Letter to the Editor
Acute myeloid leukemia (AML) is a clonal hematopoietic disease caused by both inherited and acquired genetic alterations. Conventional cytogenetic analysis reveals chromosomal aberrations in about 50% of AML cases. These chromosomal rearrangements promote leukemia by affecting cell proliferation, differentiation, and/or survival. In addition to these genetic alterations, cooperating mutations are required for leukemogenesis (1–3). Among subtypes of AML, pediatric acute megakaryoblastic leukemia (AMKL) is primarily associated with three cytogenetic subtypes: one with trisomy 21 plus mutation of GATA1, one with the t(1;22) translocation (4, 5), and a third recently reported recurrent cryptic inversion of chromosome 16 (inv(16)(p13.3q24.3)) (6, 7). AMKL with t(1;22) is typically an aggressive disease, presenting in early infancy with a median survival of just 8 months. Translocation between chromosomes 1p13 and 22q13 results in the fusion gene, RBM15-MKL1. The mechanism(s) by which the fusion protein promotes leukemia is unknown. RBM15-MKL1 knock-in mice develop AMKL, but with delayed onset (16–18 months) and incomplete penetrance (8). Induced RBM15-MKL1 overexpression in cell lines causes cell death (9). Together, these findings suggest that RBM15-MKL1 requires cooperating mutations to induce AMKL. To identify potential cooperating mutations in t(1;22) AMKL, we performed whole exome capture followed by next-generation sequencing in diagnostic and remission samples from a t(1;22)-AMKL patient. An 11 week-old boy presented with loss of appetite and fussiness. Samples of bone marrow and peripheral blood were obtained at presentation. Morphological and immunophenotypic analyses were performed at the UC Davis Pathology Department according to standard protocols. FISH and cytogenetic analyses were performed by the Mayo Clinic Laboratory (Children’s Oncology Group designated laboratory). The remaining diagnostic sample was frozen. The blood had a white blood cell count (WBC) of 38,000/mm3, hemoglobin of 6.1g/dL, and platelet count of 99,000/mm3. The peripheral blood smear showed immature blast cells (18% of WBC). Bone marrow examination revealed severe fibrosis (Figure 1A, B). Peripheral blood immunophenotyping showed 15% leukemic blasts based on CD45 expression and SSC profile. These cells expressed CD33 (75%), CD34 (44%), CD41 (63%) and CD61 (79%) and cytogenetic analyses showed 46,XY,t(1;22)(p13;q13) (Figure 1C). Based on the fibrotic marrow, megakaryocytic markers, and translocation, the diagnosis of AMKL was assigned. He responded very well to the treatment with cytarabine, etoposide, daunorubicin, mitoxantrone, L-asparaginase, and Gemtuzumab, the Children’s Oncology Group AAML0531 protocol. Because he did not have an HLA matched family donor, he did not undergo allogeneic hematopoietic stem cell transplantation. He is currently disease free 4 years after completing therapy. Peripheral blood was collected and cryopreserved 6 and 9 months after remission was achieved.
Figure 1.
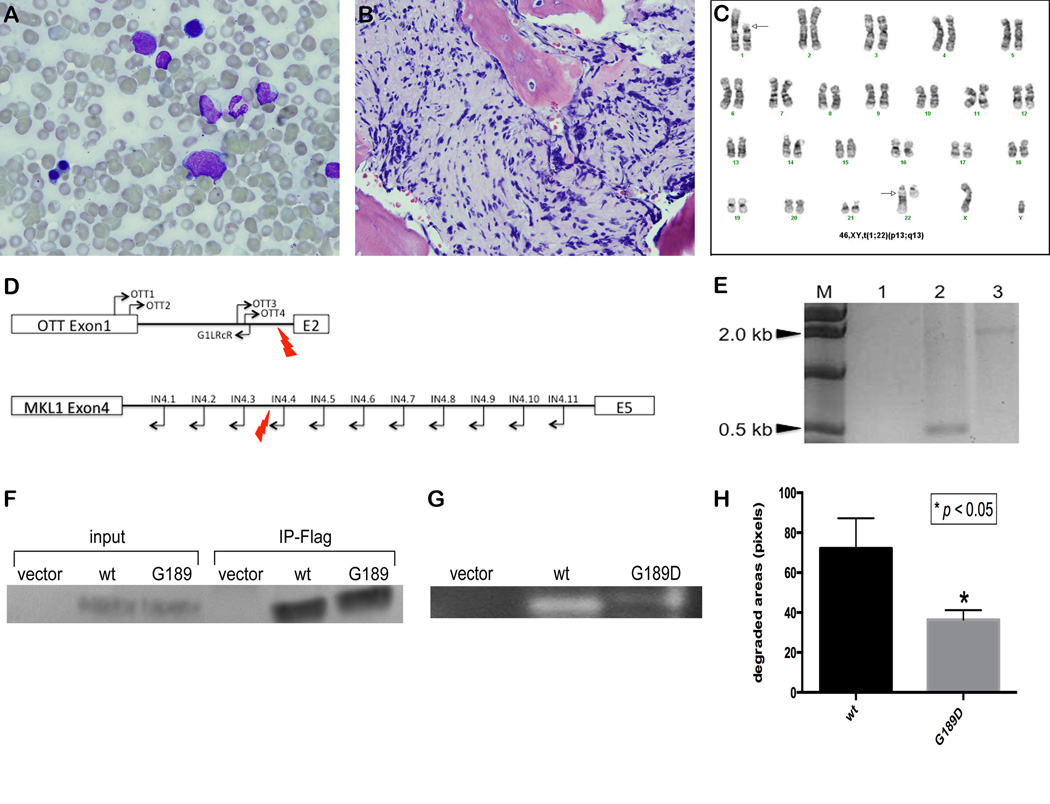
Analysis of t(1;22) sample and functional analysis of MMP8G189D. (A) and (B) show two blasts (arrows) from a touch-prepped bone marrow (original magnification 100×) and severe fibrosis in bone marrow core (original magnifications 50×), respectively. (C) shows G banded karyotype with t(1;22)(p13;q13) at diagnosis from the patient. Arrows indicate the rearranged chromosomes. (D) Diagram of primer bound locations on OTT gene and MKL1 gene. Red lightening symbols indicate the break point. (E) PCR amplicon of OTT4/IN4.3 was not observed in lane 1 but PCR amplicons of OTT4/IN4.4 and OTT4/IN4.5 were observed at around 500 bp and 2.2 kb, in lane 2 and 3, respectively. This analysis indicated that the breakpoint is between IN4.3 and IN4.4 in MKL1 intron 4. (F) Western blot was probed with anti-Flag antibody on the whole cell lysates and immunoprecipitates by flag beads of HEK 293T cells carrying with empty vector (empty), wild-type (wt) and mutant (G189D) MMP8. (G) Type 1 collagen substrate zymography showed the reduced enzymatic activity of mutant MMP8 (G189D) compared to wild-type MMP8 (wt). (H) The degraded areas by MMP8 were calculated using ImageJ.
The quality of extracted genomic DNA from the fixed cells of the diagnostic sample using the Gentra Puregen kit (Qiagen) was highly comparable to that of freshly thawed peripheral blood of the remission sample (A260/280 = 1.89 vs. 1.86, leukemia and remission, respectively) and no difference in quality was seen in the further analyses, including exome capture, PCR amplification, and sequencing. To delineate the breakpoints on chromosomes 1 and 22, PCR was performed as described (5). Multiplex PCR was performed with combinations of primers (as shown in Figure 1D) to narrow down the breakpoint region. When single primer pairs were then used for individual PCR reactions, no amplification occurred with OTT4 and In4.1-3, but 0.5 kb and 2.2 kb products were amplified with OTT4 and In4.4 and In4.5, respectively (Figure 1E) confirming that the translocation occurred between chromosome 1 and 22 with the breakpoint in intron 1 of OTT and intron 4 of MKL1.
To identify potential cooperating mutations in this patient sample, exome capture followed by deep sequencing was performed on the leukemia and remission blood samples from the patient. A total of 88,807,512 and 79,716,978 reads were obtained from the leukemic and remission samples, respectively (Supplementary Table S2). Picard software identified and removed PCR duplicates (14.0% of the leukemia and 9.4% of the blood sample). About 96% of the remaining unique reads from both samples mapped to only one location in the genome with ~ 94% of targeted bases with >10× coverage (Table S2). Varscan identified 97,926 single nucleotide variations (SNVs) with a minimum variant frequency in the sample of >0.02 and minimum average quality score >20. Of these variants, 22,326 were called somatic variations, which were filtered to require <0.2 p-value that the SNV was different in leukemia vs. remission, leaving 4,580. Of the 4,580 SNV, 2,123 were in genic regions and 95.3% were classified as coding variants, with 75.2% being non-synonymous. SIFT predicted 36.0% and 62.9% of the coding variants as damaging and tolerated variations, respectively (Supplementary Table 3). Of 1,956 coding variants, 1,876 were novel SNPs based on dbSNP. Of the 1,956, 377 SNVs remained after further filtering to require <0.05 p-value and damaging/tolerated SIFT predictions, and to exclude known dbSNPs. To overcome the low blast purity (15% of cells), we used a less stringent filter to maximize the chance of detecting variations within the leukemic cells, which resulted in false positive variants from the initial analysis. Therefore, it was necessary to carefully inspect the reads manually using IGV. After the manual inspection of the 377 SNVs for base quality displayed in the Integrated Genomics Viewer (IGV v2.1,(10)), we found 12 variants to be likely candidates and performed further validation for these 12 variants (Table 1). Neither known AML-associated mutations nor mutations in tumor suppressor genes were found. However, small indels would not be detectable in our data due to the low blast percentage.
Table 1.
Somatic mutations identified by exome sequencing and ddPCR
| Position | Gene symbol |
Ref/Mut | Substitution | p_value | # Mut Reads |
SIFT Prediction |
Allele Frequency |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Exome sequencing |
ddPCR |
|||||||||
| Leukemic | Remission | Leukemic | Remission | |||||||
| chr11:102592188 | MMP8 | C/T | G189D | 2.47E-06 | 18 | damaging | 14.40% | 0.00% | 14.50% | 0% |
| chr15:75684939 | SIN3A | G/T | S832* | 4.33E-02 | 5 | nonsense | 3.47% | 0.00% | 0% | 0% |
| chr18:52899759 | TCF4 | T/G | I544L | 4.60E-03 | 9 | tolerated | 4.46% | 0.00% | 0% | 0% |
| chr5:140230206 | PCDHA9 | G/T | S709I | 2.87E-02 | 4 | damaging | 4.76% | 0.00% | 0% | 0% |
| chr1:38022574 | DNALI1 | G/T | W15C | 4.50E-02 | 4 | tolerated | 4.35% | 0.00% | 0% | 0% |
| chr11:60638721 | ZP1 | G/T | W349L | 4.06E-02 | 4 | damaging | 4.60% | 0.00% | 0% | 0% |
| chr19:48305564 | TPRX1 | T/A | N235I | 1.79E-03 | 8 | tolerated | 7.69% | 0.00% | N/A | N/A |
| chr15:28456220 | HERC2 | G/A | L2333F | 5.05E-02 | 5 | tolerated | 4.50% | 0.00% | 50% | 50% |
| chr20:44579096 | ZNF335 | G/T | Q887K | 5.37E-02 | 3 | tolerated | 3.45% | 0.00% | 0% | 0% |
| chr3:146246571 | PLSCR1 | G/T | P48T | 4.47E-02 | 4 | tolerated | 7.14% | 0.00% | 0% | 0% |
| chr3:42730118 | KBTBD5 | G/T | E444* | 5.30E-02 | 3 | nonsense | 4.55% | 0.00% | 0% | 0% |
| chr9:130550569 | CDK9 | T/C | L170P | 5.23E-02 | 4 | damaging | 7.84% | 0.00% | 0% | 0% |
To validate the candidate mutations, Sanger sequencing was performed, but proved to be technically challenging due to the low percentage of blasts. By Sanger sequencing, the expected peak for the variant alleles was not distinguishable except for one in HERC2, which proved to exist in both the leukemic and remission samples at approximately 50%, suggesting a unique SNP in the patient (data not shown). To improve the sensitivity, 10 of the remaining 11 variants were further validated with ddPCR (1 failed in assay design due to repetitive sequences and high GC content). Despite multiple repetitions and assessment using modified primer pairs, of the 10 variants assessed using ddPCR, only MMP8 was validated (Supplementary Figure S1), while the other 9 variants were not detectable in the leukemic sample (Table 1). Given that, for example, a candidate mutation with 7% frequency was found in CDK9 using exome sequencing, we had hypothesized that this was a heterozygous mutation in the leukemic blasts. However, this was not validated with ddPCR even after analysis of over 14,000 molecules of this region from the diagnostic DNA sample (Supplementary Figure S1). While CDK9 is known to play a role in megakaryocyte maturation and to regulate both the cell cycle and transcription, further inspection of the exome data showed that the sequence quality was low at the mutant bases. Other unvalidated variant calls had low number of mutant reads, low mutant base quality, or possible mapping problems. The ddPCR validation demonstrated that more stringent p-value, mutant base quality and read number filtration is necessary when analyzing challenging low blast % samples.
The MMP8 mutation in our patient sample, a substitution from C to T on chr11:102592188, resulted in amino acid change MMP8G189D. This mutation is predicted to be damaging and deleterious by SIFT and PROVEAN (11, 12), respectively. This region is universally conserved in all vertebrates except elephant and frog (http://genome.uscs.edu/, (13)). MMP8 encodes matrix metalloproteinase-8, which breaks down type I, II and III collagens in the extracellular matrix, and requires both calcium and zinc ions for activity (14). The variation is located at a calcium ion binding site in the catalytic domain (Figure S2). To determine the effect of this mutation on enzymatic activity, we generated constructs containing either wild-type or mutant MMP8 and expressed them in HEK 293T cells. No difference was observed in expression level between the constructs (Figure 1F). Type 1 collagen substrate gel zymography revealed significantly reduced proteolytic activity of the mutant compared to wildtype MMP8 (Figure 1G and H, p-value=0.017). To further assess whether MMP8 may play a role in AMKL leukemogenesis, we evaluated MMP8 expression in primary AMKL samples. Although RNA was not available from this case, gene expression data from 41 pediatric AML cases including 16 cases of AMKL demonstrated 8 of the 16 AMKL cases to have detectable MMP8 expression (Figure S3) (7). All AMKL samples in this study had >90% purity, an important consideration given that MMP8 is known to be highly expressed in mature myeloid cells. Transcriptome sequencing was available for 14 of these AMKL cases and the sample with the highest signal by exon array also had significant levels of the MMP8 transcript by sequencing. In an independent cohort of 60 AMKL cases also analyzed by gene expression profiling, a subset of patients had evidence of MMP8 expression (15). While the purity of these samples was extremely variable (12–94%), several samples with low purity (M7122, 12%; M7026, 28%) had low levels of MMP8 expression demonstrating that the variability in MMP8 expression cannot solely be due to myeloid cell contamination. Furthermore, there are cases with elevated expression that are relatively pure (M7040, 70%; M7072, 90%). Although the same mutation was not found in any of the samples, MMP8 is aberrantly expressed in a subset of AMKL cases. In addition, MMP8 mRNA is expressed in murine megakaryocyte-erythroid progenitors based on the ENCODE database (GSE40522). Given that MMP8 was the only candidate gene with 14% variation frequency in the exome sequencing, the patient had few cooperating mutations in coding regions, which might explain the excellent response to treatment and survival. In conclusion, we provide the first data identifying a novel somatic mutation that may provide a key insight to understanding the pathogenesis of RBM15-MKL1 in AMKL.
Supplementary Material
Acknowledgements
We thank Dr. Yardena Samuels (Weizmann Institute of Science, Rehovot, Israel) for providing protocols for collagen zymography. This work was supported by Connecticut Stem Cell grant #10SCB03 (DSK), NIH R01HL106184 (PGG), a National Research Service Award from NIH/NCI F32CA165683 (YK), Hyundai Hope on Wheels Award (NS and TAG), The Eric Trump Foundation (TAG), and the Yale Stem Cell Center Core funded by the Connecticut Stem Cell Research Fund.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary information is available at Leukemia's website.
References
- 1.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fröhling S, Scholl C, Levine RL, Loriaux M, Boggon TJ, Bernard OA, et al. Identification of Driver and Passenger Mutations of FLT3 by High-Throughput DNA Sequence Analysis and Functional Assessment of Candidate Alleles. Cancer Cell. 2007;12(6):501–513. doi: 10.1016/j.ccr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Walter MJ, Ding L, Shen D, Shao J, Grillot M, McLellan M, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25(7):1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baruchel A, Daniel MT, Schaison G, Berger R. Nonrandom t(1;22)(p12-p13;q13) in acute megakaryocytic malignant proliferation. Cancer genetics and cytogenetics. 1991 Jul 15;54(2):239–243. doi: 10.1016/0165-4608(91)90213-e. [DOI] [PubMed] [Google Scholar]
- 5.Mercher T, Busson-Le Coniat M, Khac FN, Ballerini P, MauchauffÈ M, Bui H, et al. Recurrence of OTT-MAL fusion in t(1;22) of infant AML-M7. Genes, Chromosomes and Cancer. 2002;33(1):22–28. doi: 10.1002/gcc.1208. [DOI] [PubMed] [Google Scholar]
- 6.Thiollier C, Lopez CK, Gerby B, Ignacimouttou C, Poglio S, Duffourd Y, et al. Characterization of novel genomic alterations and therapeutic approaches using acute megakaryoblastic leukemia xenograft models. The Journal of Experimental Medicine. 2012 Oct 8;209(11):2017–2031. doi: 10.1084/jem.20121343. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruber Tanja A, Larson Gedman A, Zhang J, Koss Cary S, Marada S, Ta Huy Q, et al. An Inv(16)(p13.3q24.3)-Encoded CBFA2T3-GLIS2 Fusion Protein Defines an Aggressive Subtype of Pediatric Acute Megakaryoblastic Leukemia. Cancer Cell. 2012;22(5):683–697. doi: 10.1016/j.ccr.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercher T. The OTT-MAL fusion oncogene activates RBPJ-mediated transcription and induces acute megakaryoblastic leukemia in a knockin mouse model. The Journal of Clinical Investigation. 2009 Apr 01;119(4):852–864. doi: 10.1172/JCI35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Descot A, Rex-Haffner M, Courtois G, Bluteau D, Menssen A, Mercher T, et al. OTT-MAL Is a Deregulated Activator of Serum Response Factor-Dependent Gene Expression. Mol Cell Biol. 2008 Oct 15;28(20):6171–6181. doi: 10.1128/MCB.00303-08. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics. 2012 Apr 19; doi: 10.1093/bib/bbs017. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi Y. Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine. Orlando, Florida: ACM; 2012. A fast computation of pairwise sequence alignment scores between a protein and a set of single-locus variants of another protein; pp. 414–417. [Google Scholar]
- 13.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Research. 2012 Jan 1;40(D1):D918–D923. doi: 10.1093/nar/gkr1055. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massova I, Kotra LP, Mobashery S. Structural insight into the binding motifs for the calcium ion and the non-catalytic zinc in matrix metalloproteases. Bioorganic & Medicinal Chemistry Letters. 1998;8(7):853–858. doi: 10.1016/s0960-894x(98)00128-0. [DOI] [PubMed] [Google Scholar]
- 15.Bourquin JP, Subramanian A, Langebrake C, Reinhardt D, Bernard O, Ballerini P, et al. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proceedings of the National Academy of Sciences of the United States of America. 2006 Feb 28;103(9):3339–3344. doi: 10.1073/pnas.0511150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.