HIV-1 persists in a latent reservoir (LR) despite antiretroviral therapy (ART)1–5. This reservoir is the major barrier to HIV-1 eradication6,7. Current approaches to purging the LR involve pharmacologic induction of HIV-1 transcription and subsequent killing of infected cells by cytolytic T lymphocytes (CTL) or viral cytopathic effects8–10. Agents that reverse latency without activating T cells have been identified using in vitro models of latency. However, their effects on latently infected cells from infected individuals remain largely unknown. Using a novel ex vivo assay, we demonstrate that none of the latency reversing agents (LRAs) tested induced outgrowth of HIV-1 from the LR of patients on ART. Using a novel RT-qPCR assay specific for all HIV-1 mRNAs, we demonstrate that LRAs that do not cause T cell activation do not induce significant increases in intracellular HIV-1 mRNA in patient cells; only the PKC agonist bryostatin-1 caused substantial increases. These findings demonstrate that current in vitro models do not fully recapitulate mechanisms governing HIV-1 latency in vivo. Further, our data indicate that non-activating LRAs are unlikely to drive the elimination of the LR in vivo when administered individually.
HIV-1 cure is hindered by viral persistence in a small fraction (~1/106) of resting CD4+ T cells (rCD4s) that harbor latent but replication-competent proviruses1–3. Upon cellular activation, latency is reversed and replication-competent virus is produced. Although T cell activation reverses latency, global T cell activation is toxic, generating interest in small molecule latency-reversing agents (LRAs) that do not activate T cells. Due to the low frequency of latently infected rCD4s in vivo, cell models have been used to identify a number of mechanistically distinct LRAs. These include: (1) histone deacetylase (HDAC) inhibitors, thought to function through epigenetic and other mechanisms11–14; (2) disulfiram, postulated to involve nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB)15,16; and (3) the bromodomain-containing protein 4 (BRD4) inhibitor JQ1, which elicits effects through positive transcription elongation factor (P-TEFb)17–20. Acting through signaling pathways associated with T cell activation, protein kinase C (PKC) agonists such as phorbol esters, prostratin21–23 and bryostatin-112,24–26 also reverse latency in cell models.
Evidence that putative LRAs reverse latency ex vivo in primary rCD4s from HIV-1-infected individuals is limited; disulfiram and the HDAC inhibitor vorinostat have been tested in patient cells with inconsistent results11,13,16,27,28. Clinical trials in patients on ART are ongoing with disulfiram and the HDAC inhibitors vorinostat, romidepsin, and panobinostat27,29. A recent trial of disulfiram showed no consistent evidence of latency reversal30. In another clinical trial, a single dose of vorinostat modestly increased intracellular RNAs containing HIV-1 gag sequences in rCD4s of patients on ART27. Ex vivo treatment of patient cells with vorinostat induced outgrowth in some studies11,13 but no virion production in another study28. Importantly, no LRA has been shown to reduce the size of the LR.
A consistent ex vivo validation strategy has not been employed to compare putative LRAs. Given the costs and risks associated with clinical trials, such a strategy is important for HIV-1 eradication research. Therefore, we utilized three independent assays to evaluate the efficacy of LRAs in cells from HIV-1 infected individuals on suppressive ART (participant characteristics in Supplementary Table 1).
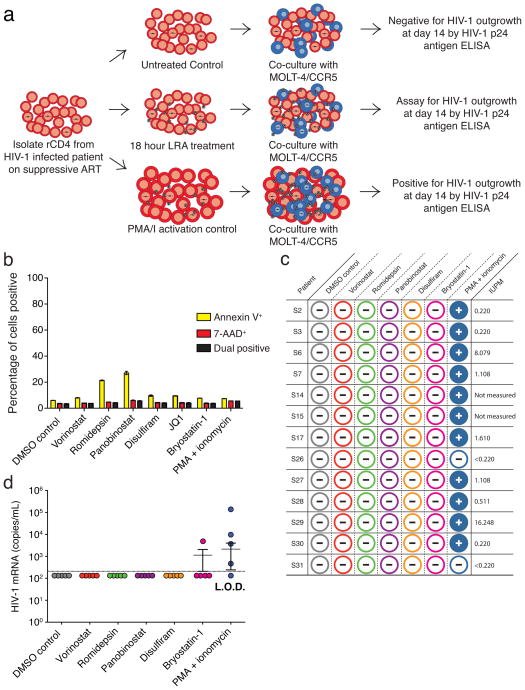
We first tested LRAs in a modified viral outgrowth assay1. In the original assay, patient-derived rCD4s were activated and co-cultured with CD4+ T lymphoblasts from healthy donors to expand released virus. Induction of outgrowth provides conclusive evidence of latency reversal. In the modified assay, T cell activation was replaced with LRA treatment. The subsequent co-culture of patient rCD4s with healthy donor lymphoblasts constitutes a mixed lymphocyte reaction, which induces background reactivation of latent HIV-131 and complicates LRA evaluation. Therefore, we treated rCD4s with LRAs and then cultured the cells with a transformed CD4+ T cell line (MOLT-4/CCR5) (Fig. 1a) that supports robust HIV-1 replication but does not induce allogeneic stimulation of rCD4s (Supplementary Fig. 1a–c). We treated five million purified rCD4s from infected individuals on ART with single LRAs for 18 h and then co-cultured the cells with MOLT-4/CCR5 cells for 14 days to permit viral outgrowth. T cell activation with phorbol 12-myristate 13-acetate + ionomycin (PMA/I) served as a positive control. We concurrently measured the frequency of latently infected cells32. We evaluated vorinostat, romidepsin, panobinostat, disulfiram and bryostatin-1 at clinically relevant concentrations that effectively reversed latency in a primary cell model (see below) and that were not toxic to rCD4s. No drug treatment induced cell death as shown by the lack of 7-AAD staining (Fig. 1b). Surprisingly, none of the LRAs induced viral outgrowth from cells from any individual tested while PMA/I-treated cultures were positive for every patient with a detectable LR (Fig. 1c).
Figure 1. LRAs do not induce outgrowth of latent HIV-1.
(a) Schematic of LRA outgrowth assay. (b) LRA-treated rCD4s were stained with Annexin-V and 7-AAD. Toxicity was defined as percent positivity by flow cytometry. (c) Viral outgrowth from LRA-treated rCD4s from infected individuals. Wells positive by ELISA for HIV-1 p24 antigen at 14 days are depicted with a positive sign. Negative wells are depicted with a negative sign. (d) Culture supernatant HIV-1 mRNA (copies mL−1) from LRA-treated rCD4s obtained from five infected individuals (S26–S30). Dotted line indicates limit of detection (208.3 copies mL−1). Error bars indicate mean ± s.e.m.
We next asked whether LRA treatment induced rapid virus release. We collected culture supernatants from rCD4s from five infected individuals (S26–S30) after 18 h of LRA treatment and prior to addition of MOLT-4/CCR5 cells for measurement of viral outgrowth. PMA/I induced virus release as detected by HIV-1 mRNA in the supernatant from four out of five individuals (S26–S29) (Fig. 1D). Bryostatin-1 treatment induced detectable supernatant HIV-1 mRNA from one infected individual (S27), whereas no other LRA had a measurable effect (Fig. 1d). None of the LRAs induced subsequent viral outgrowth from these treated cells, including the cells from the single individual (S27) that released HIV-1 mRNA after bryostatin-1 treatment (Fig. 1c).
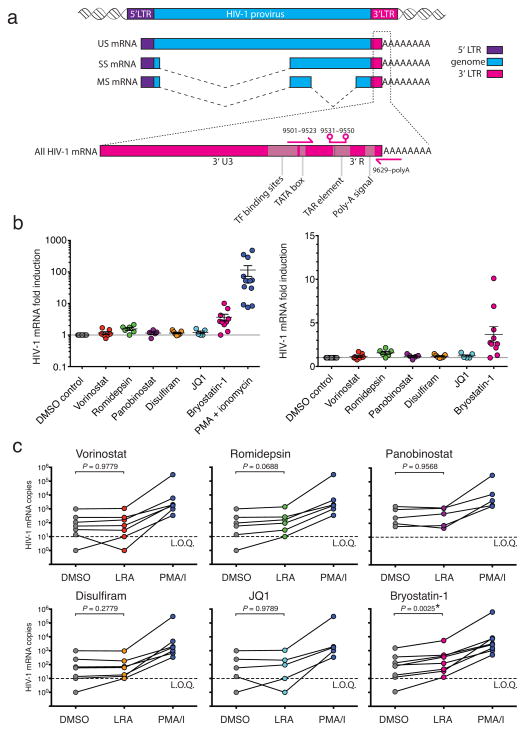
The most widely used method to detect induction of HIV-1 transcription16,27 in cells from infected individuals involves the measurement of RNAs containing HIV-1 gag sequences. Because this method lacks a stringent selection for poly-adenylated RNAs, it does not exclusively detect fully elongated and correctly processed HIV-1 mRNAs. Therefore, we devised a new assay specific for intracellular HIV-1 mRNA using a primer/probe set that detects the 3′ sequence common to all correctly terminated HIV-1 mRNAs (Fig. 2a). We detected baseline intracellular HIV-1 mRNA in rCD4s from ten out of 11 infected individuals. Stimulation with PMA/I for 18 h dramatically increased intracellular HIV-1 mRNA (mean increase = 115.5-fold, Fig. 2b). However, at clinically relevant concentrations that reverse latency in a primary cell model (Fig. 3B, C), vorinostat, romidepsin, panobinostat, disulfiram, and JQ1 failed to increase intracellular HIV-1 mRNA in rCD4s from infected individuals when used as single agents (Fig. 2b, c). Bryostatin- 1 caused significant increases in some infected individuals (Fig. 2c). We observed similar results after 6 h of LRA treatment (Supplementary Fig. 2).
Figure 2. LRAs do not consistently induce HIV-1 mRNA production in cells from HIV-1 infected individuals on ART.
(a) Schematic of HIV-1 mRNA detection by RT-qPCR. Intracellular HIV-1 mRNA from LRA-treated rCD4s obtained from infected individuals presented as (b) fold change relative to DMSO control (mean ± s.e.m.) and (c) copies of HIV-1 mRNA per million rCD4 equivalents. Data points represent mean effect of the LRA for each individual. Statistical significance was determined using a paired t-test. RT(−) controls were negative for all samples. Lines connect data points from each infected individuals. Dotted line indicates limit of quantification (L.O.Q.) of 10 copies. Detectable values below L.O.Q. were assigned 10 copies. Undetectable values were assigned 1 copy.
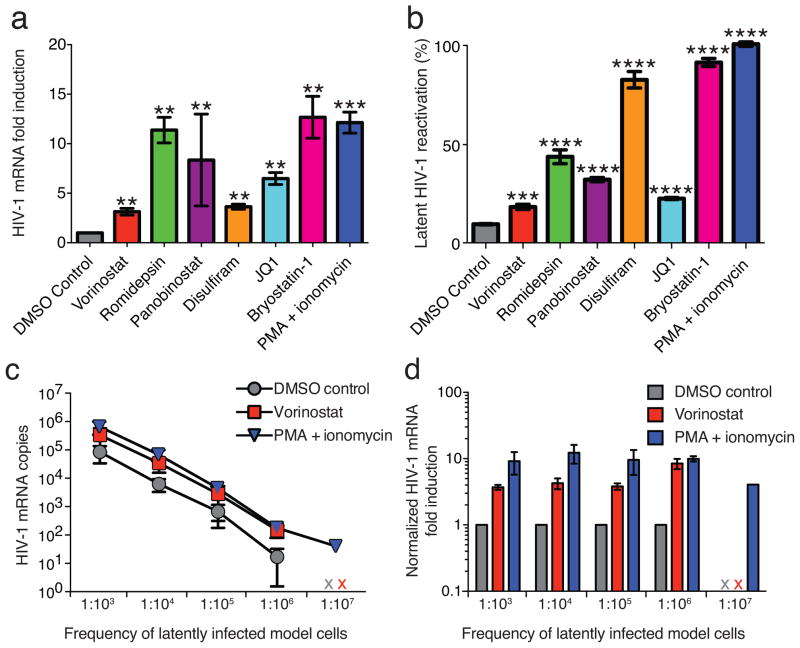
Figure 3. A primary CD4+ T cell model of HIV-1 latency is responsive to LRAs.
(a) Intracellular HIV-1 mRNA from LRA-treated BCL-2-transduced primary CD4+ T latency model cells. Changes are presented as fold induction relative to DMSO control (mean ± s.d.). (b) LRA-induced reactivation in latency models cells, defined as the percent GFP+ cells normalized to the effect of PMA/I treatment (mean ± s.d.) as measured by flow cytometry. Intracellular HIV-1 mRNA in serially diluted latency models cells, presented as (c) copies of HIV-1 mRNA per million rCD4 equivalents (mean ± s.d.) and (d) fold change relative to DMSO control (mean ± s.d.). An x indicates sample was below the limit of detection. RT(−) controls were negative for all samples. For a and b, statistical significance was determined using unpaired t-test. Asterisk indicates P value >0.05 (** indicates 10−2, *** indicates 10−3, **** indicates 10−4).
While no effect was seen in latently infected cells from infected individuals, LRA treatment increased intracellular HIV-1 mRNA in a B-cell lymphoma 2 (BCL-2) transduced primary rCD4 model of latency (Fig. 3a). LRA-induced increases in HIV-1 mRNA were consistent with measurements of the fraction of cells that up-regulate HIV-1 gene expression, as assessed by GFP reporter (Fig. 3b). The frequency of latent infection in this model is substantially higher than that observed in vivo4. To confirm that our assay effectively detects intracellular HIV-1 mRNA increases at frequencies of latent infection seen in vivo, we treated model cells with a known percentage of latent infection and then serially diluted these cells into rCD4s from uninfected individuals immediately prior to RNA isolation. We detected proportionate increases in intracellular HIV-1 mRNA in vorinostat-treated cells down to a frequency of 1/106 cells (Fig. 2d, e). Therefore, the lack of LRA efficacy in cells from HIV-1 infected individuals is not a result of assay insensitivity. Rather, our findings demonstrate that freshly isolated latently infected cells from infected individuals responded differently to LRAs than latency model cells.
RT-qPCR assays that detect gag-containing sequences in total RNA are frequently used to detect latency reversal. These sequences do not necessarily represent bona fide unspliced HIV-1 mRNA. HIV-1 integrates into host genes that are actively transcribed in rCD4s33,34, allowing for the production of chimeric host/HIV-1 primary transcripts. Such transcripts, initiated at host promoters, could contain gag sequence and would be indistinguishable from LTR-initiated transcripts by conventional gag RT-qPCR assays (Fig. 4a). We therefore designed a primer/probe set that amplifies a region of the LTR that is not transcribed during LTR-initiated and correctly terminated HIV-1 transcription. This primer/probe set is specific for transcripts containing read-through of the 5′ LTR or 3′ LTR, independent of proviral orientation (Fig. 4a). We treated ten million rCD4s from infected individuals on ART with vorinostat or PMA/I for 6 h and compared the levels of HIV-1 mRNA, read-through transcripts, and transcripts containing gag sequence (Fig. 4a, b). We detected a small increase (~2-fold) in transcripts containing gag sequence in vorinostat-treated rCD4s from four out of five infected individuals, consistent with previous reports27 (Fig. 4b). Vorinostat treatment also induced increases in read-through transcripts (Fig. 4b) comparable to the increases in transcripts containing gag sequence but had no effect on levels of HIV-1 mRNA (Fig. 4b).
Figure 4. Vorinostat induces transcripts containing HIV-1 gag sequence but not HIV-1 mRNA in cells from HIV-1 infected individuals on ART.
(a) Schematic of RT-qPCR detection of host/HIV-1 read-through transcripts (purple arrows), transcripts containing HIV-1 gag sequence (blue arrows), and HIV-1 mRNA (pink arrows). (b) Effect of vorinostat and PMA/I on intracellular HIV-1 read-through, gag-containing, and mRNA transcripts in rCD4s from five infected individuals, presented as fold change relative to DMSO control. (c) Schematic of Gag specific cDNA synthesis and qPCR detection of read-through transcripts. (d) Effect of vorinostat on read-through transcripts containing gag in from five infected individuals, presented as fold change relative to DMSO control. RT(−) controls were negative for all samples.
To prove that the read-through signal is amplified from a transcript that initiated upstream of the 5′ LTR and contains gag sequence, we primed cDNA synthesis with a gag primer (Fig. 4c). We detected comparable, statistically significant inductions of read-through and gag transcripts after 6 h of vorinostat treatment (Fig. 4d) (P = 0.027, P = 0.011, respectively; ratio paired t-test of transcript copies), indicative of read-through transcription. PMA/I induction of gag transcripts greatly exceeded that of read-through transcripts, indicative of LTR-initiated transcription (Supplementary Fig. 3). While not every potential LRA will induce read-through transcription by activating a host gene, our data show that chimeric host/HIV-1 transcripts can have a confounding effect on the RT-qPCR signal obtained with standard gag primers. Such an effect should be taken into consideration when evaluating LRAs using conventional gag RT-qPCR assays.
The novel assays presented herein facilitated the first comparative ex vivo evaluation of candidate LRAs. Our data demonstrate that none of the leading candidate non-T cell activating LRAs tested significantly disrupted the LR ex vivo. The striking discordance between the effects of non-stimulating LRAs in in vitro models of HIV-1 latency and the ex vivo effects in rCD4s from infected individuals on ART indicates that these models do not fully capture all mechanisms governing HIV-1 latency in vivo. These compounds are unlikely to drive the elimination of the LR in vivo when administered individually. The only active single agent was the PKC agonist bryostatin-1, which is likely too toxic for clinical use. Whether other PKC agonists or other compounds that stimulate signaling pathways associated with T cell activation can be safely administered remains to be seen, and further progress may depend on finding safe and active combinations of LRAs.
Methods
Cell isolation and culture
The Johns Hopkins Institutional Review Board approved this study and all research participants in this study gave written informed consent. Infected individuals were enrolled under the criteria of suppression of viremia to undetectable levels (<50 copies mL−1) on ART for at least 6 months. PBMC were purified using density centrifugation from whole blood or continuous-flow centrifugation leukapheresis product. CD4+ T lymphocytes were enriched by negative depletion (CD4+ T cell Isolation Kit, Miltenyi Biotec). Resting CD4+ T lymphocytes were further enriched by depletion of cells expressing CD69, CD25, or HLA-DR (CD69 MicroBead Kit II, Miltenyi Biotec; CD25 MicroBeads, Miltenyi Biotec; Anti-HLA-DR MicroBeads, Miltenyi Biotec). Purity of resting CD4+ lymphocytes was verified by flow cytometry and was typically greater than 95%. With the exception of experiments designed to detect viral outgrowth, cells were cultured with 10 μM T20 to prevent new infection events.
Treatment of rCD4s with LRAs
rCD4s were treated with the following concentrations: 335 nM vorinostat, 40 nM romidepsin, 30 nM panobinostat, 500 nM disulfiram, 1 μM JQ1, 10 nM bryostatin-1, or 50 ng mL−1 PMA plus 1 μM ionomycin.
MOLT-4/CCR5 outgrowth assay
Five million purified rCD4s were treated with LRA for 18 h in a volume of 1 mL RPMI + 10% FBS. Cells were then resuspended, transferred to a microcentrifuge tube and pelleted. Cells were washed with 1 mL sterile PBS to remove residual drug and pelleted. rCD4s were then cultured with MOLT-4/CCR5 cells in 8 mL RPMI + 10% FBS in individual wells in 6 well plates. After 4 days of culture, cells were resuspended and split into two wells of a 6 well plate with the media volume adjusted to 8 mL per well. After 7 days of culture, wells were resuspended and split 1:2 with the media volume adjusted to 8 mL per well. Viral outgrowth was assessed at 14 days using the Alliance HIV-1 p24 antigen ELISA kit (Perkin Elmer).
Cell lines
MOLT-4/CCR5 cells from Dr. Masanori Baba, Dr. Hiroshi Miyake, and Dr. Yuji Iizawa were obtained from the NIH AIDS Reagent Program, NIAID, NIH35.
Generation of latently HIV-1 infected BCL-2 transduced cells
Latently HIV-1 infected BCL-2 transduced cells were generated as described previously36. Briefly, primary CD4+ lymphoblasts were transduced with BCL-2 and allowed to return to a resting state in the absence of exogenous cytokines. BCL-2 transduced cells were then activated and expanded in the presence of exogenous IL-2. After expansion, cells were activated again and infected with a recombinant HIV-1 containing GFP in place of the env gene. After infection, cells were allowed to return to a resting state and GFP-negative cells were isolated via cell sorting. This population of cells includes the fraction of cells that are in vitro latently infected. Reversal of latency is assessed by flow cytometry analysis of GFP expression.
Measurement of intracellular HIV-1 RNA transcripts
Cells were treated with each LRA in triplicate in the presence of 10 μM T20 (5 × 106 cells for experiments measuring only HIV mRNA and 10 × 106 cells for experiments measuring multiple transcripts). Cells were pelleted in RNase-free low binding microcentrifuge tubes and subsequently lysed with 1 mL of TRIzol Reagent (Invitrogen). RNA was isolated using the manufacturer’s protocol. For experiments in which multiple transcripts were measured, a DNase digest was performed using TURBO DNase (Ambion). RNA was subsequently re-extracted using Acid-Phenol:Choloroform, pH 4.5 (Ambion) per manufacturer’s protocol. cDNA synthesis was performed using qScript cDNA Supermix containing random hexamers and oligo-dT primers(Quanta Biosciences). Gag specific cDNA synthesis was performed using Superscript III First-Strand Synthesis (Invitrogen) using only a gag primer (sequence listed below). A fraction of the RNA was retained for RT(−) control reactions.
Real-time PCR was performed in triplicate using TaqMan® Universal PCR Master Mix (Applied Biosystems) on an ABI7900 Real-Time PCR machine. Approximately one million cell equivalents of cDNA or RNA (for no-RT control reactions) template was used in each PCR reaction. Primers and probes are listed below. The cycling parameters were as follows: (i) 2 min at 50°C; (ii) 10 min at 95°C; and (iii) 45–50 cycles at 95°C for 15 and then 60°C for 60 s. Molecular standard curves were generated using serial dilutions of a TOPO plasmid containing the 5′ LTR, Gag, or the last 352 nucleotides of viral genomic RNA plus 30 deoxyadenosines.
Results from the triplicate samples for each drug treatment were averaged and presented as fold change relative to DMSO control (mean ± s.e.m.) or copies of HIV-1 mRNA per million rCD4 equivalents. The limit of quantification was set as the dilution point at which the Ct of the plasmid molecular standard replicates had a standard deviation > 0.5. We determined that the limit of quantification for all transcripts was 10 copies. A PCR signal of less than 10 copies (1–9 copies) was treated as 10 copies in calculations of fold change and marked as 10 copies on graphs depicting RNA copies. Undetectable PCR signal was treated as 10 copies in calculations of fold change and marked as 1 copy on graphs depicting RNA copies. Levels of RNA polymerase II (Pol2) and Glucose-6-phosphate dehydrogenase (G6PD) RNA were also measured for each sample to use as an endogenous control. Voronistat, romidepsin, panobinostat, JQ1 and PMA/I treatment consistently increased expression Pol2 and G6PD. Samples treated with the same drug had even levels of Pol2 and G6PD, indicating that the template inputs were approximately equal.
Measurement of supernatant HIV-1 mRNA
HIV-1 mRNA was extracted from 0.2mL of supernatant from five million cultured rCD4s after 18 h of LRA treatment using the ZR-96 Viral RNA kit (Zymo Research). cDNA synthesis was performed using qScript cDNA Supermix (Quanta Biosciences). Real-time PCR was performed using TaqMan Fast Advanced mastermix (Applied Biosystems) on an ABI Viia 7 Real-Time PCR machine. Primers and probes listed below. Manufacturer’s thermal cycling conditions were used. Molecular standard curve was generated as described above.
Primer and probe sequences
Nucleotide coordinates are indicated relative to HXB2 consensus sequence.
HIV-1 mRNAs were detected using the following primers and probe, modified from Shan et al.37:
Forward (5′→3′) CAGATGCTGCATATAAGCAGCTG (9501–9523)
Reverse (5′→3′) TTTTTTTTTTTTTTTTTTTTTTTTGAAGCAC (9629-poly A)
Probe (5′→3′) FAM-CCTGTACTGGGTCTCTCTGG-MGB (9531–9550)
Transcripts containing HIV-1 gag sequence were detected using the following primers and probe, described previously27.
Forward (5′→3′) ACATCAAGCAGCCATGCAAAT (1368–1388)
Reverse (5′→3′) TCTGGCCTGGTGCAATAGG (1453–1471)
Probe (5′→3′) VIC-CTATCCCATTCTGCAGCTTCCTCATTGATG-TAMRA (1401–1430)
Chimeric host/HIV-1 read-through transcripts were detected using the following primers and probe:
Forward (5′→3′) CAGATGCTGCATATAAGCAGCTG (416–438, 9501–9523)
Reverse (5′→3′) CACAACAGACGGGCACACAC (556–575, 9641–9660)
Probe (5′→3′) FAM-CCTGTACTGGGTCTCTCTGG-MGB (446–465, 9531–9550)
cDNA synthesis reaction with gag primer sequence:
Reverse (5′→3′) GTCACTTCCCCTTGG (1480–1494)
Supplementary Material
Acknowledgments
We thank Holly McHugh and Anitha Devadason for assistance with study participants. Funding: the Martin Delaney Collaboratory of AIDS Researchers for Eradication and the Delany AIDS Research Enterprise (US National Institutes of Health AI096113, 1U19AI096109), the Foundation for AIDS Research 108165-50-RGRL, the Johns Hopkins Center for AIDS Research, US National Institutes of Health 43222, the Howard Hughes Medical Institute.
Footnotes
Author Contributions:
The authors contributed as follows: GL, CKB, and RFS designed experiments; GL and CKB performed experiments; CMD obtained IRB approval and managed study participant recruitment; GL, CKB, JDS and RFS analyzed the data and wrote the manuscript.
References and Notes
- 1.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 2.Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 3.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siliciano JD, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 5.Strain MC, et al. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc Natl Acad Sci USA. 2003;100:4819–4824. doi: 10.1073/pnas.0736332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richman DD, et al. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 7.Deeks SG, et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol. 2012;12:607–614. doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karn J. The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit. Curr Opin HIV AIDS. 2011;6:4–11. doi: 10.1097/COH.0b013e328340ffbb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhary SK, Margolis DM. Curing HIV: Pharmacologic approaches to target HIV-1 latency. Annu Rev Pharmacol and Toxicol. 2011;51:397–418. doi: 10.1146/annurev-pharmtox-010510-100237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakre S, Chavez L, Shirakawa K, Verdin E. HIV latency: experimental systems and molecular models. FEMS Microbiol Rev. 2012;36:706–716. doi: 10.1111/j.1574-6976.2012.00335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Archin NM, et al. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses. 2009;25:207–212. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartholomeeusen K, Fujinaga K, Xiang Y, Peterlin BM. Histone Deacetylase Inhibitors (HDACis) That Release the Positive Transcription Elongation Factor b (P-TEFb) from Its Inhibitory Complex Also Activate HIV Transcription. J Biol Chem. 2013;288:14400–14407. doi: 10.1074/jbc.M113.464834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Contreras X, et al. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem. 2009;284:6782–6789. doi: 10.1074/jbc.M807898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirakawa K, Chavez L, Hakre S, Calvanese V, Verdin E. Reactivation of latent HIV by histone deacetylase inhibitors. Trends Microbiol. 2013;21:277–285. doi: 10.1016/j.tim.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyon G, Zerbato J, Mellors JW, Sluis-Cremer N. Disulfiram reactivates latent HIV-1 expression through depletion of the phosphatase and tensin homolog. AIDS. 2013;27:F7–F11. doi: 10.1097/QAD.0b013e3283570620. [DOI] [PubMed] [Google Scholar]
- 16.Xing S, et al. Disulfiram reactivates latent HIV-1 in a BCL-2-transduced primary CD4+ T cell model without inducing global T cell activation. J Virol. 2011;85:6060–6064. doi: 10.1128/JVI.02033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee C, et al. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol. 2012;92:1147–1154. doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehm D, et al. BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle. 2013;12:452–462. doi: 10.4161/cc.23309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Guo J, Wu Y, Zhou Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 2013;41:277–287. doi: 10.1093/nar/gks976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, et al. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep. 2012;2:807–816. doi: 10.1016/j.celrep.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korin YD, Brooks DG, Brown S, Korotzer A, Zack JA. Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J Virol. 2002;76:8118–8123. doi: 10.1128/JVI.76.16.8118-8123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulkosky J, et al. Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood. 2001;98:3006–3015. doi: 10.1182/blood.v98.10.3006. [DOI] [PubMed] [Google Scholar]
- 23.Williams SA, et al. Prostratin antagonizes HIV latency by activating NF-kappaB. J Biol Chem. 2004;279:42008–42017. doi: 10.1074/jbc.M402124200. [DOI] [PubMed] [Google Scholar]
- 24.DeChristopher BA, et al. Designed, synthetically accessible bryostatin analogues potently induce activation of latent HIV reservoirs in vitro. Nat Chem. 2012;4:705–710. doi: 10.1038/nchem.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinter AL, Poli G, Maury W, Folks TM, Fauci AS. Direct and cytokine-mediated activation of protein kinase C induces human immunodeficiency virus expression in chronically infected promonocytic cells. J Virol. 1990;64:4306–4312. doi: 10.1128/jvi.64.9.4306-4312.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehla R, et al. Bryostatin modulates latent HIV-1 infection via PKC and AMPK signaling but inhibits acute infection in a receptor independent manner. PloS One. 2010;5:e11160. doi: 10.1371/journal.pone.0011160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Archin NM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blazkova J, et al. Effect of histone deacetylase inhibitors on HIV production in latently infected, resting CD4(+) T cells from infected individuals receiving effective antiretroviral therapy. J Infect Dis. 2012;206:765–769. doi: 10.1093/infdis/jis412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen TA, Tolstrup M, Winckelmann A, Ostergaard L, Sogaard OS. Eliminating the latent HIV reservoir by reactivation strategies: Advancing to clinical trials. Hum Vacc Immunother. 2013;9 doi: 10.4161/hv.23202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spivak AM, et al. A Pilot Study Assessing the Safety and Latency-Reversing Activity of Disulfiram in HIV-1-Infected Adults on Antiretroviral Therapy. Clin Infect Dis. 2013 doi: 10.1093/cid/cit813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moriuchi H, Moriuchi M, Fauci AS. Induction of HIV replication by allogeneic stimulation. J Immunol. 1999;162:7543–7548. [PubMed] [Google Scholar]
- 32.Laird GM, et al. Rapid Quantification of the Latent Reservoir for HIV-1 Using a Viral Outgrowth Assay. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han Y, et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol. 2004;78:6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroder AR, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 35.Baba M, Miyake H, Okamoto M, Iizawa Y, Okonogi K. Establishment of a CCR5-expressing T-lymphoblastoid cell line highly susceptible to R5 HIV type 1. AIDS Res Hum Retroviruses. 2000;16:935. doi: 10.1089/08892220050058344. [DOI] [PubMed] [Google Scholar]
- 36.Yang HC, et al. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest. 2009;119:3473. doi: 10.1172/JCI39199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shan L, et al. A novel PCR assay for quantification of HIV-1 RNA. J Virol. 2013;87:6521. doi: 10.1128/JVI.00006-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.