der quantitativen Erfassung des Planktons. (Mit besondere Beriicksichtigung des Ultrapanktons). Verh Int Verein Theor Angew Limnol 5: 567–595. [Google Scholar]
- 28.EN 15204 (2006) Water quality - Guidance standard on the enumeration of phytoplankton using inverted microscopy (Utermöhl technique). Brussels, Belgium: European Committee for Standardization. 39 p. [Google Scholar]
- 29. Lund JWG, Kipling C, Le Cren ED (1958) The Inverted Microscope method of estimating algal numbers and the statistical basis of estimations by counting. Hydrobiologia 11: 143–170. [Google Scholar]
- 30. Medlin L, Elwood HJ, Stickel S, Sogin ML (1988) The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71: 491–499. [DOI] [PubMed] [Google Scholar]
- 31.Hepperle D (2004) SeqAssem. A sequence analysis tool, contig assembler and trace data visualization for molecular sequences. Win32-Version. Distributed by the author. Available: http://www.sequentix.de.
- 32. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 33. Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20: 2317–2319. [DOI] [PubMed] [Google Scholar]
- 34. Viprey M, Guillou L, Ferréol M, Vaulot D (2008) Wide genetic diversity of picoplanktonic green algae (Chloroplastida) in the Mediterranean Sea uncovered by a phylum-biased PCR approach. Environ Microbiol 10: 1804–1822. [DOI] [PubMed] [Google Scholar]
- 35. Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Romari K, Vaulot D (2004) Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol Oceanogr 49: 784–798. [Google Scholar]
- 38. Chao A (1984) Non parametric estimation of the number of classes in a population. Scand J Statist 11: 265–270. [Google Scholar]
- 39. Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis. Paleontologia Electronica 4: 1–9. [Google Scholar]
- 40. Massana R, Guillou L, Díez B, Pedro C (2002) Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the Ocean. Appl Environ Microbiol 68: 4554–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Massana R, Castresana J, Balague V, Guillou L, Romari K, et al. (2004) Phylogenetic and ecological analysis of Novel Marine Stramenopiles. Appl Environ Microbiol 70: 3528–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Charvet S, Vincent WF, Lovejoy C (2011) Chrysophytes and other protists in High Arctic lakes: molecular gene surveys, pigment signatures and microscopy. Polar Biol 35: 733–748. [Google Scholar]
- 43. Luo W, Bock C, Li HR, Padisák J, Krienitz L (2011) Molecular and microscopic diversity of planktonic eukaryotes in the oligotrophic Lake Stechlin (Germany). Hydrobiologia 661: 133–143. [Google Scholar]
- 44. Kolodziej K, Stoeck T (2007) Cellular identification of a novel uncultured marine stramenopile (MAST-12 Clade) small-subunit rRNA gene sequence from a norwegian estuary by use of fluorescence in situ hybridization-scanning electron microscopy. Appl Environ Microbiol 73: 2718–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Massana R, Terrado R, Forn I, Lovejoy C, Pedrós-Alió C (2006) Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ Microbiol 8: 1515–1522. [DOI] [PubMed] [Google Scholar]
- 46. del Campo J, Massana R (2011) Emerging diversity within chrysophytes, choanoflagellates and bicosoecids based on molecular surveys. Protist 162: 435–448. [DOI] [PubMed] [Google Scholar]
- 47. Andersen RA, Van de Peer Y, Potter D, Sexton JP, Kawachi M, et al. (1999) Phylogenetic analysis of the SSU rRNA from members of the Chrysophyceae. Protist 150: 71–84. [DOI] [PubMed] [Google Scholar]
- 48. Boenigk J, Pfandl K, Stadler P, Chatzinotas A (2005) High diversity of the “Spumella-like” flagellates: an investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions. Environ Microbiol 7: 685–697. [DOI] [PubMed] [Google Scholar]
- 49. Shi XL, Marie D, Jardillier L, Scanlan DJ, Vaulot D (2009) Groups without cultured representatives dominate eukaryotic picophytoplankton in the oligotrophic South East Pacific Ocean. PloS ONE 4(10): e7657 doi:10.1371/journal.pone.0007657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zuendorf A, Bunge J, Behnke A, Barger KJ, Stoeck T (2006) Diversity estimates of microeukaryotes below the chemocline of the anoxic Mariager Fjord, Denmark. FEMS Microbiol Ecol 58: 476–491. [DOI] [PubMed] [Google Scholar]
- 51. Bachvaroff TR, Kim S, Guillou L, Delwiche CF, Coats DW (2012) Molecular diversity of the syndinean genus Euduboscquella based on single-cell PCR analysis. Appl Environ Microbiol 78: 334–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deane JA, Strachan IM, Saunders GW, Hill DRA, Mcfadden GI (2002) Cryptomonad evolution: nuclear 18S rDNA phylogeny versus cell morphology and pigmentation. J Phycol 38 (6): 1236–1244. [Google Scholar]
- 53. Not F, Valentin K, Romari K, Lovejoy C, Massana R, et al. (2007) Picobiliphytes: A marine picoplanktonic algal group with unknown affinities to other eukaryotes. Science 315: 252–254. [DOI] [PubMed] [Google Scholar]
- 54. Seoane S, Laza A, Orive E (2006) Monitoring phytoplankton assemblages in estuarine waters: The application of pigment analysis and microscopy to size-fractionated samples. Estuar Coast Shelf Sci 67: 343–354. [Google Scholar]
- 55. Harrison PJ, Yin K, Lee JHW, Gan J, Liu H (2008) Physical–biological coupling in the Pearl River Estuary. Cont Shelf Res 28: 1405–1415. [Google Scholar]
- 56. Clay BL, Kugrens P (1999) Characterization of Hemiselmis amylosa sp. nov. and phylogenetic placement of the blue-green Cryptomonads H. amylosa and Falcomonas daucoides . Protist 150: 297–310. [DOI] [PubMed] [Google Scholar]
- 57. Savin MC, Martin JL, LeGresley M, Giewat M, Rooney-Varga J (2004) Plankton diversity in the Bay of Fundy as measured by morphological and molecular methods. Microb Ecol 48: 51–65. [DOI] [PubMed] [Google Scholar]
- 58. Laza-Martínez A (2012) Urgorri complanatus gen. et sp. nov. (Cryptophyceae), a red-tide-forming species in brackish waters. J Phycol 48: 423–435. [DOI] [PubMed] [Google Scholar]
- 59. Shalchian-Tabrizi K, Bråte J, Logares R, Klaveness D, Berney C, et al. (2008) Diversification of unicellular eukaryotes: cryptomonad colonizations of marine and fresh waters inferred from revised 18S rRNA phylogeny. Environ Microbiol 10: 2635–2644. [DOI] [PubMed] [Google Scholar]
- 60. Alpine A, Cloern J (1988) Phytoplankton growth rates in a light-limited environment, San Francisco Bay. Mar Ecol Prog Ser 44: 167–173. [Google Scholar]
- 61. Wylezich C, Jürgens K (2011) Protist diversity in suboxic and sulfidic waters of the Black Sea. Environ Microbiol 13: 2939–2956. [DOI] [PubMed] [Google Scholar]
- 62. Trigueros JM, Orive E (2000) Tidally driven distribution of phytoplankton blooms in a shallow, macrotidal estuary. J Plankton Res 22: 969–986. [Google Scholar]
- 63. Quinlan EL, Phlips EJ (2007) Phytoplankton assemblages across the marine to low-salinity transition zone in a blackwater dominated estuary. J Plankton Res 29: 401–416. [Google Scholar]
- 64. Caron DA, Countway PD, Brown MV (2004) The growing contributions of molecular biology and immunology to protistan ecology: molecular signatures as ecological tools. J Eukaryot Microbiol 51: 38–48. [DOI] [PubMed] [Google Scholar]
- 65. Šlapeta J, López-García P, Moreira D (2006) Global dispersal and ancient cryptic species in the smallest marine eukaryotes. Mol Biol Evol 23: 23–29. [DOI] [PubMed] [Google Scholar]
- 66. Liu H, Probert I, Uitz J, Claustre H, Aris-Brosou S, et al. (2009) Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans. Proc Natl Acad Sci U S A 106: 12803–12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stoeck T, Hayward B, Taylor GT, Varela R, Epstein SS (2006) A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist 157: 31–43. [DOI] [PubMed] [Google Scholar]
- 68. Zhu F, Massana R, Not F, Marie D, Vaulot D (2005) Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52: 79–92. [DOI] [PubMed] [Google Scholar]
- 69. Muylaert K, Van Mieghem R, Sabbe K, Tackx M, Vyverman W (2000) Dynamics and trophic roles of heterotrophic protists in the plankton of a freshwater tidal estuary. Hydrobiologia 432: 25–36. [Google Scholar]
- 70. Potvin M, Lovejoy C (2009) PCR-based diversity estimates of artificial and environmental 18S rRNA gene libraries. J Eukaryot Microbiol 56: 174–181. [DOI] [PubMed] [Google Scholar]
- 71. Caron DA, Countway PD, Savai P, Gast RJ, Schnetzer A, et al. (2009) Defining DNA-based operational taxonomic units for microbial eukaryote ecology. Appl Environ Microbiol 75: 5797–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Profiles of nutrient concentrations along the estuarine continuum. Silicate, phosphate and nitrate/nitrite were analyzed in the samples collected in the water column. (A logarithmic scale was used for the data representation).
(PDF)
Principal component analysis (PCA) of environmental parameters. Samples (dots) and variables (green lines) are displayed for the first two axes. Blue dots: surface samples and red dots: samples of the near-bottom water layer.
(TIFF)
Correspondence analysis (CA) based on phytoplankton composition (relative-abundance matrix). Taxa considered in the analysis accounted for >1% of total phytoplankton in at least one sample. Blue dots: surface samples and red dots: bottom samples. *Freshwater diatoms included the following pennates: Asterionella formosa, Nitzschia acicularis, and undetermined Naviculaceae. *Chlorophytes included Chlorophyceae (Scenedesmus spp., Monoraphidium contortum, Chlamydomonas sp.) and Trebouxiophyceae (Micractinium sp., Dictyosphaerium pulchellum).
(DOC)
Rarefaction curves determined for the three 18S rRNA gene libraries (VIRE River, ESTUARY and BAY). Curves were constructed at 98% sequence similarity cut-off value.
(TIFF)
Maximum likelihood (ML) tree showing the position of the non-photosynthetic stramenopiles OTUs (Bicosoecida, MASTs). OTUs were obtained from the the VIRE River (V), the ESTUARY (Es) and the BAY (B) clone libraries. Tree construction was based on an alignment of 96 partial sequences (ca 490 align positions). Number of clones per OTU is indicated in brackets. Sequences from cultured taxa appear in black, and environmental sequences in green (freshwater), blue (marine) or in brown (brackish/estuary). Bootstrap values (>50%) are indicated. The Dinophyceae Gyrodinium fusiforme and Peridinium umbonatum were used as outgroup.
(TIF)
Phytoplankton taxa identified by microscsopy. Samples corresponded to surface (As → Fs) and bottom (Bb → Eb) waters. “(SEM)” refers to additional taxa identified in the surface samples As, Cs and Es (also selected for clone library analysis, referred to as VIRE River, ESTUARY and BAY) using scanning electron microscopy.
(DOC)