Abstract
BACKGROUND
Autophagy has recently been found to play important roles in tumorigenesis and LRPPRC has been identified as an inhibitor that suppresses autophagy and mitophagy and maintains mitochondrial activity. We hypothesized that LRPPRC levels can be used as a biomarker for diagnosis and prognosis of prostate cancer.
METHODS
We performed immunochemistry analysis to evaluate the levels of LRPPRC in 112 samples collected from patients with prostate adenocarcinomas (PCa) and 38 samples from patients with benign prostate hyperplasia (BPH) enrolled in hospitals in Guangzhou city of China that have been followed up for 10 years.
RESULTS
We found that there were significant higher levels of LRPPRC in PCa than in BPH. More than three quarters of PCa patients showed high levels of LRPPRC while only 10% of BPH patients had similar levels of LRPPRC. The levels of LRPPRC protein were positively correlated with tumor grade, metastasis and serum PSA, but negatively correlated with hormone therapy sensitivity after 2 years of surgery and overall survival. The association of high levels of LRPPRC with late stage of PCa or hormone therapy insensitivity was confirmed in tissue samples collected from prostate-specific PTEN−/− mice or hormone-dependent and independent prostate cancer cell lines.
CONCLUSION
The levels of LRPPRC may be used as an independent biomarker for PCa patients at late-stage with poor prognosis.
Keywords: Autophagy, Benign prostatic hyperplasia, Biomarker, Cancer prognosis, LRPPRC, Mitochondria, Prostate adenocarcinomas, Prostatic intraepithelial neoplasia, PTEN
INTRODUCTION
Prostate cancer is the most common non-cutaneous malignancy and the second leading cause of cancer death among men in the United States.1 With the spread of the western diet, the incidence of prostate cancer is increasing in the world. It is estimated that every one in six men will develop prostate cancer in their lifetime, with incidence increasing with age.2 The use of prostate cancer specific biomarkers, such as prostate specific antigen (PSA) for example, have revolutionized the screening, detection, and prognostication of this disease.3 However, PSA screening has generated a lot of false positives.4 No increase of PSA has been detected in a substantial number of prostate cancer patients. Thus, it is indicated that new biomarkers are needed for diagnosis or prognosis of prostate cancer.5, 6
Autophagy, or self-digestion, is a process that begins with the formation of isolation membranes that engulf substrates including dysfunctional organelles, mis-folded/aggregated proteins and/or other macromolecules to form autophagosomes. Then autophagosomes fuse with lysosomes to generate autolysosomes in which substrates are degraded 7. Mitochondrion is one of the most prominent and vital type of organelles in eukaryotic cells. During cell cycling, mitochondria are constantly synthesized, used, damaged and destroyed through autophagy (here referred to as mitophagy).8, 9 Mitophagy is a highly regulated process and Parkin and Pink1, whose mutations may be counted for Parkinson’s disease in small numbers of patients, have recently been found to collaboratively regulate mitophagy to control mitochondrial activity.10–12 Interestingly, Pink1, PTEN-induced putative kinase, is one of target genes of PTEN whose deletion in mouse prostate created a well-accepted model vividly mimicking a lot of features of human prostate adenocarcinomas.13 Therefore, we hypothesized that those mitophagy regulatory proteins may also play important roles during the development of prostate adenocarcinomas.
LRPPRC, a leucine-rich pentatricopeptide repeat (PPR) motif-containing protein also named as LRP130, has been characterized as a mitochondrion-associated protein.14–16 It was suggested that mutations in the gene cause Leigh syndrome, French-Canadian type (LSFC), a human disorder characterized with neurodegeneration and cytochrome c oxidase deficiency.17 We found that LRPPRC interacts with MAP1S, a mitochondrion and microtubule-associated protein previously named as C19ORF5.14, 15, 18 As a sequence homologue of the microtubule-associated protein MAP1A and MAP1B, MAP1S similarly interacts with mammalian autophagy marker LC319–21 and bridges autophagic components with microtubules and mitochondria to affect autophagosomal biogenesis and degradation and suppress genome instability and tumorigenesis.21–23 In contrast to nuclear acid-associated functions such as transcriptional or translational regulation in either nuclei, mitochondria or endoplasmic reticulum,24–34 LRPPRC has been recently reported by us to associate with mitochondria, interact with Beclin 1 and Bcl-2 and form a ternary complex to maintain the Bcl-2 stability and suppress autophagy.16 As a mitochondrion-associated protein, LRPPRC may prevent mitochondria from being degrading through mitophagy in association with Parkin to maintain mitochondrial activity.21 Therefore, we felt enthusiastic to probe potential roles of LRPPRC in prostate adenocarcinomas.
In this study, we performed immunochemistry analysis to evaluate the levels of LRPPRC in prostate tissues from human patients with prostate adenocarcinomas (PCa) or benign prostate hyperplasia (BPH), PTEN-deficient mice and human prostate cell lines. We found that the immunestaining intensity of LRPPRC, percentage of LRPPRC positive cells and their combined score representing LRPPRC level are significantly higher in PCa than in BPH. Prostate tissues at late stage of prostate adenocarcinomas as indicated by higher tumor grade, metastasis, higher serum PSA, or PCa tissues with lower hormone therapy sensitivity after 2 years of surgery, express higher LRPPRC levels. Such association of high LRPPRC levels with late stage of PCa or hormone therapy insensitivity was confirmed in prostate PCa tissue samples collected from prostate-specific PTEN−/− mice or hormone-dependent and independent prostate cancer cell lines. PCa patients with higher intensity, frequency and level of LRPPRC exhibited shorter overall survival time after treatment and the LRPPRC level was the marker with the highest Harzard Ratio based on a multivariate analysis. Therefore, the LRPPRC level may serve as an independent biomarker for late-stage PCa patients with poor prognosis.
MATERIALS AND METHODS
Patients and Diagnosis
In the present study, a total of 150 patients including 112 patients with prostate cancer and 38 with BPH as controls were evaluated. These patients had consented to donate prostate tissue samples from surgery or by biopsy and underwent treatment including radiotherapy, prostatectomy, transurethral resection of the prostate (TURP) and hormone therapy from 1999 to 2003 at The Fifth Affiliated Hospital of Guangzhou Medical University, Sun Yat-Sen Memorial Hospital, and The First People’s Hospital of Guangzhou and The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou city, China. The patients were followed for 10 years for a complete set of clinical data. Each patient was treated according to the stage of the disease. Tumors were confirmed histopathologically and were staged according to TNM classification by World Health Organization. All primary samples taken from the patients were fixed in 10% formalin, embedded in paraffin, sectioned consecutively at 5 μm, and stained by hematoxylin and eosin. The histological types were assigned to two independent clinical pathologists in a double-blinded manner.
Immunohistochemistry Staining of Human Prostate Tissue Samples
The sections were deparaffinized and rehydrated, and endogenous peroxidase was inhibited by 0.3% H2O2 methanol. Five percent fat-free milk was used to block non-specific background for one hour. Slides were then incubated with 1:200 rabbit anti-human LRPPRC (Cat# sc-166178, Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C overnight. After washing, the slides were reacted with a second antibody of 1:5000 anti-rabbit-HRP (Santa Cruz Biotechnology, Santa Cruz, CA). Tumor cells with cytoplasmic staining were considered positive for expression of LRPPRC. The percentage of positive staining (LRPPRC frequency) was scored as 0 for 0%, 1 for 1–25%, 2 for 26–50%, 3 for 51–75% and 4 for >75% of cells with positive staining. The staining intensity (LRPPRC intensity) was scored as 0 for no staining, 1 for weakly stained, 2 for moderately stained, and 3 for strongly stained. The final scores for LRPPRC levels were the sum of the scores for LRPPRC intensity and frequency. The final scores of 4 or higher were considered as high LRPPRC levels and scores less than 4 were considered as low LRPPRC levels. The slides were scored by 2 independent clinical pathologists in a double blinded manner.
Statistical Analysis
The statistical analysis of staining scores of LRPPRC in PCa, BPH, and various clinicopathological groups were evaluated with the t-test. The overall survival was measured from the start of treatment until the end of the observation period and analyzed by the Kaplan-Meier method. Cox proportional-hazard analysis with univariate or multivariate method was used to explore the effect of variables on overall survival. The SAS software was used for all statistical analyses and a P value of <0.05 was considered significant.
Generation of Prostate Specific PTEN−/− Mice and Collection of Prostate Tissue Samples
Prostate specific PTEN knock-out (PTEN−/−) mice were obtained by crossing PTENf/f mice from Jackson Laboratory with prostate specific ARR2PBi-Cre (Pb-Cre) mice as described previously.13, 35 Normal prostate tissues were collected from 5 or 11 months old wild type mice. Tissues of prostatic intraepithelial neoplasia (PIN) or prostate adenocarcinomas (PCa) were collected from prostate-specific PTEN knockout mice at the same age of 11 months. Prostate tissues were isolated then either frozen in liquid nitrogen and stored at −80°C for immunoblot analysis or fixed in 4% paraformaldehyde at 4°C overnight, dehydrated, processed, paraffin-embedded, and then sectioned in 5-μm thickness for immuno-fluorescent staining.
Immunoblot Analyses
Frozen tissues were homogenized and proteins were isolated in RIPA buffer (150 mmol/L NaCl, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mmol/L Tris–HCl; pH 8.0). The protein concentrations of prostate tissue lysates were determined as described previously.21 After boiled, lysates were centrifuged, supernatants with 20 μg total proteins were loaded onto 5–10% polyacrylamide gels containing 0.1% SDS. Proteins were separated through electrophoresis and transferred to PVDF membranes. After blocking, the proteins were probed with 1:200–1000 primary antibodies and detected with 1:10000 horseradish peroxidase (HRP)-conjugated secondary antibodies and the ECL Western Blotting Detection Reagents. Antibodies used were mouse anti-β-actin (Cat# sc-47778, Santa Cruz Biotechnology) and mouse anti-GAPDH (Cat# sc-47724, Santa Cruz Biotechnology). HRP-conjugated secondary antibodies against mouse (Cat# 172–1011) or rabbit (Cat# 172–1019) were from Bio-Rad. ECL Western Blotting Detection Reagents and PVDF transfer membranes were purchased from GE Health. After exposure, x-ray films were developed, washed, dried and scanned into image files.
Fluorescent Confocal Microscopy
Immunofluorescence staining was performed on 5 μm sections mounted on Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA). Sections were deparaffinized in xylenes and rehydrated through a graded series of ethanol/water solutions. The antigens were retrieved by boiling in citrate buffer (10 mM sodium citrate sodium buffer, pH 6.0) for 20 minutes at 100°C. The primary antibodies were used at a 1:200 concentration. After washing, the specifically bound first antibodies were detected with FITC-conjugated anti-rabbit secondary antibodies (Cat# A21206, Invitrogen). The sections were counterstained with TOPRO-3 iodide to label nuclei and observed under a Zeiss LSM 510 confocal microscope.
RESULTS
Expression of LRPPRC in PCa and BPH Tissues
Totally 150 prostate biopsy tissues isolated from 112 patients with prostate cancer (PCa) and 38 patients with benign prostatic hyperplasia (BPH) were used for the study. Patients underwent treatment from 1999 to 2003 at The Fifth Affiliated Hospital of Sun Yat-Sen Memorial Hospital, The First People’s Hospital of Guangzhou and The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, China. Collection of prostate tissues and patient information was approved by patients who were followed for 10 years for complete clinical data. All patients were men with age ranged from 52–85 years and a mean age of 69 years, and survived for period ranged from 11 to 95 months and a median time of overall survival of 77 months. The clinical parameters of these patients are summarized in Table 1. Tumor samples were confirmed by histologists in the hospital and were staged according to TNM classification system endorsed by World Health Organization (WHO).
Table 1.
The clinical features of patients with PCa and their LRPPRC Levels.
| Clinical Features | Case Numbers | LRPPRC Levels Mean ± Stdev | P Value |
|---|---|---|---|
| Tissue Type | |||
| PCa | 112 | 4.6±1.4 | <0.001 |
| BPH | 38 | 1.7±1.6 | |
| Age (years) | |||
| < 70 | 50 | 4.5±1.4 | 0.481 |
| ≥70 | 62 | 4.7±1.5 | |
| Gleason Score | |||
| < 8 | 91 | 4.4±1.3 | 0.090 |
| ≥8 | 21 | 5.3±1.7 | |
| Preoperative PSA | |||
| <10 ng/ml | 41 | 4.2±1.1 | 0.018 |
| ≥10 ng/ml | 71 | 4.8±1.5 | |
| Tumor Grade | |||
| I/II | 61 | 4.1±1.4 | <0.001 |
| III | 51 | 5.3±1.2 | |
| Metastasis | |||
| No | 58 | 3.9±1.2 | <0.001 |
| Yes | 54 | 5.3±1.3 | |
| HTSin2* | |||
| No | 10 | 5.3±1.5 | 0.110 |
| Yes | 102 | 4.5±1.4 | |
| HTSaf2* | |||
| No | 50 | 5.3±1.2 | <0.001 |
| Yes | 62 | 3.9±1.2 | |
| Survival Time | |||
| <5 years | 51 | 5.3±1.2 | <0.001 |
| ≥5 years | 61 | 3.9±1.3 | |
| Survival Time | |||
| <10 years | 83 | 5.1±1.2 | <0.001 |
| ≥10 years | 29 | 3.2±1.1 | |
HTSin2: Hormone Therapy Sensitivity within 2 Years.
HTSaf2: Hormone Therapy Sensitivity after 2 Years.
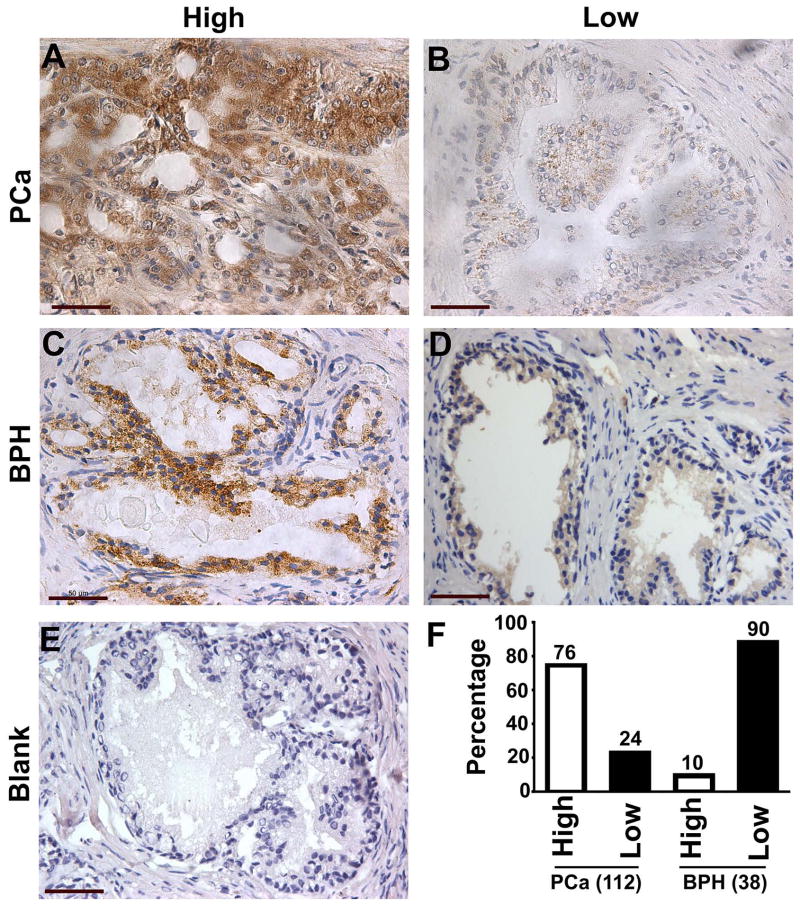
Standard immunohistochemistry protocol was used in the detection of LRPPRC in prostate tissues and the results were evaluated by two independent pathologists in a double-blinded manner. The immune-reactivity of LRPPRC was detected to be localized in the cytoplasm of epithelial cells of prostate cancer and BPH tissues, and no LRPPRC signals were detected in the prostate stromal region (Fig. 1). The staining was heterogeneous and both strong and weak staining signals were detected in prostate cancer and BPH tissues. Quantitation of the positive signals demonstrated that 85 of 112 (76%) patients with PCa exhibited overexpression of LRPPRC (LRPPRC scored ≥4) and 4 of 38 (10%) patients with BPH demonstrated high expression of LRPPRC (Fig. 1F). On average, LRPPRC levels in patients with PCa were significantly higher than in patients with BPH (Table 1). Therefore, LRPPRC is a novel marker displaying significant difference between two different types of prostate tumors with different malignancies.
Figure 1.
The expression of LRPPRC in prostate cancer (PCa) and benign prostate hyperplasia (BPH). A,B. Representative images of prostate cancer expressing high (A) or low levels of LRPPRC (B). Bar: 50 μm. C,D. Representative images of benign prostate hyperplasia expressing high (C) or low levels of LRPPRC (D). Bar: 50 μm. E. A tumor sample not stained with antibody against LRPPRC shown as a negative control. Bar: 50 μm. F. Quantitation of high or low expression levels of LRPPRC in prostate cancer and benign prostate hyperplasia. Bars represent the percentage of human patients with high or low levels of LRPPRC.
Relation of LRPPRC Levels to Clinical Features of Patients with PCa
Among PCa patients with different ages, we did not detect any significant difference among different age groups. However, significant difference was detected between different tumor grades (P<0.001), serum PSA level (P=0.018), metastatic status (P<0.001), and hormone therapy insensitivity after 2 years (P<0.001) (Table 1). Patient diagnosed with higher tumor grades expressed higher levels of LRPPRC than patients with lower tumor grades. Similarly, patients with higher serum PSA levels, with metastatic tumor and insensitive to hormone therapy after two years had higher levels of LRPPRC than those with lower serum PSA levels, with non-metastatic tumor and sensitive to hormone therapy after two year. Therefore, LRPPRC may serve as an independent diagnosis marker for patients at late stage of PCa.
Relation of LRPPRC Levels to Patient’s Survival
To track the correlation of levels of LRPPRC with patients’ overall survival, we followed the patients for more than 10 years. Higher LRPPRC intensities, higher LRPPRC frequencies and higher LRPPRC levels associated with shorter overall survival (Fig. 2). We found patient surviving longer than five or ten years had significantly lower levels of LRPPRC than patients surviving shorter than five or ten years (P<0.001) (Table 1). Hazard ratio analysis demonstrated that both high LRPPRC intensity and high LRPPRC frequency individually associated well with shorter overall survival. The combined scores of LRPPRC intensity and frequency, LRPPRC levels, became a more sensitive marker for poor prognosis. In addition, Gleason scores, metastasis states and clinical stage predicted well the patients’ overall survival by both univariate and multivariate analysis (Table 2). Thus, high LRPPRC levels may be independently used as a better marker to predict poor prognosis of PCa patients at late stage.
Figure 2.
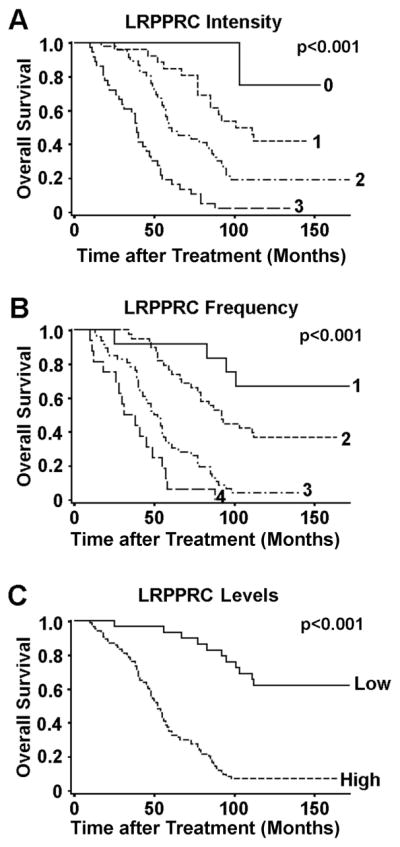
The Kaplan-Meier survival curves showing the overall survival time after treatment of PCa patients with high or low LRPPRC intensities (A), frequencies (B) and levels (C). The significance of difference between two groups was estimated by chi-square test.
Table 2.
Univariate or multivariate cox proportional hazard ratios for overall survival time
| Hazard Ratio (95%CI) | P Value | |
|---|---|---|
| Univariate Analysis | ||
| LRPPRC Intensity | 2.64 (1.95–3.57) | <0.0001 |
| LRPPRC Frequency | 2.74 (2.07–3.63) | <0.0001 |
| LRPPRC Levels | 6.54 (3.41–12.56) | <0.0001 |
| Metastasis | 4.04 (2.57–6.34) | <0.0001 |
| Clinical Stage | 4.01 (2.55–6.32) | <0.0001 |
| Gleason Score | 3.14 (1.89–5.21) | <0.0001 |
| Preoperative PSA | 1.45 (1.10–1.91) | 0.0083 |
| HTSaf2* | 0.18 (0.12–0.29) | <0.0001 |
| HTSin2* | 0.14 (0.07–0.30) | <0.0001 |
| Multivariate Analysis | ||
| LRPPRC Levels | 4.60 (2.26–9.37) | <0.0001 |
| Gleason Score | 2.99 (1.76–5.09) | <0.0001 |
| Metastasis | 2.09 (1.27–3.42) | 0.0036 |
| Clinical Stage | 1.80 (1.06–3.07) | 0.0311 |
| Preoperative PSA | 1.11 (0.82–1.49) | 0.5031 |
| HTSin2* | 0.48 (0.22–1.03) | 0.0598 |
| HTSaf2* | 0.46 (0.26–0.79) | 0.0049 |
HTSin2: Hormone Therapy Sensitivity within 2 Years.
HTSaf2: Hormone Therapy Sensitivity after 2 Years.
Expression of LRPPRC in Prostate-specific PTEN−/− Mouse Model
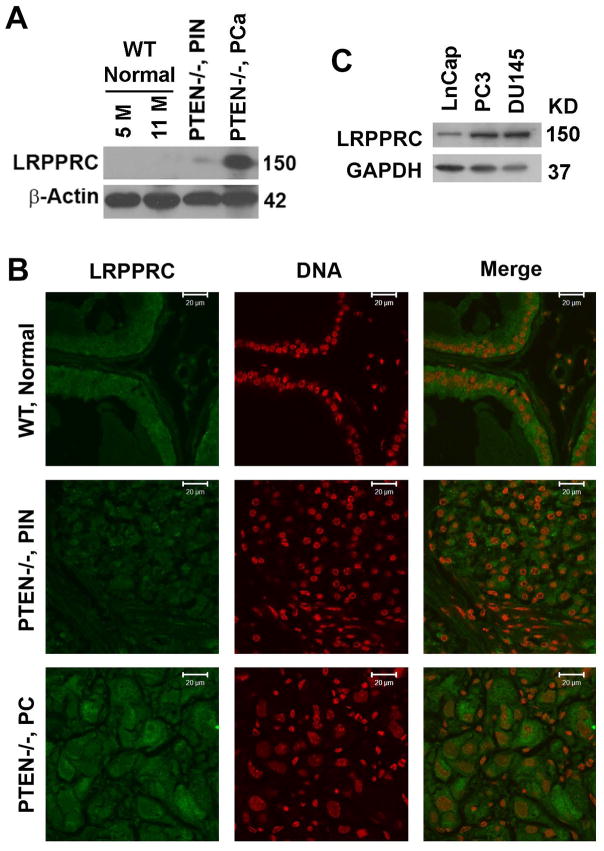
PTEN is a tumor suppressive gene and PTEN−/− mouse models have high incidence of prostate cancer development at the age of 11 months.13 In order to determine the expression of LRPPRC in mouse prostate cancer, we crossed the PTENf/f mice with ARR2PBi-Cre (Pb-Cre) mice to generate prostate specific PTEN−/− mice. We then isolated prostate tissue from 5 or 11 month old wild type and prostatic intraepithelial neoplasia and PCa tissues from 11 months old PTEN−/− mice and performed western blot and immunofluorescence assays. We found that the expression of LRPPRC was not detectable in normal prostate tissues collected from wildtype mice at different ages but significantly higher in prostate cancer tissue isolated from PTEN−/− mice than in normal prostate tissue isolated from age-matched wild type mice and PIN isolated from PTEN−/− mice at the same age (Fig. 3A,B). The data from mice models confirms that the levels of LRPPRC correlate with the stages of human prostate cancer.
Figure 3.
The expression of LRPPRC in prostate tissues from wild type and PTEN−/− mouse models and cultured human prostate cell lines. A. Immunoblot assay of LRPPRC expression in normal prostate tissue from 5 or 11 month-old wildtype mice, prostatic intraepithelial neoplasia (PIN) and prostate adenocarcinomas (PCa) from PTEN−/− mice at the same age of 11 months. B. Immunoflurescent assay of LRPPRC expression in same tissues as indicated above (A). Bar: 20 μm. C. Immunoblot analyses of the expression levels of LRPPRC in human prostate cancer cell lines.
Expression of LRPPRC in Hormone-dependent and Independent PCa Cell Lines
As the prostate is a hormone organ, early stage prostate cancers are usually hormone-dependent, and late stage prostate cancer become hormone-independent, in order to see the levels of LRPPRC in different hormone stages of prostate cancer, we extracted proteins from several human prostate cancer cell lines. We found that the levels of LRPPRC were lower in hormone dependent prostate cancer cells (LnCap) than in hormone-independent prostate cancer cells such PC3 and DU-145). This indicates that levels of LRPPRC were higher in hormone independent prostate cancers, which are usually detected in patients at late stage of prostate cancer (Fig. 3C). The data from prostate cancer cell lines further confirm that the levels of LRPPRC are negatively related to hormone therapy sensitivity after 2 years as detected in human patients.
DISCUSSION
The expression of LRPPRC is correlated with tumor grade, metastasis, serum PSA, and hormone sensitivity after 2 years. More importantly, 10-years’ following-up data for PCa patients demonstrated that the levels of LRPPRC are correlated with 5 and 10-year survival of patients. Patient groups with higher levels of LRPPRC have a significantly shorter survival time than patient groups with lower levels of LRPPRC. The LRPPRC level is an independent biomarker that predicts poor prognosis of PCa patients as detected in the hazard ratio assays. Thus, we have identified LRPPRC as a novel biomarker of prognosis for PCa patients.
Similar condition has been found in other types of cancers. Recent studies have found that LRPPRC is overexpressed in a variety of human tumors, such as lung adenocarcinoma, oesophageal squamous cell carcinoma, stomach, colon, mammary and endometrial adenocarcinoma, and lymphoma, although LRPPRC is not expressed in the surrounding non-neoplastic cells.36 Suppression of LRPPRC leads to dramatic reduction of tumor cells’ resistance to apoptosis, invasion and in vitro colony formation in lung adenocarcinoma, as well as in Hodgkin lymphoma cells.36,37 These results further strengthen the potential of LRPPRC to be used as a prognosis marker for not only prostate cancer but also other types of cancers.
Based on our results from a mouse model with the LRPPRC-interactive MAP1S deficiency, we suggest that stresses resulted from carcinogens or genome instability-driven metabolic stresses cause accumulation of aggresomes or dysfunctional organelles such as mitochondria. Autophagy is activated to remove the accumulated. If autophagy is defective, the accumulated products enhance oxidative stress triggering DNA double strand breaks and weakening mitotic checkpoint so that genome instability is caused after continuous karyotype evolution. Genome instability enhances tumor initiation and development. Autophagy intrinsically plays suppressive roles during tumor onset and development and autophagy defect leads to development of more malignant tumors and accumulation of mitochondria.22, 23 Therefore, accumulation of mitochondria in malignant tumors leads to elevation of LRPPRC levels as the consequence of autophagy defect.
Alternatively, LRPPRC primarily maintains mitochondrial potential and activity and high levels of LRPPRC suggest higher mitochondrial activity. Under stresses resulted from cancer-inducing genome instability, mitochondria in tumor cells are naturally subjected to be damaged and further degradation through autophagy. In respond to such stresses, tumor cells may evolve to increase their LRPPRC levels to protect their mitochondria from damage. LRPPRC forms a complex with Bcl-2 and the tumor suppressor Beclin 1 and inhibits autophagy.16 High levels of LRPPRC are expected to enhance the stability of Bcl-2 and protect the LRPPRC-associated mitochondria from degradation through mitophagy.16, 21, 36 It has been known that overexpression of members of the Bcl-2 family of pro-survival proteins is commonly associated with unfavorable pathogenesis in cancer.38 Specifically, high levels of Bcl-2 protein are detected in androgen-independent tumors in advanced stages of the pathology.39 Indeed, most tumor cells need more energy than their normal mature counterparts.40 Prostate cancer, like other cancers, demonstrates abnormal mitochondria activity.41, 42 Therefore, patients at late stage of prostate adenocarcinomas exhibit higher levels of LRPPRC than those at early stage of the disease because of adoption to new micro-environment in tumor cells and tissues.
The importance of mitochondria in cancer development has already been reflected by the fact that many mitochondria-targeting agents have emerged as novel means to selectively target tumors.43, 44 Many compounds have been developed to specifically target the different aspects of mitochondria in cancer cells such as mitochondrial permeability transition,45 permeability transition pore complex constituents,46 reactive oxygen species,47 mitochondrial outer membrane permeabilization,48 and mitochondrial metabolism.49 Some of these treatments have generated promising results.43 The association of high levels of autophagy inhibitor LRPPRC with malignant prostate cancer reported here provides a new potential target for prostate cancer therapy through autophagy regulation although the exact mechanism is under investigation.
In conclusion, we are the first to demonstrate the association of high levels of LRPPRC with poor prognosis of prostate cancer patients. Our study provides a novel basis for future development of diagnostic and therapeutic tools for prostate cancers in clinic.
Acknowledgments
We thank Drs. Shan Wu, Chi Zhang, Weidong Chen and Wen He for their help in collecting tumor materials and clinical data.
FUNDING SUPPORT
This study is partially supported by DOD New Investigator Award W81XWH and NCI 1R01CA142862 to Leyuan Liu.
Footnotes
CONFLICT INTEREST DISCLOSURES
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Baade PD, Youlden DR, Krnjacki LJ. International epidemiology of prostate cancer: geographical distribution and secular trends. Mol Nutr Food Res. 2009;53:171–184. doi: 10.1002/mnfr.200700511. [DOI] [PubMed] [Google Scholar]
- 3.Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 4.Velonas VM, Woo HH, Remedios CG, Assinder SJ. Current status of biomarkers for prostate cancer. Int J Mol Sci. 2013;14:11034–11060. doi: 10.3390/ijms140611034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Froehner M, Abolmaali N, Wirth MP. Prostate-specific antigen-negative prostate cancer recurrence? Urology. 2013;81:e17–18. doi: 10.1016/j.urology.2012.10.036. [DOI] [PubMed] [Google Scholar]
- 6.Bucerius J, Ahmadzadehfar H, Hortling N, Joe AY, Palmedo H, Biersack HJ. Incidental diagnosis of a PSA-negative prostate cancer by 18FDG PET/CT in a patient with hypopharyngeal cancer. Prostate Cancer Prostatic Dis. 2007;10:307–310. doi: 10.1038/sj.pcan.4500959. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L, Xie R, Nguyen S, Ye M, McKeehan WL. Robust autophagy/mitophagy persists during mitosis. Cell Cycle. 2009;8:1616–1620. doi: 10.4161/cc.8.10.8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terman A, Gustafsson B, Brunk UT. The lysosomal-mitochondrial axis theory of postmitotic aging and cell death. Chem Biol Interact. 2006;163:29–37. doi: 10.1016/j.cbi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Geisler S, Holmstrom KM, Skujat D, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 11.Matsuda N, Sato S, Shiba K, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, Gao J, Lei Q, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Amy V, Liu G, McKeehan WL. Novel complex integrating mitochondria and the microtubular cytoskeleton with chromosome remodeling and tumor suppressor RASSF1 deduced by in silico homology analysis, interaction cloning in yeast, and colocalization in cultured cells. In Vitro Cell Dev Biol Anim. 2002;38:582–594. doi: 10.1290/1543-706x(2002)38<582:ncimat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L, McKeehan WL. Sequence analysis of LRPPRC and its SEC1 domain interaction partners suggest roles in cytoskeletal organization, vesicular trafficking, nucleocytosolic shuttling and chromosome activity. Genomics. 2002;79:124–136. doi: 10.1006/geno.2001.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou J, Yue F, Jiang X, Li W, Yi J, Liu L. Mitochondrion-associated protein LRPPRC suppresses the initiation of basal levels of autophagy via enhancing Bcl-2 stability. Biochem J. 2013;454:447–457. doi: 10.1042/BJ20130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mootha VK, Lepage P, Miller K, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Vo A, Liu G, McKeehan WL. Putative tumor suppressor RASSF1 interactive protein and cell death inducer C19ORF5 is a DNA binding protein. Biochem Biophys Res Commun. 2005;332:670–676. doi: 10.1016/j.bbrc.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kayaba H, Hirokawa M, Watanabe A, et al. Serum markers of graft-versus-host disease after bone marrow transplantation. J Allergy Clin Immunol. 2000;106:S40–44. doi: 10.1067/mai.2000.106060. [DOI] [PubMed] [Google Scholar]
- 20.Schoenfeld TA, McKerracher L, Obar R, Vallee RB. MAP 1A and MAP 1B are structurally related microtubule associated proteins with distinct developmental patterns in the CNS. J Neurosci. 1989;9:1712–1730. doi: 10.1523/JNEUROSCI.09-05-01712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie R, Nguyen S, McKeehan K, Wang F, McKeehan WL, Liu L. Microtubule-associated protein 1S (MAP1S) bridges autophagic components with microtubules and mitochondria to affect autophagosomal biogenesis and degradation. J Biol Chem. 2011;286:10367–10377. doi: 10.1074/jbc.M110.206532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie R, Wang F, McKeehan WL, Liu L. Autophagy enhanced by microtubule- and mitochondrion-associated MAP1S suppresses genome instability and hepatocarcinogenesis. Cancer Res. 2011;71:7537–7546. doi: 10.1158/0008-5472.CAN-11-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, McKeehan WL, Wang F, Xie R. MAP1S enhances autophagy to suppress tumorigenesis. Autophagy. 2012;8:278–280. doi: 10.4161/auto.8.2.18939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper MP, Qu L, Rohas LM, et al. Defects in energy homeostasis in Leigh syndrome French Canadian variant through PGC-1alpha/LRP130 complex. Genes Dev. 2006;20:2996–3009. doi: 10.1101/gad.1483906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper MP, Uldry M, Kajimura S, Arany Z, Spiegelman BM. Modulation of PGC-1 coactivator pathways in brown fat differentiation through LRP130. J Biol Chem. 2008;283:31960–31967. doi: 10.1074/jbc.M805431200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu F, Addis JB, Cameron JM, Robinson BH. LRPPRC mutation suppresses cytochrome oxidase activity by altering mitochondrial RNA transcript stability in a mouse model. Biochem J. 2012;441:275–283. doi: 10.1042/BJ20110985. [DOI] [PubMed] [Google Scholar]
- 27.Mili S, Pinol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol Cell Biol. 2003;23:4972–4982. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mili S, Shu HJ, Zhao Y, Pinol-Roma S. Distinct RNP Complexes of Shuttling hnRNP Proteins with Pre-mRNA and mRNA: Candidate Intermediates in Formation and Export of mRNA. Mol Cell Biol. 2001;21:7307–7319. doi: 10.1128/MCB.21.21.7307-7319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem J. 2004;382:331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sondheimer N, Fang JK, Polyak E, Falk MJ, Avadhani NG. Leucine-rich pentatricopeptide-repeat containing protein regulates mitochondrial transcription. Biochemistry. 2010;49:7467–7473. doi: 10.1021/bi1008479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruzzenente B, Metodiev MD, Wredenberg A, et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA, Consortium L. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol Biol Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L, Sanosaka M, Lei S, et al. LRP130 protein remodels mitochondria and stimulates fatty acid oxidation. J Biol Chem. 2011;286:41253–41264. doi: 10.1074/jbc.M111.276121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gohil VM, Nilsson R, Belcher-Timme CA, Luo B, Root DE, Mootha VK. Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. J Biol Chem. 2010;285:13742–13747. doi: 10.1074/jbc.M109.098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin C, McKeehan K, Wang F. Transgenic mouse with high Cre recombinase activity in all prostate lobes, seminal vesicle, and ductus deferens. Prostate. 2003;57:160–164. doi: 10.1002/pros.10283. [DOI] [PubMed] [Google Scholar]
- 36.Tian T, Ikeda J, Wang Y, et al. Role of leucine-rich pentatricopeptide repeat motif-containing protein (LRPPRC) for anti-apoptosis and tumourigenesis in cancers. Eur J Cancer. 2012;48:2462–2473. doi: 10.1016/j.ejca.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 37.Michaud M, Barakat S, Magnard S, Rigal D, Baggetto LG. Leucine-rich protein 130 contributes to apoptosis resistance of human hepatocarcinoma cells. Int J Oncol. 2011;38:169–178. [PubMed] [Google Scholar]
- 38.Lessene G, Czabotar PE, Colman PM. BCL-2 family antagonists for cancer therapy. Nat Rev Drug Discov. 2008;7:989–1000. doi: 10.1038/nrd2658. [DOI] [PubMed] [Google Scholar]
- 39.Catz SD, Johnson JL. BCL-2 in prostate cancer: a minireview. Apoptosis. 2003;8:29–37. doi: 10.1023/a:1021692801278. [DOI] [PubMed] [Google Scholar]
- 40.Mathupala SP, Ko YH, Pedersen PL. The pivotal roles of mitochondria in cancer: Warburg and beyond and encouraging prospects for effective therapies. Biochim Biophys Acta. 2010;1797:1225–1230. doi: 10.1016/j.bbabio.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parr RL, Mills J, Harbottle A, et al. Mitochondria, prostate cancer, and biopsy sampling error. Discov Med. 2013;15:213–220. [PubMed] [Google Scholar]
- 42.Herrmann PC, Gillespie JW, Charboneau L, et al. Mitochondrial proteome: altered cytochrome c oxidase subunit levels in prostate cancer. Proteomics. 2003;3:1801–1810. doi: 10.1002/pmic.200300461. [DOI] [PubMed] [Google Scholar]
- 43.Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9:447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 44.Dias N, Bailly C. Drugs targeting mitochondrial functions to control tumor cell growth. Biochem Pharmacol. 2005;70:1–12. doi: 10.1016/j.bcp.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 45.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 46.Marzo I, Brenner C, Zamzami N, et al. Bax and adenine nucleotide translocator cooperate in the mitochondrial control of apoptosis. Science. 1998;281:2027–2031. doi: 10.1126/science.281.5385.2027. [DOI] [PubMed] [Google Scholar]
- 47.Magda D, Miller RA. Motexafin gadolinium: a novel redox active drug for cancer therapy. Semin Cancer Biol. 2006;16:466–476. doi: 10.1016/j.semcancer.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Ross K, Rudel T, Kozjak-Pavlovic V. TOM-independent complex formation of Bax and Bak in mammalian mitochondria during TNFalpha-induced apoptosis. Cell Death Differ. 2009;16:697–707. doi: 10.1038/cdd.2008.194. [DOI] [PubMed] [Google Scholar]
- 49.Simons AL, Ahmad IM, Mattson DM, Dornfeld KJ, Spitz DR. 2-Deoxy-D-glucose combined with cisplatin enhances cytotoxicity via metabolic oxidative stress in human head and neck cancer cells. Cancer Res. 2007;67:3364–3370. doi: 10.1158/0008-5472.CAN-06-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]