Abstract
IMPORTANCE
A better understanding of the etiology of obesity is a clinical priority. Obesity is highly heritable and specific genes are being identified. Discovering the mechanisms through which obesity-related genes influence weight would help pinpoint novel targets for intervention. One potential mechanism is satiety responsiveness. Lack of satiety characterizes many monogenic obesity disorders, and lower satiety responsiveness is linked with weight gain in population samples.
OBJECTIVE
We tested the hypothesis that satiety responsiveness is an intermediate behavioral phenotype associated with genetic predisposition to obesity in children.
DESIGN
Cross-sectional observational study.
SETTING
Population-based cohort of twins born 1994–1996 (Twins Early Development Study).
PARTICIPANTS
2258 unrelated children (53% female; mean age: 9.9 years, SD: 0.84); one randomly selected from each twin pair.
EXPOSURE
Genetic predisposition to obesity. We created a polygenic risk score (PRS) comprising 28 common obesity-related single nucleotide polymorphisms identified in a meta-analysis of obesity-related genome-wide association studies.
MAIN OUTCOMES AND MEASURES
Satiety responsiveness was indexed with a standard psychometric scale (the Child Eating Behavior Questionnaire). BMI standard deviation scores (BMI-SDS) and waist-SDS, using 1990 UK reference data, were calculated from parent-reported anthropometric data for the child. Information on satiety responsiveness, anthropometrics and genotype were available for 2258 children. We examined associations between the PRS, adiposity and satiety responsiveness.
RESULTS
The PRS was negatively related to satiety responsiveness (beta, −0.060; 95% CI, −0.019 to −0.101), and positively related to adiposity (BMI-SDS: beta, 0.177; 95% CI, 0.136 to 0.218; waist-SDS: beta, 0.167; 95% CI, 0.126 to 0.208), and more children in the top 25% of the PRS were overweight than in the lowest 25% (18.5% versus 7.2%, respectively; OR, 2.90; 95% CI, 1.98 to 4.25). Associations between the PRS and adiposity were significantly mediated by satiety responsiveness (BMI-SDS: P = 0.006; waist-SDS: P = 0.005).
CONCLUSIONS AND RELEVANCE
These results support the hypothesis that low satiety responsiveness is one of the mechanisms through which genetic predisposition leads to weight gain in an environment rich with food. Strategies to enhance satiety responsiveness could help prevent weight gain in genetically at-risk children.
Keywords: Genetic, appetite, satiety, children, obesity, overweight, single nucleotide polymorphism
BACKGROUND
Obesity is one of the great global health challenges1 not only increasing in prevalence but also developing earlier in life.2 Public health research is making progress in identifying environmental drivers of rising population weights, but less is known about mechanisms underlying individual differences in susceptibility to the `obesogenic' environment.
Weight is under strong genetic influence, with heritability estimates from family, adoption and twin studies averaging over 50%.3;4 Genome-wide Association Studies (GWAS) have identified more than 30 single nucleotide polymorphisms (SNPs) that collectively explain around 1.5% of the variance in adult BMI.5 The majority of these SNPs also show associations with adiposity in children,5 and combined into a polygenic risk score, explained 0.6%–3% of the variance in BMI across different ages in a large pediatric cohort.6
The value of identifying SNPs that influence the risk of complex diseases is not simply to predict disease - indeed their predictive power is often disappointingly low - but to identify causal steps on the path from gene to disease that can be targeted to reduce risk.7;8 The spotlight is most often on intermediate biological processes that could be targets for pharmacotherapy. However, intermediate behavioral processes could also serve as intervention targets.
Our understanding of body weight regulation has been greatly advanced by investigation of rare monogenic forms of obesity. The first mutation to be discovered was a homozygous mutation in the leptin gene, which results in a clinical phenotype characterized by severe early onset obesity and hyperphagia.9 Mutations in the leptin receptor gene, the melanocortin 4 receptor gene, and the pro-opiomelanocortin gene result in similar features.10 All genes associated with severe early-onset obesity are involved in regulation of leptinmelanocortin pathways in the hypothalamus, and are thought to affect body weight largely through impacting appetite.11
The first gene to be linked with common obesity (FTO) is also highly expressed in the hypothalamus,12 and its expression is responsive to short-term variation in energy balance from under- or over-feeding.13 Human studies have linked SNPs in the FTO gene with appetitive characteristics, including higher food intake,14;15 lower satiety responsiveness,16 and dysregulated neurobiological mediators of appetite,17 suggesting that common genetic variants may also influence adiposity via appetitive mechanisms, albeit with considerably smaller effect sizes than the monogenic disorders. Longitudinal studies in children have shown that lower satiety sensitivity is associated with greater weight gain, implicating a causal role in the development of adiposity.18–20 Satiety sensitivity is also highly heritable;21;22 raising the possibility that common variants other than FTO exert their effects on weight through appetitive pathways. At present, the mechanisms through which common obesity-related SNPs influence weight are largely unknown.
The present study used an established psychometric measure of appetite (the Child Eating Behavior Questionnaire) in a large sample of children, to test the hypothesis that satiety responsiveness is associated with polygenic obesity risk and could be an intermediate neurobehavioral process linking genetic risk of obesity to weight gain.
METHODS
STUDY POPULATION
Participants in this study were one randomly selected child from each twin pair from the Twins Early Development Study (TEDS). TEDS is a population-based twin birth cohort of over 16,000 families with twins born in Britain between 1994 and 1996.23 Parents provided informed consent for each part of the study before data collection. Ethical approval was provided by King's College London's Ethics Committee. The sampling frame for this analysis was 5182 families who had taken part in an appetite and weight study in 2006 when the children were approximately 10 years old,24 and 3152 children who had been genotyped for a mathematical and reading ability study in 2010.25 The children included in this analysis were the overlapping children from the two studies (n=2258). The analysis sample was somewhat higher socioeconomic status, and had a slightly lower birth weight than the full sample, but differences were small.
GENOTYPING
In 2010 genome-wide genotyping was carried out for one randomly selected child from each of 3665 TEDS families as part of the Wellcome Trust Case Control Consortium 2 (WTCCC2), to study the genetic basis of reading and mathematical abilities.25 DNA was extracted from buccal swabs, and the Affymetrix 6.0 GeneChip was used to genotype ~1 million SNPs using standard experimental protocols.26 IMPUTE v2 software27 was used to impute approximately 2 million additional SNPs from WTCCC2 controls and HapMap 2 and 3. Stringent quality control resulted in reduction of data to ~1.7 million SNPs for 3152 individuals.26 From these we selected SNPs or their proxies known to increase obesity risk. Proxy SNPs were identified using the SNAP online tool.28
GENETIC PREDISPOSITION SCORE
A polygenic risk score indexing genetic predisposition to obesity was calculated using 28 of the 34 known obesity SNPs from published meta-analyses in adults5 and children,29 of which 24 obesity-risk increasing SNPs were available on the Affymetrix 6.0 GeneChip: rs9939609 (FTO), rs2867125 (TMEM18), rs571312 (MC4R), rs10938397 (GNPDA2), rs10767664 (BDN), rs2815752 (NEGR), rs7359397 (SH2B1) rs3817334 (MTCH2), rs29941 (KCTD15), rs543874 (SEC16B), rs987237 (TFAP2B), rs7138803 (FAIM2), rs10150332 (NRXN3), rs713586 (POMC), rs12444979 (GPRC5B), rs2241423 (MAP2K5), rs1514175 (TNNI3K), rs10968576 (LRRN6), rs887912 (FANCL), rs13078807 (CADM2), rs1555543 (PTBP2), rs206936 (NUDT3), rs9568856 (OLFM4), rs9299 (HOXB5). Another 4 SNPs were indexed using proxy SNPs in high linkage disequilibrium with the original (R2>0.9): rs2112347 (FLJ35779) was indexed using rs3797580 (R2=1); rs4836133 (ZNF608) was indexed using rs6864049 (R2=1); rs4929949 (RPL27A) was indexed using rs9300093 (R2=0.97); rs3810291 (TMEM160) was indexed using rs7250850 (R2=1). For 6 of the 34 obesity-risk increasing SNPs, we neither had genotyped markers nor could find a reliable proxy SNP (R2>0.8): rs2890652 (LRP1B), rs9816226 (ETV5), rs13107325 (SLC39A8), rs4771122 (MTIF3), rs11847697 (PRKD1), rs2287019 (QPCTL).
For each SNP, each participant has a possible score of 0 (no obesity risk-increasing alleles), 1 (1 risk-increasing allele) or 2 (2 risk-increasing alleles). A mean polygenic risk score was created for each child from the 24 genotyped SNPs and 4 proxy SNPs, by summing the total number of obesity risk-increasing alleles and dividing by the total possible number. Possible scores therefore ranged from 0 to 56, with higher scores indicating a greater genetic predisposition to obesity. Weighted mean scores were calculated to take account of differences in effect size by multiplying each SNP by its beta coefficient derived from published meta-analyses.5;29 A second polygenic risk score was calculated that excluded FTO (rs9939609), and a third that excluded both FTO and MC4R (rs571312).
MEASUREMENT OF ADIPOSITY
Adiposity was indexed using BMI standardized scores (BMI-SDS) and waist circumference standardized scores (waist-SDS). In 2006, when the children were 8–11 years old, anthropometric data were collected as part of a study of appetite and adiposity.30 Questionnaires and tape measures were mailed to the parents, along with detailed instructions on measuring their children's height (to the nearest centimeter), weight (to the nearest pound or tenth of a kilogram), and waist circumference (to the nearest centimeter). Parents recorded the date of each measurement. In a subsample of 228 families, the same measurements were made by a researcher at a home visit. Correspondence between parent- and researcher-measured height, weight and waist circumference was high (0.90, 0.83, 0.92).30
Body mass index (BMI) was calculated as weight (kg)/height (m)2. BMI and waist circumference values were converted to BMI-SDS and waist-SDS using 1990 UK growth reference data31 in the program ImsGrowth.32 International Obesity Task Force (IOTF) weight categories were created based on predicted BMI at 18 years of age using UK 1990 reference data:31 `severely underweight' (predicted BMI of <16); `very underweight' (predicted BMI of 16.0 to <17.0); `underweight' (predicted BMI of 17.0 to < 18.5); `healthy weight' (predicted BMI of 18.5 to 24.9); `overweight' (predicted BMI of 25.0 to 29.9); and `obese' (predicted BMI of ≥30). Reference data31 were used to exclude implausible anthropometric values (height: <1.05m or >1.80m; weight: <12 kg or >80 kg; BMI <11 or >32; waist circumference: <44cm or >100cm). BMI-SDS and waist-SDS were residualized for age-and sex-effects prior to analyses.
MEASUREMENT OF SATIETY RESPONSIVENESS
Satiety responsiveness was assessed with a 6-item version of the combined `Satiety Responsiveness/Slowness in Eating' scale (Satiety Responsiveness) from the Child Eating Behavior Questionnaire (CEBQ);33 a parent-report measure of child appetite that has been validated using behavioral measures of food intake.34 Illustrative items are: `my child cannot eat a meal if he or she has had a snack just before', and `my child eats more and more slowly during the course of a meal'. All items were scored using a 5-point Likert scale (`never', `rarely', `sometimes', `often', `always'), and averaged to create a total score. Scores were residualized for age and sex prior to analyses.
EXCLUSIONS
Of 3152 children with genotyping data, 2381 had data on height, weight and waist circumference. All but one also had data on Satiety Responsiveness (n=2380). Children with implausible anthropometric measurements, or who were younger than 8 years of age at the time of measurement were excluded (n=86), along with 36 children with severe medical problems. The final sample for analysis was therefore 2258.
STATISTICAL ANALYSES
Associations between the polygenic risk score, adiposity, and the Satiety Responsiveness score were tested using linear regression analyses. Logistic regression was used to estimate the odds of being overweight or obese in the top 25% of the polygenic obesity risk score, compared to the bottom 25%. The Sobel test35;36 was used to assess whether Satiety Responsiveness significantly mediated the association between the polygenic risk score and adiposity (indexed using BMI-SDS and waist-SDS). Analyses were repeated using the polygenic risk score that excluded FTO, and the polygenic risk score that excluded both FTO and MC4R. All analyses were done in SPSS Version 20.
RESULTS
CHARACTERISTICS OF THE ANALYSIS SAMPLE
Characteristics of the analysis sample are presented in Table 1. The average age of the children was just under 10 years old. Consistent with population data, more were from dizygotic than monozygotic twin pairs (60.6%, 38.9%), and there were slightly more females than males (53.3%, 46.7%).
Table 1.
Characteristics of the analysis sample (n=2258 children)
| Mean (SD) or n (%) | |
|---|---|
| Age (years) | 9.90 (0.84) |
| Sex | |
| Female | 1203 (53.3) |
| Male | 1055 (46.7) |
| Zygosity* | |
| Monozygotic | 878 (38.9) |
| Dizygotic | 1369 (60.6) |
| Weight (kilograms) | 33.27 (7.28) |
| Height (meters) | 1.39 (0.08) |
| BMI †(kilograms/meters2) | 17.03 (2.58) |
| Waist (centimeters) | 62.17 (6.74) |
| BMI-SDS ‡ | −0.02 (1.12) |
| Waist-SDS § | 0.79 (0.96) |
| Weight status ∥ | |
| Severely underweight | 16 (0.7) |
| Very underweight | 41 (1.8) |
| Underweight | 237 (10.5) |
| Healthy weight | 1672 (74.0) |
| Overweight | 241 (10.7) |
| Obese | 51 (2.3) |
| Satiety Responsiveness ¶ | 2.63 (0.67) |
| Number of obesity-risk alleles ** | 21.41 (2.89) |
| Weighted polygenic obesity risk score †† | −0.03 (0.02) |
Opposite-sex twins were classified as dizygotic; the zygosity of same-sex twins was determined using a validated 20-item questionnaire and DNA markers for pairs of questionable zygosity.43
BMI, body mass index.
BMI-SDS, BMI standard deviation score: BMI adjusted for age and sex using UK 1990 reference data.31
Waist-SDS, waist standard deviation score: waist circumference adjusted for age and sex using UK 1990 reference data.31
Weight status was classified using International Obesity Task Force categories which are based on predicted BMI at 18 years of age using UK 1990 growth reference data:31 'severely underweight' (predicted BMI of <16); Very underweight' (predicted BMI of 16.0 to <17.0); 'underweight' (predicted BMI of 17.0 to < 18.5); 'healthy weight' (predicted BMI of 18.5 to 24.9); 'overweight' (predicted BMIof 25.0 to 29.9); and 'obese' ( predicted BMI of ≥30)31.
Satiety Responsiveness: assessed using a 6-item scale from the Child Eating Behavior Questionnaire.33 The possible score ranges from 1–5.
Number of obesity risk alleles from 28 single nucleotide polymorphisms (SNPs) with a possible range of 0–56 alleles.
Mean BMI-SDS (−0.02) indicated that the level of adiposity was close to the UK 1990 reference values. Consistent with this, 13% of the sample (n=294) were underweight, most children were in the healthy weight range for their age and sex (74.0%); and relatively few were overweight (10.7%) or obese (2.3%). Mean waist-SDS was slightly higher than the 1990 reference value. Waist-SDS and BMI-SDS were positively correlated (r=0.77, P<0.001).
The mean CEBQ Satiety Responsiveness score was 2.63, and scores were normally distributed. Satiety Responsiveness significantly predicted both BMI-SDS (beta, −0.229; 95% CI, −0.190 to −0.268) and waist-SDS (beta, −0.244; 95% CI, −0.205 to −0.283).
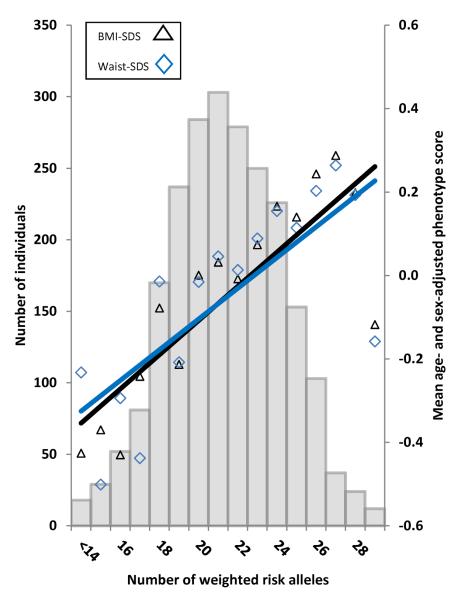
The number of obesity-increasing risk alleles was normally distributed with a mean of 21.41 (range, 11 to 32). The distribution of the polygenic obesity risk scores is shown in Figure 1.
Figure 1.
Regression of mean age- and sex-adjusted BMI-SDS and waist-SDS values across the risk-allele scores. The histogram shows that the number of weighted obesity risk alleles was normally distributed in the sample. The black triangles show the mean age- and sex-adjusted BMI-SDS values across the weighted risk-allele scores; the blue diamonds show the mean age- and sex-adjusted waist-SDS values across the weighted risk-allele scores. The solid black line shows the regression line for age- and sex-adjusted BMI-SDS predicted from the polygenic obesity risk score (R2, 0.031; beta, 0.177; beta 95% CI, 0.136 to 0.218). The solid blue line shows the regression line for age- and sex-adjusted waist-SDS predicted from the polygenic risk score (R2, 0.028; beta, 0.167; beta 95% CI, 0.126 to 0.208).
GENETIC PREDISPOSITION AND ADIPOSITY
As expected, the polygenic obesity risk score showed a linear association with BMI-SDS (beta, 0.177; 95% CI: 0.136 to 0.218) and waist-SDS (beta, 0.167; 95% CI: 0.126 to 0.208) (see Figure 1). The polygenic obesity risk score explained 3.1% of the variance in BMI-SDS and 2.8% of the variance in waist-SDS. More of the children in the top 25% of the polygenic obesity risk score were overweight or obese than in the lowest 25% of the score (18.5% versus 7.2%, respectively; OR, 2.90; 95% CI, 1.98 to 4.25).
GENETIC PREDISPOSITION AND SATIETY SENSITIVITY
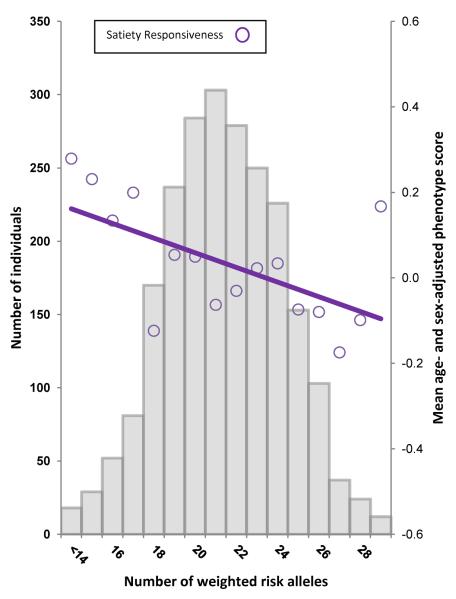
The polygenic obesity risk score showed a linear negative association with Satiety Responsiveness (beta, −0.060; 95% CI, −0.019 to −0.101); explaining 0.4% of the variance in scores (Figure 2).
Figure 2.
Regression of the mean age- and sex-adjusted Satiety Responsiveness values across the risk-allele scores. The histogram shows that the number of weighted obesity risk alleles was normally distributed in the sample. The purple circles show the mean age- and sex-adjusted Satiety Responsiveness scores across the weighted risk-allele scores. The solid purple line shows the regression line for age- and sex-adjusted Satiety Responsiveness predicted from the polygenic obesity risk score (R2, 0.004; beta, −0.060; beta 95% CI, −0.101 to −0.019).
Including Satiety Responsiveness in a multiple regression model to predict BMI-SDS from the polygenic obesity risk score, attenuated the relationship between the polygenic obesity risk score and BMI-SDS (beta from a model without Satiety Responsiveness: 0.177; 95% CI, 0.136 to 0.218; beta from a model including Satiety Responsiveness: 0.164; 95% CI, 0.125 to 0.203; beta Δ −0.013); indicating that Satiety Responsiveness partially mediated the association between genetic obesity risk and adiposity (Figure 3). The Sobel test confirmed significant mediation of the association between polygenic risk and BMI-SDS by Satiety Responsiveness (P=0.006).
Figure 3.
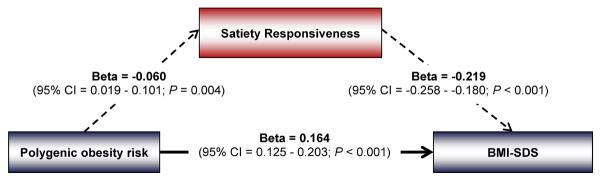
Path diagram showing that Satiety Responsiveness significantly mediates the association between polygenic risk of obesity and BMI-SDS. The path diagram shows the simple association between the polygenic risk score (PRS) and Satiety Responsiveness (beta, −0.060; 95% CI, −0.101 to −0.019), the association between the PRS and BMI-SDS adjusted for Satiety Responsiveness (beta, 0.164; 95% CI, 0.125 to 0.203), and the association between Satiety Responsiveness and BMI-SDS adjusted for the PRS (beta, −0.219; 95% CI, −0.258 to −0.180). The simple association between the PRS and BMI-SDS (beta, 0.177; 95% CI, 0.136 to 0.218) was slightly higher than the association between the PRS and BMI-SDS adjusted for Satiety Responsiveness (beta Δ 0.013), indicating that Satiety Responsiveness mediated part of the association. The Sobel test confirmed that Satiety Responsiveness significantly mediated the association between the PRS and BMI-SDS (P=0.006).
The results were virtually the same for waist-SDS. Including Satiety Responsiveness in the model attenuated the association between the polygenic obesity risk score and waist-SDS (beta from a model without Satiety Responsiveness: 0.169; 95% CI, 0.130 to 0.208; beta from a model including Satiety Responsiveness: 0.153; 95% CI, 0.114 to 0.192; beta Δ −0.016) (Figure 4). Mediation analyses confirmed that Satiety Responsiveness significantly mediated the association between the polygenic obesity risk score and waist-SDS (P=0.005).
Figure 4.

Path diagram showing that Satiety Responsiveness significantly mediates the association between polygenic risk of obesity and waist-SDS. The path diagram shows the simple association between the polygenic obesity risk score (PRS) and Satiety Responsiveness (beta, −0.060; 95% CI, −0.101 to −0.019), the association between the PRS and waist-SDS adjusted for Satiety Responsiveness (beta, 0.153; 95% CI, 0.114 to 0.192), and the association between Satiety Responsiveness and waist-SDS adjusted for the PRS (beta, −0.235; 95% CI, −0.274 to −0.196). The simple association between the PRS and waist-SDS (beta, 0.167; 95% CI, 0.126 to 0.208) was slightly higher than the association between the PRS and waist-SDS adjusted for Satiety Responsiveness (beta Δ 0.016), indicating that Satiety Responsiveness mediated part of the association. The Sobel test confirmed that Satiety Responsiveness significantly mediated the association between the PRS and waist-SDS (P=0.005).
POLYGENIC RISK SCORE WITHOUT FTO
The results were very similar for the polygenic risk score excluding FTO. Associations between the polygenic obesity risk score and BMI-SDS (beta, 0.159; 95% CI, 0.118 to 0.200), waist-SDS (beta, 0.149; 95% CI, 0.108 to 0.190) and Satiety Responsiveness (beta, −0.050; 95% CI, −0.091 to −0.009) were slightly smaller, but remained significant. Mediation analyses confirmed that Satiety Responsiveness also significantly mediated the associations between the polygenic obesity risk score excluding FTO and both BMI-SDS (P=0.019) and waist-SDS (P=0.019).
POLYGENIC RISK SCORE WITHOUT FTO AND MC4R
The results using the polygenic obesity risk score that excluded both FTO and MC4R were also similar. Associations between the polygenic obesity risk score and BMI-SDS (beta, 0.141; 95% CI, 0.010 to 0.182), waist-SDS (beta, 0.135; 95% CI, 0.094 to 0.176; P) and Satiety Responsiveness (beta, −0.042; 95% CI, −0.083 to −0.008) were smaller, but remained significant. However, Satiety Responsiveness just missed the significance level in the mediation analyses for both BMI-SDS (P=0.056) and waist-SDS (P=0.057).
DISCUSSION
In this large sample of 10 year-old children, we confirmed that a polygenic risk score indexing genetic predisposition to obesity was associated with adiposity, but also showed for the first time a significant negative relationship between the polygenic risk score and Satiety Responsiveness. Satiety Responsiveness significantly mediated the association between genetic predisposition to obesity and the two measures of adiposity.
These results are consistent with the hypothesis that one of the mechanisms through which obesity-risk genes influence adiposity is via the appetite regulatory system. This fits with evidence from the monogenic obesity disorders, which, without exception, involve disturbances of appetite leading to severe early-onset obesity.10 The present findings suggest that common obesity-risk SNPs may also exert their effects on weight via appetitive mechanisms.
There is already evidence for an appetitive pathway for FTO's effects on weight14–17 but little is known about the other identified variants. However, some of the risk-increasing SNPs are located in or near genes that regulate neural or peripheral appetitive processes (e.g. MC4R, BDNF, SH2B1, POMC, GIPR), or are linked to genes in which major mutations cause monogenic obesity disorders (e.g. MC4R and POMC).10 Importantly, the association observed in this sample was not explained entirely by FTO, because Satiety Responsiveness also significantly mediated the association between adiposity and a polygenic obesity risk score that excluded FTO, and the effects were very similar excluding both FTO and MC4R. The observed linear association between the polygenic obesity risk score and Satiety Responsiveness supports the hypothesis that each variant contributes a small but additive amount to the individual's level of satiety responsiveness.
A substantial evidence base of prospective studies links impaired satiety mechanisms to excessive weight gain18–20, and bivariate twin analyses are consistent with common genetic pathways underlying satiety responsiveness and weight in infancy.37 This suggests that genetically susceptible individuals have lower satiety responsiveness from very early in life, making them vulnerable to the abundance of highly palatable food in the modern `obesogenic' environment.
The polygenic obesity risk score in this sample explained nearly double the amount of variance in adiposity (~3%) than reported for adults (~1.5%)5; similar to another pediatric study.6 This is consistent with evidence for higher heritability of BMI in pediatric than adult twin analyses,3 and with higher molecular heritability in Genome-wide Complex Trait Analyses.38–40 Genetic tendencies towards weight gain may be more strongly expressed in children because they are less likely than adults to be making deliberate attempts at weight control.
The association between the polygenic obesity risk score and Satiety Responsiveness was small, but this is expected from the size of the association between genetic risk and adiposity itself. As highlighted recently, the value of establishing associations between disease risk variants and intermediate phenotypes lies in illuminating potential causal mechanisms that provide novel intervention targets.7;8 Breakthroughs have been made into Crohn's disease, Type 2 Diabetes and Coronary Heart Disease despite small associations with the intermediate phenotypes identified.8 The present results suggest that satiety responsiveness might be a useful target for obesity prevention or treatment; highlighting the importance of developing methods to upregulate satiety responsiveness.
This study has a number of strengths. Analyzing a pediatric sample with relatively low rates of obesity makes it less likely that lower satiety responsiveness was a result of long-standing obesity. Having two indices of adiposity – BMI and waist circumference -- strengthened the case that the association was with fat rather than lean tissue.
There are also limitations. The data are cross-sectional so it is not possible to draw conclusions about the causal direction for the association between satiety sensitivity and adiposity. However, evidence from longitudinal studies supports a stronger association from satiety sensitivity and subsequent weight gain,18;20 than the reverse pattern. Use of a twin cohort meant that the children were relatively lean, with lower prevalence of overweight and obesity, and higher rates of underweight than contemporary UK national statistics.41;42 However, there was still a good range of adiposity, and lower than average body weight should not impact relationships between genetic risk and adiposity. Anthropometric data in this study were measured by parents so may be less reliable than researcher-measured data; however they were found to be highly reliable in a sub-sample of families where measures were also taken by researchers.30
In summary, these findings support the hypothesis that common obesity-risk genes influence adiposity partly via appetitive mechanisms. This helps explain how environments and genes combine to determine weight gain: individuals who are less responsive to internal satiety cues by virtue of their genetic blueprint may be more likely to eat to excess when confronted by the multiple eating opportunities of the modern `obesogenic' environment, and consequently gain more weight. Satiety responsiveness is therefore a potential target for behavioral or pharmacological interventions.
ACKNOWLEDGEMENTS
Funding/support The Twins Early Development Study (TEDS) is supported by a program grant to Robert Plomin from the UK Medical Research Council (G0901245; and previously G0500079), with additional support from the US National Institutes of Health (HD044454; HD059215). The anthropometric data were collected as part of a grant to Jane Wardle from the UK Biological and Biotechnology Research Council (D19086) with additional support from program grant funding from Cancer Research UK. Genome-wide genotyping was made possible by grants from the Wellcome Trust Case Control Consortium 2 project (085475/Z/08/Z).
Role of the Funder None of the funders had any role in the following: design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions Clare Llewellyn and Maciej Trzaskowski had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Llewellyn, Wardle. Acquisition of data: Plomin, Wardle. Analysis and interpretation of data: Llewellyn, Trzaskowski, van Jaarsveld. Drafting of the manuscript: Llewellyn, Trzaskowski, Wardle. Critical revision of the manuscript for important intellectual content: Llewellyn, Trzaskowski, van Jaarsveld, Plomin, Wardle. Statistical analysis: Llewellyn, Trzaskowski, van Jaarsveld. Obtained funding: Plomin, Wardle. Study supervision: Plomin, Wardle.
Conflict of interest All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest and none were reported.
REFERENCES
- (1).Finucane MM, Stevens GA, Cowan MJ, et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet. 2011;377:557–567. doi: 10.1016/S0140-6736(10)62037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lee JM, Pilli S, Gebremariam A, et al. Getting heavier, younger: trajectories of obesity over the life course. Int J Obes (Lond) 2010;34:614–623. doi: 10.1038/ijo.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Elks CE, den HM, Zhao JH, et al. Variability in the heritability of body mass index: a systematic review and meta-regression. Front Endocrinol (Lausanne) 2012;3:29. doi: 10.3389/fendo.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- (5).Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Belsky DW, Moffitt TE, Houts R, et al. Polygenic risk, rapid childhood growth, and the development of obesity: evidence from a 4-decade longitudinal study. Arch Pediatr Adolesc Med. 2012;166:515–521. doi: 10.1001/archpediatrics.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Fugger L, McVean G, Bell JI. Genomewide association studies and common disease--realizing clinical utility. N Engl J Med. 2012;367:2370–2371. doi: 10.1056/NEJMp1212285. [DOI] [PubMed] [Google Scholar]
- (8).Hingorani AD, Shah T, Kumari M, Sofat R, Smeeth L. Translating genomics into improved healthcare. BMJ. 2010;341:c5945. doi: 10.1136/bmj.c5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Montague CT, Farooqi IS, Whitehead JP, et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387:903–908. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- (10).Barsh GS, Farooqi IS, O'Rahilly S. Genetics of body-weight regulation. Nature. 2000;404:644–651. doi: 10.1038/35007519. [DOI] [PubMed] [Google Scholar]
- (11).O'Rahilly S, Farooqi IS. Human obesity as a heritable disorder of the central control of energy balance. Int J Obes (Lond) 2008;32(Suppl 7):S55–S61. doi: 10.1038/ijo.2008.239. [DOI] [PubMed] [Google Scholar]
- (12).Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Gerken T, Girard CA, Tung YCL, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CNA. An Obesity-Associated FTO Gene Variant and Increased Energy Intake in Children. New Engl J Med. 2008;359:2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- (15).Wardle J, Llewellyn C, Sanderson S, Plomin R. The FTO gene and measured food intake in children. Int J Obes (Lond) 2009;33:42–45. doi: 10.1038/ijo.2008.174. [DOI] [PubMed] [Google Scholar]
- (16).Wardle J, Carnell S, Haworth CM, Farooqi IS, O'Rahilly S, Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. J Clin Endocrinol Metab. 2008;93:3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- (17).Karra E, O'Daly OG, Choudhury AI, et al. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J Clin Invest. 2013 doi: 10.1172/JCI44403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Parkinson KN, Drewett RF, Le Couteur AS, Adamson AJ. Do maternal ratings of appetite in infants predict later Child Eating Behaviour Questionnaire scores and body mass index? Appetite. 2010;54:186–190. doi: 10.1016/j.appet.2009.10.007. [DOI] [PubMed] [Google Scholar]
- (19).van Jaarsveld CHM, Boniface D, Llewellyn C, Wardle J. Appetite and growth: a longitudinal sibling analysis. JAMA Pediatr. 2013 doi: 10.1001/jamapediatrics.2013.4951. In Press. [DOI] [PubMed] [Google Scholar]
- (20).van Jaarsveld CH, Llewellyn CH, Johnson L, Wardle J. Prospective associations between appetitive traits and weight gain in infancy. Am J Clin Nutr. 2011;94:1562–1567. doi: 10.3945/ajcn.111.015818. [DOI] [PubMed] [Google Scholar]
- (21).Carnell S, Haworth CM, Plomin R, Wardle J. Genetic influence on appetite in children. Int J Obes (Lond) 2008;32:1468–1473. doi: 10.1038/ijo.2008.127. [DOI] [PubMed] [Google Scholar]
- (22).Llewellyn CH, van Jaarsveld CH, Johnson L, Carnell S, Wardle J. Nature and nurture in infant appetite: analysis of the Gemini twin birth cohort. Am J Clin Nutr. 2010;91:1172–1179. doi: 10.3945/ajcn.2009.28868. [DOI] [PubMed] [Google Scholar]
- (23).Haworth CM, Davis OS, Plomin R. Twins Early Development Study (TEDS): a genetically sensitive investigation of cognitive and behavioral development from childhood to young adulthood. Twin Res Hum Genet. 2012;16:117–125. doi: 10.1017/thg.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Carnell S, Wardle J. Appetite and adiposity in children: evidence for a behavioral susceptibility theory of obesity. Am J Clin Nutr. 2008;88:22–29. doi: 10.1093/ajcn/88.1.22. [DOI] [PubMed] [Google Scholar]
- (25).Barrett JC, Lee JC, Lees CW, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Trzaskowski M, Eley TC, Davis OS, et al. First Genome-Wide Association Study on Anxiety-Related Behaviours in Childhood. PloS one. 2013 doi: 10.1371/journal.pone.0058676. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Bradfield JP, Taal HR, Timpson NJ, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44:526–531. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wardle J, Carnell S, Haworth CM, Plomin R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am J Clin Nutr. 2008;87:398–404. doi: 10.1093/ajcn/87.2.398. [DOI] [PubMed] [Google Scholar]
- (31).Cole TJ, Freeman JV, Preece MA. Body-Mass Index Reference Curves for the Uk, 1990. Arch Dis Child. 1995;73:25–29. doi: 10.1136/adc.73.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).LMSGrowth PC. 2009. Software for LMS method. [Google Scholar]
- (33).Wardle J, Guthrie CA, Sanderson S, Rapoport L. Development of the Children's Eating Behaviour Questionnaire. J Child Psychol Psych. 2001;42:963–970. doi: 10.1111/1469-7610.00792. [DOI] [PubMed] [Google Scholar]
- (34).Carnell S, Wardle J. Measuring behavioural susceptibility to obesity: validation of the child eating behaviour questionnaire. Appetite. 2007;48:104–113. doi: 10.1016/j.appet.2006.07.075. [DOI] [PubMed] [Google Scholar]
- (35).Mackinnon DP, Fairchild AJ, Fritz MS. Mediation analysis. Annu Rev Psychol. 2007;58:593–614. doi: 10.1146/annurev.psych.58.110405.085542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Preacher KJ, Hayes AF. SPSS and SAS procedures for estimating indirect effects in simple mediation models. Behav Res Methods Instrum Comput. 2004;36:717–731. doi: 10.3758/bf03206553. [DOI] [PubMed] [Google Scholar]
- (37).Llewellyn CH, van Jaarsveld CH, Plomin R, Fisher A, Wardle J. Inherited behavioral susceptibility to adiposity in infancy: a multivariate genetic analysis of appetite and weight in the Gemini birth cohort. Am J Clin Nutr. 2012;95:633–639. doi: 10.3945/ajcn.111.023671. [DOI] [PubMed] [Google Scholar]
- (38).Llewellyn CH, Trzaskowski M, Plomin R, Wardle J. Finding the missing heritability in paediatric obesity: the contribution of genome-wide complex trait analysis. Int J Obes (Lond) 2013 doi: 10.1038/ijo.2013.30. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Vattikuti S, Guo J, Chow CC. Heritability and genetic correlations explained by common SNPs for metabolic syndrome traits. PLoS Genet. 2012;8:e1002637. doi: 10.1371/journal.pgen.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Yang J, Manolio TA, Pasquale LR, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Whitaker KL, Jarvis MJ, Beeken RJ, Boniface D, Wardle J. Comparing maternal and paternal intergenerational transmission of obesity risk in a large population-based sample. Am J Clin Nutr. 2010;91:1560–1567. doi: 10.3945/ajcn.2009.28838. [DOI] [PubMed] [Google Scholar]
- (42).Whitaker KL, Jarvis MJ, Boniface D, Wardle J. The intergenerational transmission of thinness. Arch Pediatr Adolesc Med. 2011;165:900–905. doi: 10.1001/archpediatrics.2011.147. [DOI] [PubMed] [Google Scholar]
- (43).Price TS, Freeman B, Craig I, Petrill SA, Ebersole L, Plomin R. Infant zygosity can be assigned by parental report questionnaire data. Twin Res. 2000;3:129–133. doi: 10.1375/136905200320565391. [DOI] [PubMed] [Google Scholar]