Abstract
Following its use in the initial characterization of an acute myeloid leukemia (AML) genome, next generation sequencing (NGS) has continued to molecularly refine the disease. Here we review the spectrum of NGS applications that have subsequently delineated the prognostic significance and biological consequences of these mutations. Further, we discuss the technology’s role in providing a high-resolution glimpse of AML clonal heterogeneity, which may inform future choice of targeted therapy. Though obstacles remain in applying these techniques clinically, they have already impacted patient care.
Keywords: Acute Myeloid Leukemia, Genomics, High-Throughput Nucleotide Sequencing, Epigenomics, Gene Expression Profiling
Introduction
Most acute myeloid leukemia (AML) patients are cytogenetically normal (CN-AML). Their variable overall survival suggests clinical and biologic heterogeneity and a need for additional biomarkers. As many of these patients have no detectable copy number alterations, mutations in the diploid genome are likely pathogenic. Hence, lesions driving disease progression in patients without known recurrent mutations1 may be discovered by whole-genome (WGS) and whole-exome (WES) sequencing. Indeed, the decade following the sequencing of the first cancer genome,2 that of an individual presenting with CN-AML, has seen an explosion of this technology3 that has advanced our understanding of AML through the discovery of mutations in DNMT3A4 and IDH15 and the recent finding6 that nearly all patients harbor at least one likely pathogenic mutation.
The prognostic significance and functional consequences of these and other recently discovered mutations are being elucidated. Translating these results to the clinic and assessing the impact of “targeted therapies” may further require determining whether mutations reside in subclones and how the latter evolve over time or following treatment. Just as WGS and WES have facilitated mutation discovery, these subsequent challenges are being addressed by a diverse array of next-generation sequencing technologies, including transcriptome, methylome, and targeted, custom capture sequencing.
Mutation Discovery
The capillary electrophoresis of individual fluorescently-labeled Sanger reaction products used to originally sequence the human genome over ten years at an expense of several billion dollars have since given way to “next-generation” sequencing (NGS) platforms. These can resequence the human genome at a fraction of the original cost (~$10,000 per tumor germ line pair) in 4–6 weeks. These platforms require a library preparation phase, in which synthetic DNA (adapters) are ligated onto the ends of the fragmented DNA to be sequenced. The fragments are then subjected to a polymerase-mediated reaction, whose intermediate products can be monitored in real time by the platform’s optical instruments. For example, the Illumina platform detects the incorporation of fluorescently-labeled nucleotides during an amplification reaction, with noninterference between contiguous nucleotides ensured by a step-wise reaction in which an incorporated nucleotide’s 3′ blocking group prevents further extension until its dye has been detected and its blocking group removed by chemical cleavage. These platforms provide a nucleotide resolution view of the genome that, for example, yields precise breakpoints of chromosomal rearrangements coarsely detected by less sensitive approaches, such as spectral karyotyping. The reads have characteristic lengths [generally from 25 to 100 base pairs (bp)] and error profiles (often reflecting the error-prone polymerase) that are platform dependent. These impact a platform’s suitability for a particular application [e.g., the short reads and greater depth of the Illumina platform are suited for quantitating single nucleotide variants (SNVs), while the longer reads of the Pacific Biosciences RS platform facilitate discovery of structural variants (SVs)].
Whole-Genome Sequencing (WGS)
WGS provides comprehensive DNA sequencing of the entire genome. As such, Ley and colleagues chose7 it to elucidate the unknown initiating event in tumors from two CN-AML patients through a series of studies.2, 4, 5 These led to the discovery of mutations in IDH1 and DNMT3A and have been extensively reviewed elsewhere.7–11 Additionally, they established several paradigms that have guided subsequent genomics studies in hematological12, 13 and solid tumors, including validation of sequencing results using an orthogonal platform and comparison of tumor and matched normal samples from the same patient to discover acquired, somatic variants. When matched normals are not available, putative variants may instead be filtered if they occur in a cohort of (unmatched) normal samples or are annotated in single nucleotide polymorphism (SNP) databases.
In the latest of the trio of studies5, paired-end sequencing from both ends of a DNA fragment, as opposed to single reads, provided greater genomic context and facilitated alignment of reads to the reference genome. This, coupled with maturing variant calling algorithms14 that analyze mapped reads to infer SNVs in the presence of sequencing errors, (mis) alignment artifacts, and tumor/normal contamination, dramatically improved variant-calling false positive rates. These advances accentuate the innate capability of WGS to characterize the full range of mutations, including intronic and exonic SNVs, insertions and deletions (indels), copy number alterations (CNAs), and SVs (including fusions/translocations).15
Whole-Exome Sequencing (WES)
Since mutations effecting protein function are likely within coding exons, sequencing the exome (i.e., the coding exons of annotated genes) via WES is a cost-effective alternative to WGS. WES targets exons via a capture-based library preparation phase using probes whose length, number, and exonic targets vary across platforms.16 This approach captures regions flanking the probes (e.g., ~100bp, depending on fragment length), including those in introns and untranslated regions (UTRs). However, promoters, enhancers, and intronic spicing silencers or enhancers far outside these targeted regions will not be sequenced. Further, some regions, particularly those with extreme GC content.6, 16, 17 are difficult to capture and hence are underrepresented. Nevertheless, WES is attractive in limiting analysis to the ~1–2% of the genome most likely to be of pathogenic interest.
Several groups have applied WES to rationally-selected genotypes to minimize inter-sample heterogeneity and to enrich for subtype-specific mutations. This approach identified mutations in DNMT3Ain acute monocytic leukemia,18 in BCOR in a CN-AML case molecularly screened to be free of known oncogenic mutations in NPM1, CEBPA, FLT3, or MLL,19 and in GATA2 in CN-AML patients with biallelic CEBPA mutations.20 WES and WGS have also discovered mutations in the splicing factors SF3B1, SRSF2, and U2AF1in MDS (reviewed in21). Recurrent mutations of the spliceosome, including in U2AF1, have subsequently been discovered in AML using both platforms.6
Transcriptome Sequencing (RNA-seq)
Unbiased sequencing of the transcriptome (RNA-seq) offers several advantages22 with respect to WGS and WES: it facilitates discovery of novel transcripts and of alternative splicing events and trans-splicing23 or read-through15 fusion events that can not be detected from genomic DNA. It reduces false positives by enriching for expressed transcripts and their variants, which are more likely to be pathogenic, and quantitates this expression digitally. However, it is ineffective in discovering mutations that destabilize transcripts [e.g., by inducing nonsense mediated decay (NMD)24] or are rarely expressed. Further, RNA-seq involves additional complexity during the preparation of cDNA sequencing libraries from RNA, 25 including the need to cope with potential degradation of unstable RNA and the sequence and structural dependence of cDNA synthesis and hybridization.22 Variations in this step accommodate different downstream analyses. For example, polyadenylation fractionation enriches for expressed mRNA relative to non-coding RNA, whereas size selection of unfractionated total RNA enriches for microRNA (miRNA).
Several groups26–30 have used RNA-seq to discover somatic mutations in AML at considerably reduced cost and effort relative to WGS. Greif et al.26 found mutations in RUNX1, TLE4, and SHKBP1; McNerney et al.27 discovered that CUX1 on chromosome 7q was expressed at haploinsufficient levels in monosomy 7/del(7q) de novo and therapy-related AML samples; Wen et al.28 found seven novel fusions specific to CN-AML and an additional CIITA-DEXI fusion that occurred in 48% (14/29) of CN-AML samples; Masetti et al.29 discovered a recurrent CBFA2T3-GLIS2 fusion in three of seven childhood CN-AML patients; and Walter et al.30 identified a novel ITGA5 splice variant as a potential relapse risk factor by RNA sequencing of relapsed patients who had been classified as low risk based on known cytogenetic and molecular markers.
Ramsingh et al.31 characterized miRNA expressed in a CN-AML patient by sequencing size-selected cDNA using the SOLiD platform and discovered outlying expression of miR-233 beyond the dynamic range of miRNA microarrays and RT-PCR. Subsequent miRNA-seq studies have detected six miRNA biomarkers in circulating blood that differentiate AML patients from controls32 and have uncovered two miRNAs whose loss leads to leukemia-related diseases in mice.33
Frequency and Prognostic Significance of Mutations
Discovered somatic variants are frequently validated on an orthogonal platform, e.g., mutations detected on the Illumina platform may be validated using custom primer amplification followed by direct sequencing2 or NGS on the Roche GS FLX system.5 Alternatively, deep read count scan provide validation. Deep amplicon sequencing, i.e., amplification using custom-designed PCR primers followed by deep NGS, is one such approach. Another involves liquid hybridization capture using custom sequence probes designed to cover the region of interest (e.g., spanning an SNV, an indel, or all exons within a gene) and subsequent deep NGS.34, 35 The scalability of custom probe approaches is attractive when validating many variants.
Both amplicon- and capture-based strategies are useful in defining mutation frequencies across a large cohort and in clinical correlation studies.1 Ampliconpyro sequencing has been extensively used36–38 to determine the clinical significance of TET2 mutations,39 though the findings are inconsistent.40 One study found little correlation between outcome and TET2 mutations in MDS,36 while another revealed their correlation with inferior event-free survival in de novo CN-AML cases37, 38 and particularly in the European Leukemia Net favorable-risk subgroup.38 Further, amplicon sequencing using theIllumina41 and Roche42–44 platforms has associated SF3B1 mutations with MDS characterized by ring sideroblasts and a good clinical outcome.
Many groups have demonstrated frequent recurrent mutations in multiple genes that impact prognosis. These include mutations and deletions of TP53 in cytogenetically complex AML,45 MLL fusions,46 and FLT3-ITD (internal tandem duplication), FLT3-TKD (SNVs in tyrosine kinase domain),47 DNMT3A48–53 and ASXL154, 55 mutations in intermediate risk and/or CN-AML, all of which have been associated with poor outcomes in these cytogenetic subsets. However, even when large-scale patient populations are studied by different investigators, consistent correlations are not always observed. For example, not all retrospective studies have associated either the most common R882 and/or non-R882 DNMT3A mutations with poor outcome.53
Frequently conflicting results between correlation studies may be attributed to several factors. For example, secondary mutations may modulate the effect of another mutation, as frequently seen in patients with mutations in both SF3B1 and DNMT3A that have improved survival relative to patients with DNTM3A mutations alone.56 Further, Damm et al.57 note that their finding that SF3B1 mutations had no effect on OS or leukemic progression, in contrast to other studies, may reflect the low-risk cohort of their study. Finally, additional heterogeneity between or within studies may be introduced by different treatment regimens.57 Reliable clinical associations thus require multivariate analyses incorporating diverse mutations and other prognostic indicators or analyses restricted to molecularly homogeneous populations. Such studies could be conveniently accommodated by custom capture-based approaches.
Functional and Biological Consequences of Mutations
Bioinformatic analysis is useful in narrowing the sea of mutations discovered from large-scale genomic surveys to “driver” mutations likely significant in disease onset and progression. This winnowing out of irrelevant “passenger” mutations is partially achieved by the etiology of AML. Sequencing studies across cancer types have revealed that AML has a relatively low mutation burden (Fig 1), which increases likelihood of discovering pathogenic mutations in AML. Since “cancer genes” are often mutated across multiple cancer types, their facilitated discovery has relevance beyond AML.
Fig 1.
AML has a reduced mutation burden relative to other cancer types. Labels indicate TCGA cancer codes (https://tcga-data.nci.nih.gov/datareports/codeTablesReport.htm). Mbp: million base pairs.
Genes58, 59 and pathways60 mutated at a statistically significant rate above background mutation rates may be inferred from large-scale studies. Other approaches search for recurrently mutated subnetworks within protein-protein interaction networks61, 62 or integrate genome-wide expression and mutation data to probabilistically infer perturbations in annotated pathways.63 Candidate mutations for subsequent experimental study also include those that occur at high frequency in other cancers,64 are within annotated functional domains65 or conserved regions,66 or are predicted to disrupt protein function.67 These may be further prioritized through integrated exploration of multidimensional cancer genomics data, including that describing SNVs, CNAs, DNA methylation, and mRNA, protein, and phosphoprotein expression.68
Whether a mutated gene should be overexpressed or knocked down, for example, to assess its functionality is dependent on whether the lesion is likely a gain-of-function mutation in an oncogene or a loss-of-function mutation in a tumor suppressor. This distinction may be informed by the type and distribution of mutations within the gene and of the distribution of the mutation within the genome. Several types of mutations introduce premature termination codons, including nonsense point mutations and inopportune frame-shift indels, which are likely to destabilize the transcript. The scattering of these and/or consensus splice site mutations throughout a gene’s coding region, as has been detected in BCOR19, TET2,40 and the splicing factor ZRSR2,41, 69 are a likely indicator that it acts a tumor suppressor. Homozygous deletions or the detection of the mutation within a recurrently deleted chromosomal region, as in the CUX1 study,27 provide further evidence of a tumor suppressor role. In contrast, the clustering of missense mutations within hotspots, particularly within known functional domains or conserved regions, is more suggestive of a gain-of-function mutation. These characteristics are shared by mutations in U2AF1 (preferentially targeting residues within zinc finger domains) and SF3B1 (preferentially targeting residues within conserved70 HEAT domains). An oncogenic role is further supported for a gene having a missense mutation within a recurrently amplified region. For example, to enrich for potential oncogenes and tumor suppressors, Dolnik et al.71 designed custom DNA capture probes targeting the coding exons of 1000 genes detected within minimally deleted/gained regions via SNP analysis and used subsequent NGS to reveal mutations in RAD21 within amplified regions.
The above approaches are particularly useful in planning experimental studies for a gene with unknown function. However, several classes of AML-relevant mutations occur in genes with ascribed function, including in epigenetic regulators and splicing factors. For these, NGS approaches can conveniently assay downstream effects of the mutations.
Effects of Mutations in Epigenetic Modifiers
The epigenome is frequently dysregulated in cancer.72 Indeed, epigenetic modifiers are recurrently mutated in AML and MDS,73 including the DNA methyltransferase DNMT3A, the methylcytosine dioxygenase TET2, and the isocitrate dehydrogenases IDH1 and IDH2discussed above, as well as the polycomb-associated ASXL1 and the methyltransferases EZH2 and MLL.74 Gain-of-functionIDH1/2 mutations75 antagonize TET2 function76 and mutations in TET2, IDH1, and IDH2 are mutually exclusive.40 These mutations have clinical significance74 and the DNA methyltransferase inhibitors azacytidine and decitabine induce clinically significant complete and partial responses in patients with MDS and low-blast count AML.77 As such, array-based strategies have been used to characterize their downstream effects genome-wide and have detected a global, predominantly hyper-, methylation pattern induced by IDH1/26, 76 and TET276 mutations and largely shared76 between them.
Recent methods replace the array-based detection of earlier techniques with NGS for nucleotide resolution and reduced bias.78 and are based on one of three approaches: methylcytosine-sensitive restriction digestion, bisulfite conversion (possibly preceded by methyl-insensitive digestion), or immunoprecipitation (IP).78 DREAM, 79 an example of the first technique, performs serial digestion with methylation-sensitive SmaI and –insensitive XmaI restriction enzymes. Both target the same CCCGGG sequence, though the former is blocked by CpG methylation and leaves 5′-GGG blunt ends, while the latter cuts any remaining target sequences and leaves a 5′-CCGGG overhang, which acts as a unique signature for methylated sites. The second approach, sodium bisulfite treatment, converts unmethylated cytosine to uracil, but leaves methylated cytosine intact. To enrich for DNA that can be (differentially) methylated, Reduced Representation Bisulfite Sequencing (RRBS)80 cuts bisulfite-treated DNA at CCGG sequences using the methylation-insensitive MspI restriction enzyme and size selects fragments to ensure the presence of at least two such sites within a defined (e.g., 300bp) sequence span. Finally, IP-based approaches are typified by MeDIP-seq, 81 which uses an antibody directed against 5mC to immunoprecipitate methylated genomic regions. MethylCap-seq is a related approach that enriches for methylated DNA through capture with methyl-binding protein (MBD2).82 In all four techniques, the resulting DNA libraries are characterized by NGS.
Leukemic subtypes segregate according to differentially methylated regions, detected by MeDIP-seq, not only in promoters, as might be anticipated, but also in gene bodies, CpG islands (CGIs; inside and outside promoters) and CGI shores.83 These subtypes also clustered according to differential methylation in satellites, long terminal repeats, and in short (SINEs) and long (LINEs) interspersed nuclear elements. A toggling of methylation status in these repetitive regions between normal and leukemic blood cells was observed using DREAM analysis: sites with significant hypermethylation in normal cells tended to show significant hypomethylation in leukemia samples and vice versa.79 Mutation- and drug-specific effects have also been described. Loss of DNTM3A in hematopoietic stem cells (HSCs) induced hyper- and hypo-methylation in CpG dinucleotides, as detected by RRBS.84 This loss instead leads to predominant hypomethylation following differentiation to B cells, as assayed by DREAM.82 IDH1/2mutations lead to a marked increase in hyper- relative to hypo-methylated sites in mutants relative to controls, with an enrichment in promoter regions and CpG islands near transcription start sites (TSSs) detected by an enhanced RRBS.85 Finally, decitabine treatment was found using MethylCap-seq to significantly reduce global methylation, particularly in chromosome subtelomeric regions, possibly suggesting a region-specific mechanism of drug action.82
Mutations to epigenetic modifiers are anticipated to affect transcription and chromatin state, consequences which have been investigated using chromatin IP followed by NGS (ChIP-seq). ChIP-seq using antibodies targeting H3K27me3 revealed a significant reduction in genome-wide H3K27me3 TSS occupancy following ASXL1 knockdown.86 Specific loss of H3K27me3 at the posterior HOXA cluster, which is known to contribute to myeloid transformation, suggest that ASXL1 mutations promote transformation by relieving gene repression. Saeed et al., 87 also using ChIP-seq, identified accessible genome regions to which the oncofusion proteins AML1-ETO and PML-RARA bind.
Effects of Mutations in Splicing Factors
Minigene reporter assays of splicing mutations88 have been extended genome-wide using gene expression arrays in expectation that the mutations’ impaired ability to recognize the 3′ splice site will perturb gene expression. For example, splicing-sensitive arrays indicated that exons were significantly downregulated and introns were significiantly upregulated (i.e., unspliced) following U2AF1 mutant expression.41 RNA-seqprovides a more comprehensive and quantitative alternative to gene and exon arrays. Yoshida et al.41 validated their above finding by observing increased read counts in likely intronic regions within U2AF1 mutant samples, while Makishima et al.89 found these mutations perturbed TET2 splicing. To determine the role of modulated splicing in AML, Przychodzen et al.90 analyzed publically available RNA-seq data from The Cancer Genome Atlas (TCGA; https://tcga-data.nci.nih.gov/tcga) in their comparison of six samples with U2AF1 mutations in tAML or sAML and 14 WT samples. 35 exons found to have significantly altered expression were predominantly skipped, including genes involved in mitosis and RNA processing. In contrast, SF3B1 mutations assayed by RNA-seqinduced a high percentage of exon retention in RARS patients.91
Clonal Evolution
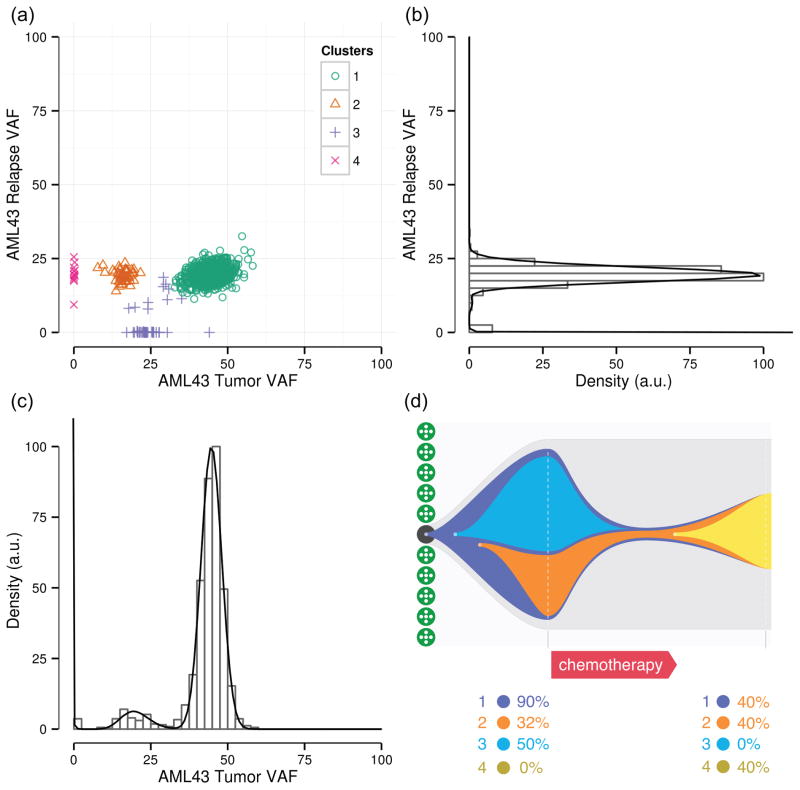
NGS has revealed that most AML tumors are oligoclonal. Variant discovery by WES and their subsequent capture-based targeted deep sequencing in hematopoietic stem/progenitor cells (HSPCs) from healthy donors indicate that the cells accumulate random mutations during aging.35 If a HSPC transforms to a leukemic blast, these passenger mutations are “captured” in its progeny as it clonally expands and serve as a genetic signature identifying the clone. In particular, the variant allele frequency (VAF, or ratio of reads supporting the variant to total reads at the locus) acts as a molecular clock indicating when in the clonal hierarchy the mutation was acquired: heterozygous, clonal mutations within a pure sample have a VAF of ~50%, whereas subclonal mutations, acquired later, are present in fewer cells and have lower VAFs. Aggregations of VAFs thus reflect clones (Fig. 2). Since, both passenger and driver mutations are valuable clonal markers, the most comprehensive perspective on clonal architecture is provided by first discovering variants using WGS and then quantifying VAFs using deep, targeted sequencing.34, 92, 93 Discovering clonal makers with WES35, 94, 95 or even through limited candidate gene resequencing96 have also revealed subclonal architecture, though their more limited number relative to WGS almost certainly (further) underestimates clonal heterogeneity. Hence, WGS-based discovery is particularly important for diseases with few protein coding mutations, such as AML, whereas WES may suffice to assess clones in diseases with high mutation burden, such as melanoma and chronic lymphocytic leukemia (CLL).95. Additionally, sensitivity in detecting low-frequency clones is improved with increased sequencing depth. For example, a sequencing depth of 100× is likely to detect VAFs as low as ~4% (95% binomial confidence interval).
Fig 2.
Subclone escapes therapy to expand into relapse clone. (a) Simultaneous (SciClone clustering93) analysis of tumor and post-treatment relapse samples34 reveals a subclone that was eradicated by therapy (cluster 4) and another that evaded therapy and became dominant during relapse. (b) Analysis of relapse only VAFs can not disambiguate subclones corresponding to clusters 3 and 5 from founding clone (cluster 2). (c) Analysis of tumor only VAFs. (d) Clonal evolution inferred from (a).
A study of tumor/relapse pairs has shown two patterns of clonal evolution during AML relapse: (1) the founding clone in the primary tumor gained additional mutations and evolved into the relapse clone or (2) a subclone escaped therapy, gained additional mutations, and expanded into the relapse clone (Fig 2).34 A likely model describing this subclonal evolution, in which AML develops through serial acquisition of mutations in HSPCs, has been supported by isolating and sequencing preleukemic HSPCs, found to have a subset of mutations of their leukemic progeny.94 A similar persistence of mutations from an antecedent disease accompanied by acquisition of additional mutations during progression to AML has been observed in serial studies tracking evolution from severe congenital neutropenia97 or MDS92 to AML. However, recurrently mutated genes display a wide range of VAFs across sAML samples, indicating that none are consistently associated with the founding clone and that the disease progresses through a variety of acquired mutations across patients.93
Clonal heterogeneity is frequently discernible from a single tumor sample. However, multiple samples derived from a single patient are often required to disambiguate subclones overlapping in the original sample and to appreciate the tumor’s complexity (Fig 2). These may be relapse samples, physically-isolated biopsies taken at the same time point, or tumor cells exposed to some manipulation (e.g., passage in culture or through immunodeficient mice) that could induce a distinct (fitness) phenotype in a subpopulation of cells. Characterizing98 this intra-tumor heterogeneity and clonal architecture is important as they may have clinical implications99 and contribute to therapy resistance.100 For example, subclonal mutations may be correlated with poor clinical outcome in CLL.95
Clinical Application of NGS
Current genetic testing in AML is inadequate to detect the clinically-relevant mutations of this heterogeneous disease.101 Metaphase cytogenetics and FISH lack resolution, while Sanger-based sequencing is cost and time prohibitive. Mass spectrometry genotyping identifies mutations at specific residues (e.g., in N/KRAS and IDH1/2), but is unable to detect mutations scattered throughout the gene body (e.g., in tumor suppressors TP53, TET2, and ASXL1). Further, genes such as N/KRAS102 and DNMT3A,4 which are expected to be targeted at well-characterized residues, have exhibited noncanonical, but oncogenic, mutations that would have been missed by SNP-directed approaches.102 In the latter case, up to 40% of patients with DNMT3A mutations harbor non-R882 mutations, which are also associated with poor outcome.4 Comprehensive WGS is an attractive diagnostic platform, but suffers from high cost and analysis time and moderate- to low-coverage insensitive to low-frequency, subclonal mutations. Deep-coverage, targeted NGS of panels of candidate genes ameliorates these concerns and leverages the community’s investment in large-scale sequencing efforts, particularly through TCGA, in cataloging somatic cancer mutations. For example, a “pan-cancer” panel assays the entire coding sequence and selected introns of 236 cancer-related genes (www.foundationone.com). A single hybrid-capture NGS platform improves efficiency and scalability over a variety of disparate methods otherwise required to discover the full spectrum of mutations active in AML, including translocations, SNVs, and indels.103 Further, the deep read counts of targeted NGS are well suited to minimal residual disease monitoring.104
Technical challenges remain, however, including the inefficient capture of GC-imbalanced targets such as CEBPA103, 105 and potentially limited genomic DNA, though whole-genome amplification may address the latter without introducing appreciable bias.106 Bioinformatic analysis14 is a bottleneck in the clinical sequencing pipeline and software tools need to be validated.107 For example, alignment is computationally demanding, potentially sensitive to noisy reads, and problematic in repetitive regions.108 Fully leveraging NGS clinically will require carefully considered informed consent. For example, WGS of a patient with multiple primary tumors revealed the presence of a cancer susceptibility, germline mutation in TP53 with clinical implications for the patient’s children.109 Because the informed-consent document included a provision to communicate clinically relevant information to family members, the treating physician contacted the next of kin to inform them of the mutation and to encourage genetic counseling. In a second case, WGS detected a cryptic fusion within a clinically relevant timeframe, which impacted the patient’s treatment plan.110 Diagnosis was facilitated by a “movable firewall” within the IRB-approved protocol that maintained the patient’s anonymity, yet allowed the research team to communicate relevant findings to the treating physician.
Despite the remaining obstacles, the two above cases demonstrate the clinical impact of NGS. These successes and earlier studies have significantly advanced the state of the art in clinical diagnosis, with at least ten open clinical trials utilizing NGS (clinicaltrials.gov; keywords: “next generation sequencing” and “cancer”). Further, large, clinically-annotated data sets have already been accumulated by prior studies and offer a rich opportunity for retrospective analysis. As technological trends continue to reduce sequencing cost, integrated6 clinical analysis and sequencing of the genome, transcriptome, and methylome, coupled with queries to drug-gene interaction databases (http://dgidb.genome.wustl.edu), will become a practical approach to comprehensively interrogating and treating leukemia.
Acknowledgments
Funding source: NIH/NCI P01 CA101937 (JFD); NIH/NCI R01 CA152329 (JFD); Barnes-Jewish Hospital Foundation, Award #7603-55 (BSW)
The authors wish to thank Joshua McMichael for assistance with figure design.
Footnotes
Finanical disclosures: None
References
- 1.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–89. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mardis ER. A decade’s perspective on DNA sequencing technology. Nature. 2011;470(7333):198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 4.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter MJ, Graubert TA, Dipersio JF, Mardis ER, Wilson RK, Ley TJ. Next-generation sequencing of cancer genomes: back to the future. Per Med. 2009;6(6):653. doi: 10.2217/pme.09.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao AV, Smith BD. Are results of targeted gene sequencing ready to be used for clinical decision making for patients with acute myelogenous leukemia? Curr Hematol Malig Rep. 2013;8(2):149–55. doi: 10.1007/s11899-013-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graubert TA, Mardis ER. Genomics of acute myeloid leukemia. Cancer J. 2011;17(6):487–91. doi: 10.1097/PPO.0b013e31823c5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Link DC. Molecular genetics of AML. Best Pract Res Clin Haematol. 2012;25(4):409–14. doi: 10.1016/j.beha.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welch JS, Link DC. Genomics of AML: clinical applications of next-generation sequencing. Hematology Am Soc Hematol Educ Program. 2011;2011:30–5. doi: 10.1182/asheducation-2011.1.30. [DOI] [PubMed] [Google Scholar]
- 12.Merker JD, Valouev A, Gotlib J. Next-generation sequencing in hematologic malignancies: what will be the dividends? Ther Adv Hematol. 2012;3(6):333–9. doi: 10.1177/2040620712458948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braggio E, Egan JB, Fonseca R, Stewart AK. Lessons from next-generation sequencing analysis in hematological malignancies. Blood Cancer J. 2013;3:e127. doi: 10.1038/bcj.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shyr D, Liu Q. Next generation sequencing in cancer research and clinical application. Biol Proced Online. 2013;15(1):4. doi: 10.1186/1480-9222-15-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maher CA, Palanisamy N, Brenner JC, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106(30):12353–8. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sulonen AM, Ellonen P, Almusa H, et al. Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol. 2011;12(9):R94. doi: 10.1186/gb-2011-12-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark MJ, Chen R, Lam HY, et al. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29(10):908–14. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan XJ, Xu J, Gu ZH, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43(4):309–15. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 19.Grossmann V, Tiacci E, Holmes AB, et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood. 2011;118(23):6153–63. doi: 10.1182/blood-2011-07-365320. [DOI] [PubMed] [Google Scholar]
- 20.Greif PA, Dufour A, Konstandin NP, et al. GATA2 zinc finger 1 mutations associated with biallelic CEBPA mutations define a unique genetic entity of acute myeloid leukemia. Blood. 2012;120(2):395–403. doi: 10.1182/blood-2012-01-403220. [DOI] [PubMed] [Google Scholar]
- 21.Ogawa S. Splicing factor mutations in myelodysplasia. Int J Hematol. 2012;96(4):438–42. doi: 10.1007/s12185-012-1182-y. [DOI] [PubMed] [Google Scholar]
- 22.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12(2):87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Walsh CE. Spliceosome-mediated RNA trans-splicing. Mol Ther. 2005;12(6):1006–12. doi: 10.1016/j.ymthe.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13(11):700–12. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greif PA, Eck SH, Konstandin NP, et al. Identification of recurring tumor-specific somatic mutations in acute myeloid leukemia by transcriptome sequencing. Leukemia. 2011;25(5):821–7. doi: 10.1038/leu.2011.19. [DOI] [PubMed] [Google Scholar]
- 27.McNerney ME, Brown CD, Wang X, et al. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood. 2013;121(6):975–83. doi: 10.1182/blood-2012-04-426965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen H, Li Y, Malek SN, et al. New fusion transcripts identified in normal karyotype acute myeloid leukemia. PLoS One. 2012;7(12):e51203. doi: 10.1371/journal.pone.0051203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masetti R, Pigazzi M, Togni M, et al. CBFA2T3-GLIS2 fusion transcript is a novel common feature in pediatric, cytogenetically normal AML, not restricted to FAB M7 subtype. Blood. 2013;121(17):3469–72. doi: 10.1182/blood-2012-11-469825. [DOI] [PubMed] [Google Scholar]
- 30.Walter RB, Laszlo GS, Alonzo TA, et al. Significance of expression of ITGA5 and its splice variants in acute myeloid leukemia: A report from the children’s oncology group. Am J Hematol. 2013 doi: 10.1002/ajh.23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsingh G, Koboldt DC, Trissal M, et al. Complete characterization of the microRNAome in a patient with acute myeloid leukemia. Blood. 2010;116(24):5316–26. doi: 10.1182/blood-2010-05-285395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhi F, Cao X, Xie X, et al. Identification of circulating microRNAs as potential biomarkers for detecting acute myeloid leukemia. PLoS One. 2013;8(2):e56718. doi: 10.1371/journal.pone.0056718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starczynowski DT, Morin R, McPherson A, et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood. 2011;117(2):595–607. doi: 10.1182/blood-2010-03-277012. [DOI] [PubMed] [Google Scholar]
- 34.Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–10. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–78. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith AE, Mohamedali AM, Kulasekararaj A, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116(19):3923–32. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 37.Bacher U, Weissmann S, Kohlmann A, et al. TET2 deletions are a recurrent but rare phenomenon in myeloid malignancies and are frequently accompanied by TET2 mutations on the remaining allele. Br J Haematol. 2012;156(1):67–75. doi: 10.1111/j.1365-2141.2011.08911.x. [DOI] [PubMed] [Google Scholar]
- 38.Weissmann S, Alpermann T, Grossmann V, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2012;26(5):934–42. doi: 10.1038/leu.2011.326. [DOI] [PubMed] [Google Scholar]
- 39.Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell. 2011;9(3):193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdel-Wahab O, Patel J, Levine RL. Clinical implications of novel mutations in epigenetic modifiers in AML. Hematol Oncol Clin North Am. 2011;25(6):1119–33. doi: 10.1016/j.hoc.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–9. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 42.Papaemmanuil E, Cazzola M, Boultwood J, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384–95. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malcovati L, Papaemmanuil E, Bowen DT, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118(24):6239–46. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mian SA, Smith AE, Kulasekararaj AG, et al. Spliceosome mutations exhibit specific associations with epigenetic modifiers and proto-oncogenes mutated in myelodysplastic syndrome. Haematologica. 2013;98(7):1058–66. doi: 10.3324/haematol.2012.075325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rucker FG, Schlenk RF, Bullinger L, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119(9):2114–21. doi: 10.1182/blood-2011-08-375758. [DOI] [PubMed] [Google Scholar]
- 46.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12(9):599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 47.Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer. 2003;3(9):650–65. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- 48.Shivarov V, Gueorguieva R, Stoimenov A, Tiu R. DNMT3A mutation is a poor prognosis biomarker in AML: Results of a meta-analysis of 4500 AML patients. Leuk Res. 2013 doi: 10.1016/j.leukres.2013.07.032. [DOI] [PubMed] [Google Scholar]
- 49.Ribeiro AF, Pratcorona M, Erpelinck-Verschueren C, et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119(24):5824–31. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]
- 50.Marcucci G, Metzeler KH, Schwind S, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2012;30(7):742–50. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renneville A, Boissel N, Nibourel O, et al. Prognostic significance of DNA methyltransferase 3A mutations in cytogenetically normal acute myeloid leukemia: a study by the Acute Leukemia French Association. Leukemia. 2012;26(6):1247–54. doi: 10.1038/leu.2011.382. [DOI] [PubMed] [Google Scholar]
- 52.Ostronoff F, Othus M, Ho PA, et al. Mutations in the DNMT3A exon 23 independently predict poor outcome in older patients with acute myeloid leukemia: a SWOG report. Leukemia. 2013;27(1):238–41. doi: 10.1038/leu.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaidzik VI, Schlenk RF, Paschka P, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG) Blood. 2013;121(23):4769–77. doi: 10.1182/blood-2012-10-461624. [DOI] [PubMed] [Google Scholar]
- 54.Metzeler KH, Becker H, Maharry K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118(26):6920–9. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schnittger S, Eder C, Jeromin S, et al. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an adverse outcome. Leukemia. 2013;27(1):82–91. doi: 10.1038/leu.2012.262. [DOI] [PubMed] [Google Scholar]
- 56.Bejar R, Stevenson KE, Caughey BA, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30(27):3376–82. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Damm F, Thol F, Kosmider O, et al. SF3B1 mutations in myelodysplastic syndromes: clinical associations and prognostic implications. Leukemia. 2012;26(5):1137–40. doi: 10.1038/leu.2011.321. [DOI] [PubMed] [Google Scholar]
- 58.Dees ND, Zhang Q, Kandoth C, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22(8):1589–98. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Getz G, Hofling H, Mesirov JP, et al. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science. 2007;317(5844):1500. doi: 10.1126/science.1138764. [DOI] [PubMed] [Google Scholar]
- 60.Wendl MC, Wallis JW, Lin L, et al. PathScan: a tool for discerning mutational significance in groups of putative cancer genes. Bioinformatics. 2011;27(12):1595–602. doi: 10.1093/bioinformatics/btr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012;22(2):398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol. 2011;18(3):507–22. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 63.Vaske CJ, Benz SC, Sanborn JZ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26(12):i237–45. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41(Database issue):D43–7. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 68.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Damm F, Kosmider O, Gelsi-Boyer V, et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood. 2012;119(14):3211–8. doi: 10.1182/blood-2011-12-400994. [DOI] [PubMed] [Google Scholar]
- 70.Rossi D, Bruscaggin A, Spina V, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118(26):6904–8. doi: 10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dolnik A, Engelmann JC, Scharfenberger-Schmeer M, et al. Commonly altered genomic regions in acute myeloid leukemia are enriched for somatic mutations involved in chromatin remodeling and splicing. Blood. 2012;120(18):e83–92. doi: 10.1182/blood-2011-12-401471. [DOI] [PubMed] [Google Scholar]
- 72.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 73.Graubert T, Walter MJ. Genetics of myelodysplastic syndromes: new insights. Hematology Am Soc Hematol Educ Program. 2011;2011:543–9. doi: 10.1182/asheducation-2011.1.543. [DOI] [PubMed] [Google Scholar]
- 74.Fathi AT, Abdel-Wahab O. Mutations in epigenetic modifiers in myeloid malignancies and the prospect of novel epigenetic-targeted therapy. Adv Hematol. 2012;2012:469592. doi: 10.1155/2012/469592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hatzimichael E, Crook T. Cancer epigenetics: new therapies and new challenges. J Drug Deliv. 2013;2013:529312. doi: 10.1155/2013/529312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gronbaek K, Muller-Tidow C, Perini G, et al. A critical appraisal of tools available for monitoring epigenetic changes in clinical samples from patients with myeloid malignancies. Haematologica. 2012;97(9):1380–8. doi: 10.3324/haematol.2011.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jelinek J, Liang S, Lu Y, et al. Conserved DNA methylation patterns in healthy blood cells and extensive changes in leukemia measured by a new quantitative technique. Epigenetics. 2012;7(12):1368–78. doi: 10.4161/epi.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Down TA, Rakyan VK, Turner DJ, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26(7):779–85. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yan P, Frankhouser D, Murphy M, et al. Genome-wide methylation profiling in decitabine-treated patients with acute myeloid leukemia. Blood. 2012;120(12):2466–74. doi: 10.1182/blood-2012-05-429175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saied MH, Marzec J, Khalid S, et al. Genome wide analysis of acute myeloid leukemia reveal leukemia specific methylome and subtype specific hypomethylation of repeats. PLoS One. 2012;7(3):e33213. doi: 10.1371/journal.pone.0033213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Challen GA, Berg JS, Goodell MA. Loss of De Novo DNA Methylation Causes Expansion of the Mouse Hematopoietic Stem Cell Pool. Blood. 2010;116(21):364–64. [Google Scholar]
- 85.Akalin A, Garrett-Bakelman FE, Kormaksson M, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8(6):e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abdel-Wahab O, Adli M, LaFave LM, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22(2):180–93. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saeed S, Logie C, Francoijs KJ, et al. Chromatin accessibility, p300, and histone acetylation define PML-RARalpha and AML1-ETO binding sites in acute myeloid leukemia. Blood. 2012;120(15):3058–68. doi: 10.1182/blood-2011-10-386086. [DOI] [PubMed] [Google Scholar]
- 88.Graubert TA, Shen D, Ding L, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2012;44(1):53–7. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Makishima H, Visconte V, Sakaguchi H, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119(14):3203–10. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Przychodzen B, Jerez A, Guinta K, et al. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood. 2013 doi: 10.1182/blood-2013-01-480970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Visconte V, Rogers HJ, Singh J, et al. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood. 2012;120(16):3173–86. doi: 10.1182/blood-2012-05-430876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Walter MJ, Shen D, Ding L, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366(12):1090–8. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Walter MJ, Shen D, Shao J, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia. 2013 doi: 10.1038/leu.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4(149):149ra18. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–26. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Conte N, Varela I, Grove C, et al. Detailed molecular characterisation of acute myeloid leukaemia with a normal karyotype using targeted DNA capture. Leukemia. 2013 doi: 10.1038/leu.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Beekman R, Valkhof MG, Sanders MA, et al. Sequential gain of mutations in severe congenital neutropenia progressing to acute myeloid leukemia. Blood. 2012;119(22):5071–7. doi: 10.1182/blood-2012-01-406116. [DOI] [PubMed] [Google Scholar]
- 98.Ding L, Raphael BJ, Chen F, Wendl MC. Advances for studying clonal evolution in cancer. Cancer Lett. 2013 doi: 10.1016/j.canlet.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108(3):479–85. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4(127):127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 101.Abdel-Wahab O. Molecular genetics of acute myeloid leukemia: clinical implications and opportunities for integrating genomics into clinical practice. Hematology. 2012;17 (Suppl 1):S39–42. doi: 10.1179/102453312X13336169155411. [DOI] [PubMed] [Google Scholar]
- 102.Tyner JW, Erickson H, Deininger MW, et al. High-throughput sequencing screen reveals novel, transforming RAS mutations in myeloid leukemia patients. Blood. 2009;113(8):1749–55. doi: 10.1182/blood-2008-04-152157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Duncavage EJ, Abel HJ, Szankasi P, Kelley TW, Pfeifer JD. Targeted next generation sequencing of clinically significant gene mutations and translocations in leukemia. Mod Pathol. 2012;25(6):795–804. doi: 10.1038/modpathol.2012.29. [DOI] [PubMed] [Google Scholar]
- 104.Thol F, Kolking B, Damm F, et al. Next-generation sequencing for minimal residual disease monitoring in acute myeloid leukemia patients with FLT3-ITD or NPM1 mutations. Genes Chromosomes Cancer. 2012;51(7):689–95. doi: 10.1002/gcc.21955. [DOI] [PubMed] [Google Scholar]
- 105.Grossmann V, Schnittger S, Schindela S, et al. Strategy for robust detection of insertions, deletions, and point mutations in CEBPA, a GC-rich content gene, using 454 next-generation deep-sequencing technology. J Mol Diagn. 2011;13(2):129–36. doi: 10.1016/j.jmoldx.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rinke J, Schafer V, Schmidt M, et al. Genotyping of 25 Leukemia-Associated Genes in a Single Work Flow by Next-Generation Sequencing Technology with Low Amounts of Input Template DNA. Clin Chem. 2013 doi: 10.1373/clinchem.2013.204099. [DOI] [PubMed] [Google Scholar]
- 107.Spencer DH, Abel HJ, Lockwood CM, et al. Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data. J Mol Diagn. 2013;15(1):81–93. doi: 10.1016/j.jmoldx.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 108.Wang W, Wei Z, Lam TW, Wang J. Next generation sequencing has lower sequence coverage and poorer SNP-detection capability in the regulatory regions. Sci Rep. 2011;1:55. doi: 10.1038/srep00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Link DC, Schuettpelz LG, Shen D, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA. 2011;305(15):1568–76. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Welch JS, Westervelt P, Ding L, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305(15):1577–84. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]