Abstract
Recent studies stress the importance of antimicrobial peptides in protecting the urinary tract from infection. Previously, we have shown that ribonuclease 7 (RNase 7) is a potent antimicrobial peptide that has broad-spectrum antimicrobial activity against uropathogenic bacteria. The urothelium of the lower urinary tract and intercalated cells of the kidney produce RNase 7 but regulation of its antimicrobial activity has not been well defined. Here we characterize the expression of an endogenous inhibitor, ribonuclease inhibitor (RI), in the urinary tract and evaluate its effect on RNase 7’s antimicrobial activity. Using RNA isolated from non-infected human bladder and kidney tissue, quantitative real-time PCR showed that RNH1, the gene encoding RI, is constitutively expressed throughout the urinary tract. With pyelonephritis, RNH1 expression and RI peptide production significantly decrease. Immunostaining localized RI production to the umbrella cells of the bladder and intercalated cells of the renal collecting tubule. In vitro assays showed that RI bound to RNase 7 and suppressed its antimicrobial activity by blocking its ability to bind the cell wall of uropathogenic bacteria. Thus, these results demonstrate a new immunomodulatory role for RI and identified a unique regulatory pathway that may affect how RNase 7 maintains urinary tract sterility.
Keywords: Ribonuclease Inhibitor, Ribonuclease 7, Antimicrobial Peptide, Urinary Tract Infection, Pyelonephritis, Intercalated Cells, Innate Immunity
Introduction
Urinary tract infections are one of the most common and serious bacterial diseases in humans. Although the urine is considered sterile, little is known how the body maintains sterility. Recent evidence suggests that antimicrobial peptides (AMP) have an important role in maintaining urinary tract sterility.1–3 AMPs have been identified as key components of the innate host defense that are important contributors to maintaining health at mucosal barriers. AMPs, which serve as natural antibiotics, are cationic molecules expressed by leukocytes and epithelial cells. AMPs may be constitutively expressed and/or induced by invading pathogens. AMPs possess broad-spectrum antimicrobial activity against gram-positive and gram-negative bacteria, enveloped viruses, fungi, and some protozoa.
Recently, our research group has shown that Ribonuclease 7 (RNase 7) is an epithelial-derived AMP that helps maintain sterility in the human kidney and urinary tract.4–6 RNase 7 is a member of the Ribonuclease A superfamily that has broad-spectrum antimicrobial activity against uropathogenic bacteria at micromolar concentrations.5, 7 The uroepithelium of the lower urinary tract and the intercalated cells of the renal collecting tubules constitutively express RNase 7 and secrete it into the urinary stream.5, 6 In addition to maintaining urinary tract sterility, RNase 7 contributes to sterility in other organ systems – including the skin, hair follicles, and oral cavity.8–12 It has been stated that, on a per molar basis, RNase 7 is the most potent human AMP.13
The mechanisms that regulate RNase 7’s antimicrobial properties are not well defined. The bactericidal activity of RNase 7 has been linked to its capacity to bind and permeate the bacterial cell membrane, independent of its ribonuclease activity.14, 15 Recently, Abtin et al identified an endogenous ribonuclease inhibitor (RI) in human epidermal keratinocytes that suppresses the antimicrobial activity of RNase 7 against yeast and E. faecium.16 RI is a 49-kDa cytosolic protein that was first isolated from human placenta.17 It has also been identified by Northern blot in other human tissues – including brain, liver, testis, and peripheral blood leukocytes.18 RI is a horseshoe-shaped peptide that binds with femtomolar affinity to members of the Ribonuclease A superfamily. Its unique structure, consisting of a repetitive amino acid sequence that is rich in leucine residues, helps foster protein-protein interactions.19, 20 Complexes formed between RI and target ribonuclease are among the tightest biomolecular interactions. When complexed with RI, ribonucleases cannot bind or degrade RNA. RI’s affinity to ribonucleases has been shown to be the primary determinant of ribonuclease cytotoxicity.21, 22 To date, the biological role of RI is not entirely understood.
In this study, we provide the initial evaluation of RI expression in the human kidney and urinary tract during states of sterility and infection. We investigate whether RI binds RNase 7 or influences RNase 7’s antimicrobial properties against uropathogenic bacteria. This work provides the basis for a better understanding of the potential roles of the RI-RNase 7 complex in maintaining urinary tract sterility.
Results
With pyelonephritis, RNH1 mRNA expression decreases concomitantly with increased RNASE7 mRNA expression
Quantitative real-time PCR demonstrates that RNH1, the gene encoding RI peptide, is basally expressed in all tested samples (Figure 1A). In the human bladder (n=4), mean RI expression was 1 692 ± 283 transcripts per 10ng RNA. RNH1 expression was significantly greater in the bladder than within the kidney (p=0.0018). In human kidney tissue (n=4), RNH1 expression was analyzed separately in the cortex, medulla, and pelvis (Figure 1A). RNH1 expression was greatest in the renal pelvis with a mean expression of 1 157 ± 208 transcripts per 10ng RNA.
Figure 1. RNH1 and RNASE7 mRNA expression in the human urinary tract during sterility and infection.
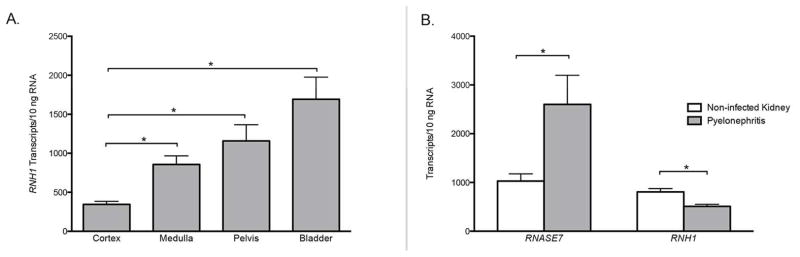
(A) RNH1, the gene encoding RI, transcript levels were quantified by real-time PCR in non-infected human bladder and kidney tissue. RNH1 mRNA expression was significantly greater in the bladder than the kidney. (B) RNASE7 and RNH1 mRNA transcript levels in non-infected human kidney tissue and in kidney tissue with pyelonephritis. RNH1 expression significantly decreased with pyelonephritis (p=0.0015) while RNASE7 expression increased (p=0.0317). Shown are the results of three independent experiments and the SEM.
In human kidney specimens with pyelonephritis, RNH1 mRNA expression was significantly lower as compared to non-infected kidney (Figure 1B). Non-infected kidney tissue (n=7) had a mean RNH1 expression of 806 ± 68 transcripts per 10 ng RNA. Kidneys with the histological diagnosis of pyelonephritis (n=8) expressed 509 ± 42 transcripts RNH1 per 10 ng RNA (p=0.0015). In contrast, with pyelonephritis, kidney RNASE7 expression increased from 1 028 ± 150 transcripts per 10 ng RNA (n=7) to 2 602 ± 595 transcripts per 10 ng RNA (n=7, p=0.035) (Figure 1B).
RI peptide production decreases with pyelonephritis while RNase 7 peptide production increases
To assess for concurrent decreases in kidney RI peptide production and increases in RNase 7 peptide production with pyelonephritis, ELISA assay and immunoblot blot analysis were performed on the same kidney tissues used for quantitative real-time PCR analysis. Results confirm the mRNA expression results. Specifically, RI peptide production decreases and RNase 7 peptide production increases in kidney tissues with pyelonephritis (n=6) compared to non-infected kidney tissue (n=6, Figure 2 and Supplemental Figure 1).
Figure 2. Kidney RI and RNase 7 peptide production during sterility and pyelonephritis.
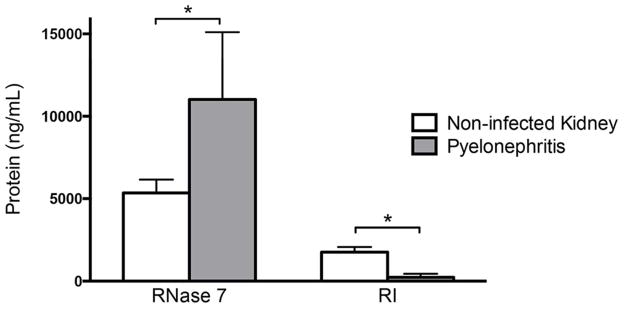
To confirm the mRNA expression of RNH1 and RNASE7 pattern is accompanied by alterations in peptide production, ELISA quantitated RI and RNase 7 peptide from the same non-infected kidney tissue and kidney tissue with pyelonephritis used in real-time PCR analysis. RI production significantly decreased with pyelonephritis (p=0.0068) while RNase 7 expression significantly increased (p=0.0386).
RI peptide is produced by the uroepithelium of the lower urinary tract and renal collecting tubules
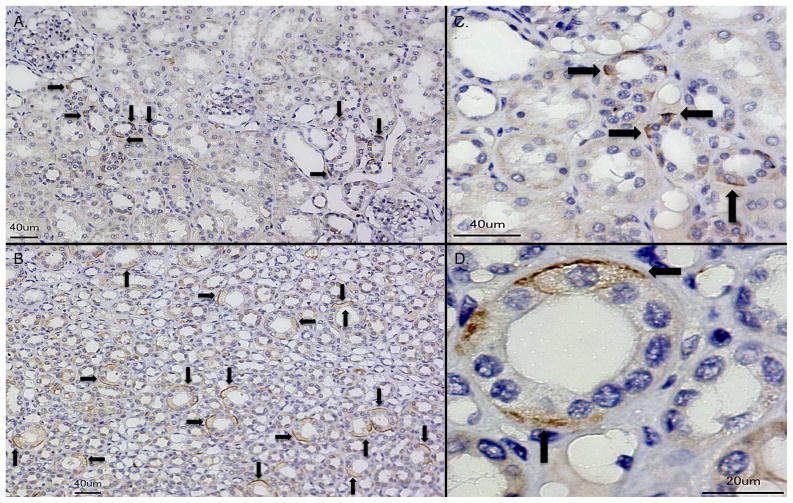
Immunohistochemistry (IHC) showed that RI immunoreactivity was greatest in the superficial umbrella cells lining the uroepithelium of the ureter and bladder. RI was not detected in the mesenchymal compartments of the ureter or bladder of all investigated specimens (n = 4, Figure 3). In the kidney, IHC demonstrated that RI was produced in tubules of the renal cortex and medulla of non-infected and infected kidney tissue. RI showed cell-specific expression in the cortical and medullary collecting tubules. RI positive cells were more readily detectable in the inner medulla than the cortex (n=4, Figure 4, Supplemental Figure 2). In both states of sterility and infection, RI was not routinely detected in the glomeruli, proximal nephron, or interstitium. RI expression was noted in erythrocytes and infiltrating leukocytes (not shown). Negative controls showed no RI immunoreactivity (Supplemental Figure 2).
Figure 3. RI is expressed throughout the urothelium of the ureter and bladder.
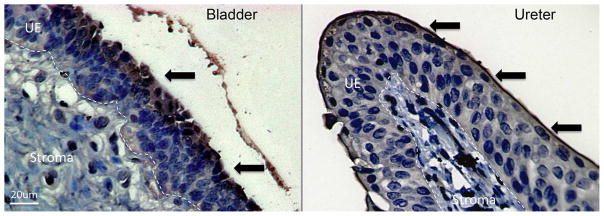
Immunohistochemistry demonstrates RI expression (brown/arrows) in the superficial uroepithelial layers of bladder and ureter. Dashed lines demarcate the border between the uroepithelial layers (UE) and the stromal layers of the bladder and ureter. Magnification 40x.
Figure 4. RI shows cell-specific expression throughout the human kidney.
Immunohistochemistry demonstrates isolated, cell specific RI expression (brown/arrows) in the collecting tubule of human kidney cortex (A) and medulla (B). Magnification 20x. (C–D) RI expression in renal collecting tubules at higher magnification (40x and 100x, respectively).
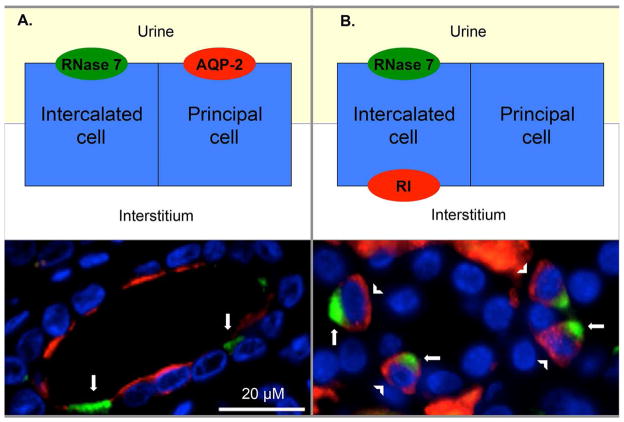
Intercalated cells express RI
Double labeling immunofluorescence (IF) allowed us to localize RI production in the collecting tubules. Principal cells of the collecting tubules were identified by apical expression of aquaporin-2 (AQP-2). As previously demonstrated, intercalated cells were identified by RNase 7 staining that was greatest on the apical cell surface.6 RI showed cell-specific, cystosolic expression that was greatest on the basolateral surfaces of the intercalated cells (Figure 5). Negative controls showed no immunoreactivity (not shown).
Figure 5. Intercalated cells express RI in the renal collecting tubule.
(A) Human kidney was labeled RNase 7 (green/arrows), aquaporin-2 (AQP-2/red), and nuclei (blue). Principal cells, identified by AQP-2 positive staining, were negative for RNase 7. (B) Human kidney was labeled with RNase 7 (green/arrows), RI (red/arrowheads), and nuclei (blue). RNase 7 positive cells/intercalated cells express RI. Magnification 100x.
Infected human urine contains full-length and degraded RI
Immunoblot analysis did not routinely detect RI in sterile human urine samples (n=10). However, full-length RI (~50kDa) and degraded RI (~25 kDa) were frequently detected in unspun urine samples infected with Escherichia coli (E. coli) (n=8) (Figure 6A, Supplemental Figure 3). In addition, full-length and degraded RI was also detected in unspun urine samples from patients with nephritis, hematuria, and sterile pyuria (n=5, not shown). RI was not detected in bacterial isolates of uropathogenic E. coli (PEDUTI-89/CFT073) or Enterococcus faecalis (E. faecalis) (PEDUTI-983).
Figure 6. RI is detected in the urinary sediment.
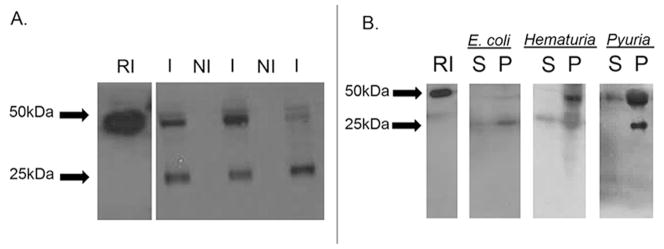
(A) Cationic peptides from unspun non-infected urine and urine infected with E. coli were subjected to SDS-PAGE followed by Western immunoblot analysis. Immunoblot analysis demonstrates that infected urine samples (I) contained full length RI (~50kDa) and degraded RI (~25kDa). RI was not detected in all non-infected (NI) urine samples. 25ng recombinant RI served as control.
(B) Cationic peptides from the supernatant and pellet fractions of centrifuged urine samples with gross hematuria, sterile pyuria, and infected urine with E.coli were subjected to SDS-PAGE followed by Western immunoblot analysis. Full-length RI was readily detected in the pellet/cellular fraction (P) compared to the supernatant (S). 25ng recombinant RI served as control.
To assess whether the urine sediment is the predominant source of urinary RI, immunoblot analysis was performed on centrifuged urine samples from patients with E. coli UTI, gross hematuria, or sterile pyuria (n=6). Full-length RI was not routinely detected in the supernatant fraction. It was routinely detected in the insoluble pellet fraction, suggesting that urinary RI originates from epithelial cells, erythrocytes, and/or infiltrating leukocytes of the urinary sediment (Figure 6B).
Neutrophil proteases degrade RI
Because infected urine contains degraded RI, we evaluated whether neutrophil proteases could participate in RI processing. We incubated recombinant RI with the neutrophil proteases proteinase 3, human neutrophil elastase, and trypsin.23, 24 To assess the specificity of neutrophil proteases on RI degradation, we also incubated recombinant RI with urokinase, an endogenous urinary protease. Urokinase and trypsin did not cause significant RI proteolysis (not shown), while RI was readily degraded by the neutrophil proteases proteinase 2 and human neutrophil elastase (Figure 7A). To assess if bacterial proteases are the source of RI proteolysis, recombinant RI was incubated with clinical bacterial isolates of uropathogenic E. coli (as outlined in Supplemental Table 1). Significant RI proteolysis was not detected (Supplemental Figure 4).
Figure 7. Neutrophil proteases degrade RI.
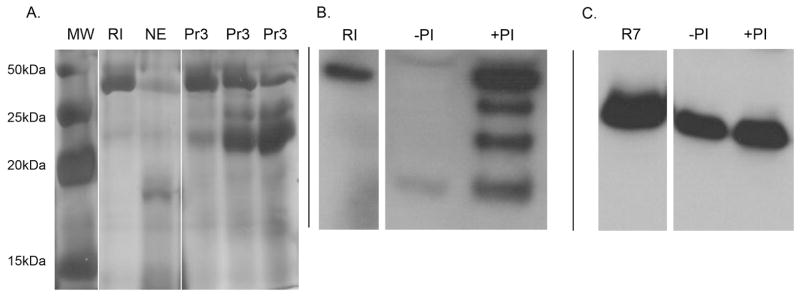
(A) Representative silver stained SDS-PAGE gels demonstrating recombinant RI proteolysis by the neutrophil proteases neutrophil elastase (NE) and proteinase 3 (Pr3). Lane 1: molecular weight marker (MW), Lane 2: 3 μg recombinant RI without proteases, Lane 3: RI incubated with 100ng NE, Lane 4–6: recombinant RI incubated with 6ng, 30ng, and 60ng Pr3, respectively. (B/C) Clinical urine isolates infected with E. coli were incubated with and without protease inhibitor cocktail (PI) and subjected to SDS-PAGE followed by Western immunoblot analysis using a monoclonal antibody directed against RI or a polyclonal antibody directed against RNase 7. (B) Results demonstrate that urine proteases degrade urinary RI and protease inhibitor cocktail prevents RI proteolysis. (C) Urinary proteases did not cause significant RNase 7 degradation. 50ng recombinant RI or RNase 7 served as control.
The addition of protease inhibitor cocktail to clinical urine samples infected with E. coli blocked RI proteolysis (n=3, Figure 7B). In contrast, urinary RNase 7 from the same infected urine isolates was not degraded in the absence of protease inhibitor cocktail (Figure 7C).
Recombinant RI binds RNase 7
Binding of recombinant RNase 7 to RI was evaluated by native gel electrophoresis and Western immunoblot analysis. Silver stained native gels demonstrate that recombinant RI binds RNase 7 (Figure 8A). Due to the different isoelectric points of RI (pI=4.7) and RNase 7 (pI=10.5), native gel electrophoresis demonstrates a shift in the mobility of RI upon complexing with RNase 7. To confirm peptide identity, immunoblot analysis was also performed using an anti-RNase 7 polyclonal antibody (Sigma-Aldrich, St Louis, MO, USA) and an anti-RI monoclonal antibody (Abcam) (not shown). Pre-incubation of RI with 5mM p-hydroxymercuribenzoate (p-HMB), a thiol-blocking agent that irreversibly inactivates RI, prevents RI from binding RNase 7 (Figure 8A).17
Figure 8. RI binds RNase 7.
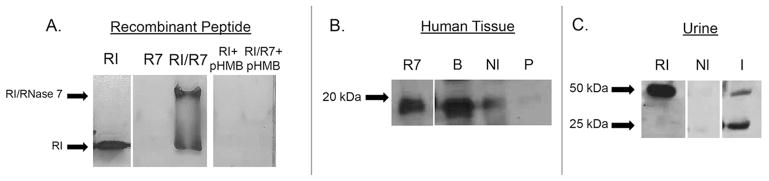
(A) Binding of recombinant RNase 7 to RI was confirmed by native gel electrophoresis. Representative native gels demonstrate a mobility shift in recombinant RI upon RNase 7 binding as visualized by silver-staining. Results also show that the interaction of RI with RNase 7 can be blocked by pre-incubation with the RI inhibitor p-HMB.
(B) Binding of endogenous RNase 7 to RI was confirmed using co-immunoprecipitation assays. Lysates of human bladder (B), non-infected kidney tissue (Nl), and kidney tissue with pyelonephritis (P) were immunoprecipitated with anti-RI monoclonal antibody, subjected to SDS-PAGE, followed by Western immunoblot analysis using anti-RNase 7 polyclonal antibody. Results indicate that RNase 7 complexes with RI in human bladder and kidney tissue. 25ng recombinant RNase 7 served as control.
(C) Urinary RI from non-infected urine (NI) and urine infected with E. coli (I) was pulled down using recombinant RNase 7 and subjected to SDS-PAGE followed by Western immunoblot analysis using anti-RI monoclonal antibody. Results demonstrate that urinary RI in infected urine binds recombinant RNase 7. 50ng recombinant RI served as control.
Endogenous RI binds RNase 7 in human kidney and urine
Binding of endogenous RNase 7 to RI was assessed by co-immunoprecipitation studies in human kidney and bladder tissue using an anti-RI monoclonal antibody (Abcam) followed by Western immunoblot analysis using an anti-RNase 7 polyclonal antibody (Sigma-Aldrich). Immunoblot analysis confirmed that endogenous RNase 7 complexes with RI in human kidney and bladder tissue. RI-RNase 7 binding decreased in kidney tissue with pyelonephritis (Figure 8B). No RNase 7 signal was detected by immunoblot when anti-RI antibody was not added in the immunoprecipitation step.
Urinary RI binding to RNase 7 was assessed by incubating sterile and infected urine samples with His-tagged recombinant RNase 7 that was complexed to nickel-nitrilotriacetic acid (Ni-NTA) affinity resin (Millipore, Billerica, MA, USA). Western immunoblot analysis, using anti-RI monoclonal antibody (Abcam), demonstrates that infected urine contains active, full-length RI that binds RNase 7. In contrast, sterile urine does not contain active RI (Figure 8C).
RI blocks the antimicrobial activity of RNase 7 in vitro
To determine if RI affects the antimicrobial activity of RNase 7, colony count reduction assays were performed. Uropathogenic bacteria were exposed to recombinant RNase 7 with and without RI. In the absence of RI, RNase 7 showed potent antimicrobial activity against uropathogenic E. coli and E. faecalis. Pre-incubation of recombinant RNase 7 with equal micromolar concentrations of RI strongly suppressed RNase 7’s antimicrobial activity. The addition of RI alone had no effect on bacterial survival (Figure 9A and Supplemental Figure 5).
Figure 9. RI inhibits the antimicrobial activity of RNase 7 against E. coli.
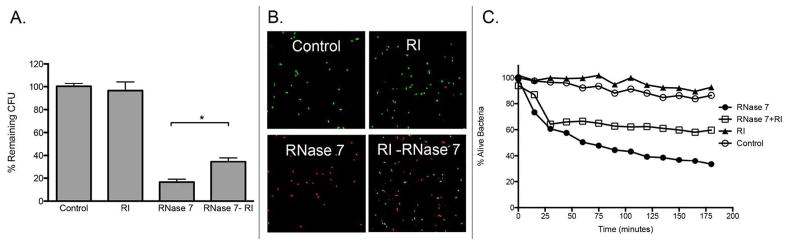
(A) Uropathogenic E. coli was exposed to 2 μM RNase 7, equal concentrations of recombinant RNase 7 pre-incubated with RI (RNase 7-RI), or 2 μM of RI. Untreated bacteria served as the control. The results are displayed as the percentage of remaining CFUs in relation to untreated controls. The antimicrobial activity of RNase 7 was significantly reduced in the presence of RI. Data represent the mean of triplicates ± SEM. (B) E. coli were stained using a 1:1 mixture of SYTO9 and propidium iodide. The SYTO9-stained cells (green) represent live cells and the propidium iodide-stained cells (red) represent killed cells. Bacterial viability was visualized after exposure to RNase 7 with and without RI at 180 minutes. Magnification 63x. (C) E. coli were stained using a 1:1 mixture of SYTO9 and propidium iodide and incubated with 2 μM RNase 7, equal concentrations of recombinant RNase 7 pre-incubated with RI (RNase 7-RI), or 2 μM of RI. Bacterial viability over time was analyzed integrating fluorescent changes in SYTO9 and propidium iodide dye. Values are the average of three replicates.
To visualize bacterial membrane integrity after incubation with recombinant RNase 7 in the presence and absence of RI, uropathogenic E. coli and E. faecalis were labeled using a Live/Dead bacterial viability assay (BacLight™, Molecular Probes, Carlsbad, CA, USA) and visualized with confocal microscopy. The Live/Dead bacterial viability assay uses a 1:1 mixture of STYO®9 and propidium iodide. SYTO®9 labels live bacteria while propidium iodide penetrates bacteria with damaged membranes. Our results confirm that RI abrogates the antimicrobial activity of RNase 7 (Figure 9B and Supplemental Figure 5B).
To evaluate how RI blocks the antimicrobial activity of RNase 7 over time, uropathogenic E. coli and E. faecalis were labeled using the Live/Dead bacterial viability assay and incubated with recombinant RNase 7, RNase 7 pre-incubated with RI, or RI alone. Changes in bacterial viability were monitored every 15 minutes by measuring the change in fluorescence SYTO®9 and propidium iodide. The ratio of SYTO®9 to propidium iodide fluorescence provides an estimate for the percentage of viable bacteria for monitoring the kinetics of bacterial death.5 Our results confirm that RI abrogates the antimicrobial activity of RNase 7 (Figure 9C and Supplemental 5C).
RI blocks urinary RNase 7 antimicrobial activity
To evaluate if RI blocks the antibacterial activity of urinary RNase 7, we inoculated human urine samples (n=4) from healthy individuals with uropathogenic E. coli strains associated with cystitis or pyelonephritis (PEDUTI-89 or CFT073, respectively).25, 26 We then determined whether the addition of recombinant RI would neutralize the antimicrobial activity of RNase 7 and lead to enhanced bacterial growth. The addition of recombinant RI inhibited the activity of RNase 7 as bacterial growth increased (Figure 10A and 10B, Supplemental Figure 6). As a positive control, the addition of antibody directed RNase 7 also neutralized the antimicrobial action of urinary RNase 7. The addition of RI storage buffer had no effect on bacterial growth (not shown).
Figure 10. RI neutralizes the antimicrobial activity of urinary RNase 7.
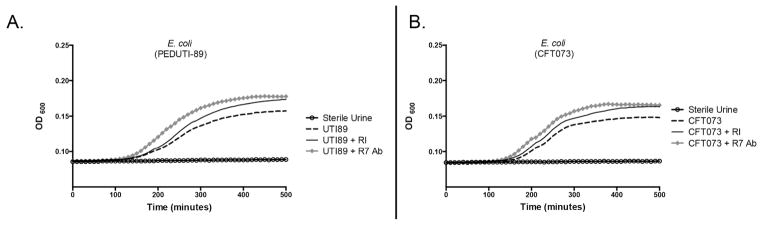
The antimicrobial properties of urinary RNase 7 were measured as changes in turbidity of cultured human urine using the absorbance at 600 nm (OD600). Human urine samples were inoculated with E. coli (PEDUTI-89 or CFT073) as shown by the dashed line. Addition of RI (solid black line) or RNase 7 monoclonal antibody (diamond studded line) blocked the antimicrobial activity of RNase 7, resulting in increased bacterial growth. The open circles represent non-inoculated urine samples.
RI blocks RNase 7 bacterial cell wall binding
To determine if RI affects RNase 7’s binding to the bacterial cell wall, we incubated uropathogenic E. coli and E. faecalis with recombinant RNase 7, RI, or equal micromolar concentrations of RI and RNase 7. Our results indicate that RNase 7 binds the cell wall of both gram-positive and gram-negative uropathogens as it was routinely recovered in the cell pellet fraction, which contains bound peptide (Figure 11A and Supplemental Figure 7A). When bacteria were incubated with the RI-RNase 7 complex, RNase 7’s ability to bind to uropathogens was decreased, as it was not as abundant in the pellet fraction. RI was routinely detected in the supernatant fraction, which contains unbound protein – indicating that RI cannot bind bacteria.
Figure 11. RI blocks RNase 7 binding to lipopolysaccharide.
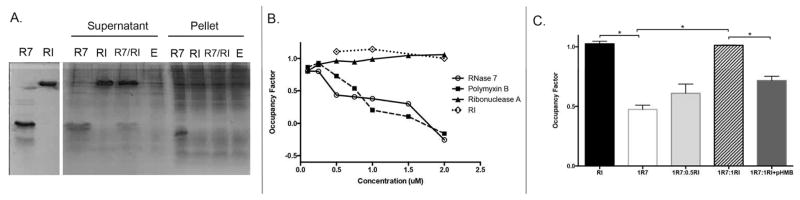
(A) To determine if RI alters RNase 7 bacterial binding, E. coli were incubated with 2 μM RNase 7 (R7), RNase 7 pre-incubated with RI (R7/RI), or 2 μM RI alone. After centrifugation, the supernatant and pellet fraction were subjected to SDS-PAGE and visualized by Coomassie Blue staining. Supernatant represents the soluble fraction that contains unbound protein while the pellet fraction contains the E.coli-bound peptides. Results demonstrate that RNase 7 binds uropathogenic E. coli (E) and that binding is reduced in the presence of RI. (B) Displacement of LPS-bound Bodipy TR cadavarine with increasing concentrations of RNase 7, Polymyxin B, RNase A, and RI. Results confirm that RNase 7 binds LPS while RI does not. (C) Displacement of LPS-bound Bodipy TR cadavarine with increasing concentrations of RI pre-incubated with 1 μM RNase 7. Results confirm that the addition of RI reduces RNase 7 binding to LPS. The addition of p-HMB to RI improves RNase 7 binding to LPS.
RI blocks RNase 7 binding to lipopolysaccharide and peptidoglycan
To gain additional insight into how RI affects RNase 7 binding to uropathogenic bacteria, binding studies on lipopolysaccharide (LPS) and peptidoglycan (PGN) were preformed.27 Binding to LPS, the main cell wall component of gram-negative bacteria, was assessed using the Bodipy TR cadavarine probe (BC) (Invitrogen, Carlsbad, CA, USA). BC binds the lipid A portion of LPS. The assay measures the competitive displacement of BC by other LPS-binding molecules.27–29 Our results show that RNase 7 has high-affinity binding toward LPS at micromolar concentrations. RNase 7 binding was comparable to polymyxin B, a powerful LPS binder (Figure 11B).27 In contrast, RI and Ribonuclease A did not bind LPS. When increasing concentrations of RI were incubated with 1μM of recombinant RNase 7, RNase 7’s binding to LPS significantly decreased (Figure 11C). Pre-incubation of RI with p-HMB prior to RNase 7 addition did not affect RNase 7-LPS binding.
We also assessed whether RI blocks RNase 7 binding to PGN, the main cell wall component of gram-positive bacteria. Our results demonstrate that RNase 7 binds to PGN as most of the peptide was recovered with the insoluble PGN pellet fraction. When RNase 7 was complexed with RI, PGN binding decreased (Supplemental Figure 7B).
Discussion
RNase 7 is a potent AMP that has an important antimicrobial role in maintaining urinary tract sterility.4–6 To date, the regulation of RNase 7 is not well defined. In this study, we begin to elucidate a regulatory mechanism of RNase 7’s antimicrobial activity. In doing so, we provide the initial characterization of RI in the human kidney, ureter, and bladder. We demonstrate that the urothelium of the lower urinary tract and intercalated cells of the collecting tubules express RI. We also show that endogenous RI binds RNase 7 and suppresses its antimicrobial action against uropathogenic bacteria. These are important, novel findings as they characterize a new immunomodulatory role for RI and identify a unique regulatory mechanism that affects RNase 7’s activity to help maintain sterility in the urinary tract. The presented data and conclusions only begin to define the role of RI in the urinary tract. However, they do provide a solid foundation for novel approaches and future studies that evaluate the cellular role of RI in the innate immune defense of the kidney and urinary tract.
Quantitative real-time PCR results demonstrate that RNH1, the gene encoding RI peptide, is expressed in the human kidney and urinary tract. Like RNASE7, RNH1 expression is greater in the bladder than the kidney.6 Immunostaining demonstrates that the differentiated, superficial umbrella cells of the ureter and bladder produce RI peptide. In addition, the basal stem cells of the uroepithelium only show limited RI production. This expression pattern mirrors RI peptide expression in the skin, where RI expression is greatest in differentiated keratinocytes when compared to proliferating keratinocytes.16 We hypothesize that this expression pattern provides a protective effect from unbound ribonucleases at the epithelial-urinary interface.
In the kidney, quantitative real-time PCR demonstrates that RNH1 expression complements RNASE7 expression in that it is greatest in the renal medulla and pelvis.6 IHC confirmed these results. With infection, RNase 7 and RI expression patterns considerably differ. RNASE7 expression and peptide production increase with pyelonephritis.5 In contrast, quantitative real-time PCR and ELISA indicate that RNH1 expression and RI peptide production decrease with pyelonephritis. These results suggest that, with infection, RNase 7 expression increases while RI expression decreases to enhance the antimicrobial activity of RNase 7 and facilitate bacterial clearance.
The decrease in RI peptide production may relate to how RI handles the oxidative stress that is generated with infection. Oxidative stress and the corresponding production of reactive nitrogen intermediates and reactive oxygen species is a fundamental mechanism in renal parenchymal inflammatory processes – including tubulointerstitial damage, pyelonephritis, and renal scarring.30, 31 RI is sensitive to these oxidant molecules and is degraded with oxidation.32, 33 Human RI contains 32 reduced cysteine residues. All of these cysteine residues must remain reduced for RI to maintain activity and bind ribonuclease peptides. Oxidation of a single cysteine residue causes rapid oxidation of the remaining cysteine residues through an “all-or-none” mechanism and induces a rapid conformational change that results in the inactivation of RI peptide and release of bound ribonucleases.32–34
Our results demonstrate that pyelonephritis and the corresponding production of oxidative stress, may impact the interactions of RI and RNase 7. In the kidney, immunoprecipitation assays demonstrate decreased RI-RNase 7 binding during infection/pyelonephritis. In vitro studies demonstrate that when RI’s sulfhydryl groups are modified by the addition of p-HMB, a process that closely resembles the effects of oxidative stress, RI is inactivated and cannot bind RNase 7.32, 35 These results, when used in conjunction with previously published studies, suggest that oxidation may be a mechanism by which the activity of RI, and the subsequent antimicrobial activity of RNase 7, is regulated in the urinary tract.
Alternatively, the decrease in RI peptide production may also relate to endogenous proteolysis during infection. Previously, Abtin et al demonstrated that stratum corneum serine proteases participate in the antimicrobial defense of the skin as they are responsible for RI proteolysis.16 Additionally, proteases have been implicated in the direct processing of epithelial and neutrophil antimicrobial prepropeptides.36–38 In the urethra, Porter et al demonstrate that neutrophil proteases are involved in the activation of human alpha defensin 5 (HD5).24 Our results also suggest that neutrophil proteases are involved in the antimicrobial defense of the urinary tract, as they are responsible for RI degradation. We hypothesize that during states of kidney health, RI binds endogenous RNase 7 as a safeguard, limiting its antimicrobial action against resident flora and the urinary microbiome.39 During the neutrophil influx upon infection, proteases degrade RI thereby liberating RNase 7 from RI and promote RNase 7 antimicrobial activity. The role of specific proteases in the urinary tract is ongoing. The processing of RI by site-specific proteases could result in alterations in RNase 7’s antimicrobial profile – increasing its functional versatility and optimizing the balance between protecting the host microbiome and site-specific pathogen-directed activity.
Immunostaining demonstrates that both RI and RNase 7 are expressed in the cytosol of intercalated cells. However, within the intercalated cell, their expression patterns are significantly different. RI expression is greatest on the basolateral surfaces of intercalated cells while RNase 7 is greatest on the apical surfaces. These results indicate that RNase 7 is ideally positioned to defend against an ascending infection. Alternatively, they suggest RI is ideally positioned to serve as a “sentry” to safeguard intercalated cells from oxidative stress or the potentially damaging activity of adventitious, extracellular ribonucleases in the bloodstream and/or renal interstitium.
Immunoblot studies routinely detect RI in infected urine samples. However, RI is not considered a secreted peptide. Although RI inhibits secretory ribonucleases, it has not been detected in extracellular fluids.18, 22, 40 Its labile nature and sensitivity to thiol oxidation prevent it from maintaining activity outside the cytoplasm.21 Immunoblot studies confirm that RI is not secreted, as the urine sediment is the primary source of RI since it is routinely detected in the pellet/cellular fraction of centrifuged urine samples. Erythrocytes, infiltrating leukocytes, or bladder uroepithelia – cell types known to express RI – are the likely source of urinary RI expression.16, 33, 41 Although it is possible that some urinary RI peptide originates from filtration, RI is a rather large peptide (~49 kDa) so only a small portion should pass through the glomerular membrane. Moreover, previous evidence suggests that RI is not detected in plasma.18, 21 To persist in the urine, RI would also need to escape the efficient peptide absorption mechanisms of the proximal tubule.
Abtin et al show that RI suppresses the ribonucleolytic activity of RNase 7 and limits its ability to degrade cellular RNA.16 Previously published studies also demonstrate that RNase 7’s bactericidal mechanism is independent of its ribonuclease enzymatic. Instead, its bactericidal properties are dependent upon bacterial binding and direct membrane disruption.5, 14, 27 Our results indicate that RI abrogates the antimicrobial activity of RNase 7 by limiting its ability to bind gram-negative and gram-positive bacteria. Specifically, bacterial binding assays indicate that RI complexes with RNase 7 and blocks its ability to bind to LPS and PGN of the bacterial cell wall. As shown by Huang et al, RNase 7 possesses a flexible cluster of N-terminal cationic lysine residues that are critical for its bactericidal activity.14 Therefore, we postulate that RI masks the N-terminal amino acid residues that are critical for RNase 7’s antimicrobial action. Prior studies demonstrate that RI complexes to the N-terminal entity of ribonuclease A peptides.42 Alternatively, our recently published work suggests that alterations in RNase 7’s secondary structure markedly reduce its antimicrobial activity.7 Thus, RI may suppress RNase 7’s bactericidal effects by altering its secondary or tertiary structure. When RI complexes with RNase 1, it induces conformational changes in RNase 1 that alter its protein function.43 Current studies are ongoing by our research group to evaluate the structural/functional relationship of RNase 7 and RI.
Because this work focuses on the use of human specimens, there are some inherent weaknesses. First, sample size is limited due to availability of kidney samples with pyelonephritis. Moreover, obtaining serial kidney samples from the same patient or kidney samples in an acutely infected patient is not routinely feasible. Finally, due to patient privacy and protection, we do not have access to the bacterial etiology causing pyelonephritis for the kidney specimens utilized in this manuscript.
In conclusion, this is the first study to identify and quantitate the expression of RI in the human urinary tract during sterility and infection. Our results suggest that RI plays an important role in the regulation of the antimicrobial activity of RNase 7. Our results show that during pyelonephritis, RI expression decreases, which may occur in response to oxidative stress or proteolysis resulting from neutrophil recruitment. Furthermore, activation of the host innate immune system during infection may promote functional and/or quantitative down-regulation of RI. When RI is degraded it does not complex with RNase 7, thereby improving the antimicrobial activity of RNase 7. Studies to compare how RI regulates the antimicrobial activity of RNase 7 within the intercalated cell and during states of cystitis and pyelonephritis are warranted. Moreover, studies that evaluate how RI regulates the antimicrobial action of RNase 7 in vivo are warranted. Ongoing evaluation of the factors that regulate RI production and/or RNase 7’s antimicrobial activity may lend insight into the pathogenesis of UTIs and the development of novel treatment strategies for patients at risk for UTIs or chronic infections.
Materials and Methods
Study approval
Informed written consent was obtained from all patients participating in this study. For subjects less than 18 year of age, written parental/guardian consent was obtained. The Nationwide Children’s Hospital (NCH) Institutional Review Board approved this study along with the consent process and documents (IRB07-00383). Uropathogenic E. coli (PEDUTI-89) was recovered from a patient with a UTI.25 E. coli CFT073 was isolated from the blood and urine of a woman with pyelonephritis.26 Additional uropathogenic bacterial isolates were obtained from patients seeking treatment for UTIs at NCH as approved by the Institutional Review Board (IRB-06-00603).
Human Samples
Non-infected kidney samples (n=7) were obtained from adults undergoing nephrectomy for renal tumors. Tissue samples were free of microscopic signs of disease or inflammation. Infected kidney tissue was provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute (Supplemental Table 2).44 Due to patient privacy and confidentiality, the Cooperative Human Tissue Network does not provide clinical information regarding the patient’s clinical history of pyelonephritis – including presenting symptoms, microbial etiology, laboratory evaluation (i.e. leukocyte count, c-reactive protein), or imaging modalities (including renal ultrasound, dimercaptosuccinic acid scan, or voiding cystourethrogram). However, these adult patients underwent nephrectomy due to the clinical diagnosis of pyelonephritis (n=8). Two independent pathologists confirmed the histopathologic diagnosis of pyelonephritis. Non-infected (n=10) and infected urine (n=10) samples were obtained from children presenting to NCH. The diagnosis of UTI was made by a positive urine culture according to the American Academy of Pediatrics Guidelines.45 All infected urine samples had >104 CFU/mL of E. coli and the presence of pyuria (Supplemental Table 1). Protease inhibitor cocktail was added to unspun urine samples, centrifuged urine samples, and the urinary pellet fraction that was isolated after centrifugation.
Identification of E. coli Serotype and Virulence Factors
In addition to standard culture techniques, uropathogenic E. coli isolates were serologically typed using the previously published multiplex PCR method.46 This multiplex PCR method simultaneously detects fourteen E. coli serogroups (O1, O2, O4, O6, O7, O8, O15, O16, O18, O21, O22, O25, O75, and O83). Additional E. coli virulence factor genes (including papAH, papC, fimH, hlyA, cnf1) were also identified using multiplex PCR as previously published (Supplemental Table 2).47
Ribonucleic Acid Isolation and Reverse Transcription
Total RNA was isolated from frozen tissue using the Promega Total RNA Isolation System (Promega, Madison, WI, USA). For cDNA synthesis, 4–8 μg of total RNA was reverse transcribed with Superscript III reverse transcriptase using an oligo-(dT)12–18 primer according to the supplier’s protocol (Invitrogen, Carlsbad, CA, USA).
Cloning of Gene Specific Plasmids for Standard Curves
cDNA encoding RNASE7, RNH1, and GAPDH were cloned into a 4-Topo plasmid vector (Invitrogen) according to the manufacturer’s instructions. Plasmids were sequenced to confirm that the correct constructs were obtained. Serial dilutions of gene specific plasmids were quantitated and used in real-time PCR experiments to generate standard curves.6, 48
Quantitative Real-time PCR
Real-time PCR was performed using single-stranded cDNA from human kidney tissue with specific oligonucleotide primer pairs using the 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). All reactions were performed in triplicate. PCR primers were selected using previously published standards and sequences were confirmed using DNAstar® Laser Gene SeqBuilder. Previously published RNase 7 and GAPDH primers were used.5 RI forward primer: 5′-CAT CAG CTC TGC ACT TCG AG-3′ and RI reverse primer 5′-CAA GAG GTT GTC GCT GAG GT-3′.16 Gene specific plasmid standards were included with every set of reactions.
Briefly, cDNA corresponding to 10 ng RNA served as the template in a 25 μL reaction of 0.1 μmol/L of each primer and 1x Light-Cycler-Fast Start DNA Master SYBR green mix. The PCR conditions were: initial denaturation at 95°C for 10 min, followed by 40 cycles with each cycle consisting of denaturation at 95°C for 30 seconds, annealing at 65°C for 30 seconds, and extension at 72°C for 30 seconds. Representative products were subjected to agarose gel electrophoresis with ethidium bromide staining to confirm the specificity of the PCR reactions. A melting temperature profile curve was performed with each reaction.
Expression and Purification of Recombinant RNase 7
The human RNASE7 sequence was generated from a kidney cDNA library and cloned into E. coli expression vector pDEST17 (Invitrogen, Carlesbad, CA, USA), which adds a six-residue histidine tag at the N-terminus.5, 7 Protein expression in the E. coli BL21 AI strain (Invitrogen), folding of the peptide from inclusion bodies, and the purification steps were carried out as described previously.5, 7
Enzyme-linked immunosorbent assay
A RNase 7 ELISA assay was used as previously described with the exception that a monoclonal anti-RNase 7 antibody was used as the detecting antibody (Atlas Antibody, Stockholm, Sweden).6 No cross reactivity was noted with 150 ng recombinant Ribonuclease 3, 5, 6, or 8. To quantitate kidney RNase 7 peptide production, 200 ng of frozen kidney tissue was homogenized in phosphate buffered saline with protease inhibitor cocktail and diluted 1:10. Samples were measured in triplicate. Results using our RNase 7 ELISA were comparable to that of a commercially available RNase 7 ELISA kit (MyBioSource, San Diego, CA, USA). Kidney RI concentrations were also measured in triplicate using Elabscience RI ELISA (Donghu, China). RNase 7 and RI peptide concentrations were standardized to kidney GAPDH production.
SDS-PAGE and Native Gel Immunoblot
For Western immunoblot analysis on human specimens, samples were prepared and processed as previously described.5 Equal concentrations of kidney tissue or urinary protein were loaded onto 15% sodium dodecyl sulfate (SDS) gels and subjected to electrophoresis. After electrophoretic separation, proteins were transferred onto a polyvinylidene difluoride membrane. The membranes were blocked and incubated with mouse monoclonal anti-human RI antibody (Abcam). After washing, a horseradish peroxidase-conjugated anti-mouse antibody was applied for detection (Cell Signaling Technology, Danvers, MA, USA). Proteins were visualized using and ECL detection system. To confirm antibody specificity and ensure no cross reactivity to human immunoglobulin, which may be present during UTI, human sera (15 ng–160ng, Sigma) served as a negative control (Supplemental Figure 3).
Binding of RNase 7 to RI was evaluated by native gel electrophoresis. Recombinant RI (Ambion/Life Technologies, Grand Island, NY, USA), RNase 7, or equal micromolar concentrations of RI plus RNase 7 were loaded onto a 12% Tris-HCl (pH 6.8) gel and subjected to electrophoresis. In addition, RI was pre-incubated with 5mM p-HMB at 37°C for 30 minutes before incubation with RNase 7 at 37°C for 60 minutes. Peptides were visualized by silver staining.
Immunohistochemistry
Following deparaffinization, rehydration, and antigen retrieval, a biotin block and a serum-free protein block were performed (Superblock, ScyTek Laboratories, Logan, UT, USA). The slides were incubated overnight at 4°C with monoclonal mouse RI antibody (Abcam), washed, and incubated with anti-polyvalent biotinylated antibody and UltraTek Streptavidin/HRP (ScyTek Laboratories). Sections were developed using 0.1% diaminobenzidine tetrachloride with 0.02% hydrogen peroxide and counterstained with hematoxylin. Negative controls sections were incubated with non-immune serum in place of RI antibody.
Immunofluorescence
Double-labeled immunofluorescence was performed to localize RI expression in the kidney. Sections were incubated with a mixture of antisera against RI (1:250) and antisera against AQP-2 (1:500) or RNase 7 (1:100). Principal cells were identified with a goat polyclonal anti-human AQP-2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and intercalated cells with a rabbit polyclonal anti-human RNase 7 antibody (Sigma-Aldrich).6 Rhodamine donkey polyclonal anti-goat (Jackson ImmunoResearch Laboratories, West Grove, PA, USA), rhodamine goat anti-mouse (Jackson ImmunoResearch Laboratories), and FITC donkey polyclonal anti-rabbit (Santa Cruz) served as the secondary antibodies. The slides were examined with a Leica DM4000B microscope and digitally photographed using Spot RT camera/software (Diagnostic Instruments, Sterling Heights, MI, USA).
Co-Immunoprecipitation Assay
To evaluate whether endogenous RI binds RNase 7 in human kidney and bladder tissue, co-immunoprecipitation assays were performed using Dynabeads® Protein G resin (Invitrogen). 10 μg of mouse monoclonal RI antibody (Abcam) was complexed to the Dynabeads resin for 30 minutes at room temperature. The Dynabeads-anti RI antibody complex was washed and incubated with 400 μg of sterile human kidney, 400 μg of human bladder lysates, or 800 μg of pyelonephritis kidney lysates for one hour. After washing the Dynabeads-anti RI antibody-antigen complex, the anti RI antibody-antigen complex was eluted from the Dynabeads resin and suspended in 1x electrophoresis loading buffer. Samples were loaded on SDS-PAGE followed by Western immunoblot analysis using rabbit anti-RNase 7 antibody (Sigma-Aldrich). The RNase 7 positive signal detects RNase 7 peptide that was endogenously complexed to RI in tissue lysates. Samples processed without the addition of the anti-RI antibody in the immunoprecipitation reaction served as the control.
Urine Peptide Binding Assay
To assess if urinary RI binds RNase 7, urinary RI-recombinant RNase 7 binding was evaluated using immobilized metal affinity chromatography. Histidine-tagged recombinant RNase 7 (10 μg) was complexed to 8 μL Ni-NTA affinity resin (Millipore) for one hour at room temperature. The resin was washed and resuspended in 1x Binding Buffer. Sterile and infected urinary proteins with protease inhibitor were concentrated using Amicon Ultra Centrifugal Filters with a 3 kDa cutoff (Millipore) and adjusted to pH 6.8 in 5x binding buffer (250 mM NaH2PO4, pH 6.8, 1.5 M NaCl, 25 mM imidazole). To each urine concentrate, equal volumes of bound recombinant RNase 7-Ni-NTA resin were added. Samples incubated for one hour, were washed, and centrifuged. The pellet fraction was resuspended in 1x SDS-Lamelli buffer, boiled, and subjected SDS-PAGE and Western immunoblot as described above using anti-RI monoclonal antibody (Abcam). Sterile urine, which does not contain RI, was used as the control.
Proteolysis Assays
3 μg recombinant RI was incubated with increasing concentrations of human neutrophil elastase (100 ng–500 ng) (Abcam), proteinase 3 (6 ng–60 ng) (Abcam), trypsin (5μg–25 μg, Mediatech Inc. Manassas, VA, USA), and urokinase from human kidney cells (50 U-2500 U, Sigma-Aldrich) for 90 minutes at 37°C. Samples were subjected to SDS-PAGE. Proteolysis was evaluated by silver stained gels.
Clinical urinary isolates infected with E. coli were incubated at 37°C for 90 minutes with and without protease inhibitor cocktail. Samples were subjected to SDS-PAGE followed by Western Immunoblot analysis using a monoclonal anti-RI (Abcam) antibody or a polyclonal anti-RNase 7 antibody (Sigma). To evaluate if bacterial proteases are responsible for RI degradation, recombinant RI (1.1 μg) was incubated with uropathogenic E. coli isolates (Supplemental Table 2) that were grown to OD600 = 0.8 in 1% peptone buffer for 90 minutes at 37°C. Samples were then subjected to SDS-PAGE followed by Western Immunoblot analysis using a monoclonal anti-RI antibody.
Antimicrobial Activity Assay
The antimicrobial activity of recombinant RNase 7 in the presence and absence of RI was estimated using colony count reduction assays with uropathogenic E. coli (PEDUTI-89) and E. faecalis (PEDUTI-983). Single cell colonies were grown to mid-exponential growth phase in a 37°C shaking incubator. Working dilutions of 104 bacteria/mL were prepared.5
Test uropathogens were incubated with recombinant RNase 7, equal micromolar concentrations of RNase 7 plus RI, or RI alone for three hours at 37°C. Before incubation with the uropathogens, RNase 7 was incubated with RI at 37°C for thirty minutes.16 Because RI is stored in 20 mM Hepes-KOH (pH 7.6), 50 mM KCI, 5 mM dithiothreitol and 50% (v/v) glycerol, the same buffer without RI was used in control reactions. The antimicrobial activity of RNase 7 was analyzed by plating the incubation mixtures on Laria-Bertani agar, incubating the plates overnight at 37°C, and determining the CFUs the following day. The growth inhibitory activities of the substances were calculated as follows: [(CFU after incubation with substance)/(CFU after incubation without substance)] x 100, which represents the percentage of remaining CFUs after treatment.16
Live/Dead Bacterial Viability Assay
Bacterial viability assays were also performed using a Live/Dead BacLight™ bacterial viability kit (BacLight™, Molecular Probes, Carlsbad, CA).5 Stained E. coli or E. faecalis were incubated with RNase 7, RNase 7 plus equal micromolar concentrations of RI, RI alone, or the RI storage buffer (see above). Changes in fluorescent intensity were measured using the Spectramax M2 multi-mode microplate reader (Molecular Devices, Sunnyvale, California, USA).5 These results were compared to a standard curve that was generated using increasing concentrations of live:dead bacteria. Samples were performed in duplicate.
Confocal Microscopy
Bacterial viability assays were performed using a Live/Dead BacLight™ bacterial viability kit (BacLight™, Molecular Probes, Carlsbad, CA, USA).5 Uropathogenic E. coli (UTI-89) and E. faecalis (PEDUTI-983) were incubated with recombinant RNase 7, recombinant RNase 7 plus RI, or RI alone as described above. 10 μL of this mixture was added to poly-l-lysine coated microscope slides (Polysciences, Inc, Warrington, PA, USA) and confocal images of the bacteria were obtained using a Zeiss LSM 710 confocal laser-scanning microscope (Carl Zeiss LLC, Thornwood, NY, USA). Samples were imaged in triplicate.
Urinary Antimicrobial Neutralization Assay
The ability of RI to neutralize the antimicrobial activity of urinary RNase 7 against uropathogenic E. coli (PEDUTI-89 or CFTO73) was evaluated in human urine (n=4) as previously described.6 In brief, bacteria were cultured in Luria-Bertaini broth to an OD670 = 0.08. Bacteria (2 μL) were added to 100 μL of culture-negative human urine in a 96-well flat bottom plate. Because culture-negative urinary RNase 7 concentrations range from (0.03–0.3 μmol/L), 0.3 μmol/L of RI, 2 μL (0.6 μg) of a monoclonal RNase 7 antibody (Atlas Antibody), or 2 μL of RI storage buffer was added to each test isolate. Bacterial growth (OD600) was monitored using a Synergy HT-multimode microplate reader (BioTek Instruments, Winooski, VT).
Bacterial Binding Assay
Uropathogenic E. coli or E. faecalis cells were grown to OD600 = 0.8 and 0.4, respectively in 1% peptone buffer and incubated with 2.5 μM recombinant RNase 7, equal micromolar concentrations of recombinant RNase 7 plus RI, or 2.5 μM RI alone for one hour at 37°C. After incubation, the cells were centrifuged; proteins from the pellet fraction were extracted in electrophoresis loading buffer and supernatant fractions were concentrated using Amicon Ultra Centrifugal Filters with a 3kDa cutoff (Millipore) before adding loading buffer. Samples were analyzed by SDS-PAGE (15%) and visualized by Coomassie Blue staining.
Affinity Binding Assay for LPS
LPS binding was assessed using the fluorescent probe Bodipy TR cadavarine.27, 29, 49 Bodipy TR cadavarine (BC) strongly binds to native LPS, specifically recognizing the lipid A portion. When a peptide that interacts with LPS is added, the BC probe is displaced and its fluorescence increases.
The displacement assay was performed in a quartz cuvette by adding increasing concentrations of Polymyxin B (Sigma-Aldrich), RNase A (Sigma-Aldrich), RNase 7, RI, or RNase 7 plus RI to 1 mL of a continuously stirred mixture of LPS (1 μg/mL) labeled with BC (10 μM) in 5 mM Hepes buffer at pH 7.5. LPS was from the E.coli serotype 0111:B4 (Sigma-Aldrich). Polymyxin B (Sigma-Aldrich), a potent LPS binder was chosen as the positive control and RNase A was the negative control. Fluorescence measurements were performed in the Spectramax M2 multimode reader (Molecular Devices, Sunnyvale, CA, USA). The BC excitation wavelength and emission wavelength were 584 nm and 620 nm, respectively. Final values represent the mean of three replicates.
Quantitative effective displacement values (ED50) were calculated.28, 29 The ED50 was computed at the midpoint of the fluorescent signal versus the peptide concentration of the displacement curve by curve-fitting the data to the equation: OF = (F0−F)/(F0−Fmax). OF is the occupancy factor, F0 the fluorescence intensity of BC alone, Fmax the BC intensity in the presence of LPS at saturation, and F is the intensities of the LPS/BC mixtures at each peptide concentration.
Affinity Binding Assay for PGN
Peptide binding to PGN was analyzed by SDS-PAGE electrophoresis as described previously.27, 49 PGN was from Staphylococcus aureus (Sigma-Aldrich). Lysozyme (Sigma-Aldrich), BSA, RNase 7, RI, or RNase 7 plus RI were incubated 0.4 mg/mL PGN in 10 mM Tris-HCl (pH 7.5). Lysozyme and BSA were chosen as the positive and negative controls. Samples were kept at 4°C for 2 hours with gentle shaking and then centrifuged at 13 000g for 10 min to separate the soluble and insoluble fractions. Samples were resuspended in electrophoresis loading buffer, evaluated by SDS-PAGE, and visualized by Coomassie Blue staining.
Supplementary Material
Acknowledgments
We would like to acknowledge the Human Cooperative Human Tissue Network for providing the human samples. JDS is supported by the National Institute of Health Grant K08 DK094970-01. ALS and DSH are supported by the National Institute of Health Grant 1RC4DK090937-01.
Abbreviations
- RNase 7
Ribonuclease 7
- RI
Ribonuclease Inhibitor
- AMP
Antimicrobial Peptide
- IHC
Immunohistochemistry
- IF
Immunofluorescence
- AQP-2
Aquaporin-2
- p-HMB
p-hydroxymercuribenzoate
- Ni-NTA
nickel-nitrilotriacetic acid
- NCH
Nationwide Children’s Hospital
- CFU
Colony Forming Unit
- LPS
Lipopolysaccharide
- PGN
Peptidoglycan
- BC
Bodipy TR Cadavarine
Footnotes
Disclosure:
All the authors in this manuscript declared no competing interests.
Financial Disclosure and Conflict of Interest: The authors have no conflicts of interest or financial relationships relevant to this article to disclose.
References
- 1.Zasloff M. Antimicrobial peptides, innate immunity, and the normally sterile urinary tract. J Am Soc Nephrol. 2007;18:2810–2816. doi: 10.1681/ASN.2007050611. [DOI] [PubMed] [Google Scholar]
- 2.Ali AS, Townes CL, Hall J, et al. Maintaining a sterile urinary tract: the role of antimicrobial peptides. The Journal of urology. 2009;182:21–28. doi: 10.1016/j.juro.2009.02.124. [DOI] [PubMed] [Google Scholar]
- 3.Spencer JD, Schwaderer AL, Becknell B, et al. The innate immune response during urinary tract infection and pyelonephritis. Pediatr Nephrol. 2013 doi: 10.1007/s00467-013-2513-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zasloff M. The antibacterial shield of the human urinary tract. Kidney international. 2013;83:548–550. doi: 10.1038/ki.2012.467. [DOI] [PubMed] [Google Scholar]
- 5.Spencer JD, Schwaderer AL, Wang H, et al. Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney international. 2013 doi: 10.1038/ki.2012.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spencer JD, Schwaderer AL, Dirosario JD, et al. Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney international. 2011 doi: 10.1038/ki.2011.109. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Schwaderer AL, Kline J, et al. Contribution of Structural Domains to Ribonuclease 7’s Activity Against Uropathogenic Bacteria. Antimicrob Agents Chemother. 2012 doi: 10.1128/AAC.01378-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harder J, Schroder JM. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. The Journal of biological chemistry. 2002;277:46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 9.Simanski M, Dressel S, Glaser R, et al. RNase 7 protects healthy skin from Staphylococcus aureus colonization. J Invest Dermatol. 2010;130:2836–2838. doi: 10.1038/jid.2010.217. [DOI] [PubMed] [Google Scholar]
- 10.Koten B, Simanski M, Glaser R, et al. RNase 7 contributes to the cutaneous defense against Enterococcus faecium. PloS one. 2009;4:e6424. doi: 10.1371/journal.pone.0006424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reithmayer K, Meyer KC, Kleditzsch P, et al. Human hair follicle epithelium has an antimicrobial defence system that includes the inducible antimicrobial peptide psoriasin (S100A7) and RNase 7. Br J Dermatol. 2009;161:78–89. doi: 10.1111/j.1365-2133.2009.09154.x. [DOI] [PubMed] [Google Scholar]
- 12.Eberhard J, Menzel N, Dommisch H, et al. The stage of native biofilm formation determines the gene expression of human beta-defensin-2, psoriasin, ribonuclease 7 and inflammatory mediators: a novel approach for stimulation of keratinocytes with in situ formed biofilms. Oral microbiology and immunology. 2008;23:21–28. doi: 10.1111/j.1399-302X.2007.00385.x. [DOI] [PubMed] [Google Scholar]
- 13.Boix E, Nogues MV. Mammalian antimicrobial proteins and peptides: overview on the RNase A superfamily members involved in innate host defence. Molecular bioSystems. 2007;3:317–335. doi: 10.1039/b617527a. [DOI] [PubMed] [Google Scholar]
- 14.Huang YC, Lin YM, Chang TW, et al. The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity. The Journal of biological chemistry. 2007;282:4626–4633. doi: 10.1074/jbc.M607321200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Dyer KD, Rosenberg HF. Human RNase 7: a new cationic ribonuclease of the RNase A superfamily. Nucleic acids research. 2003;31:602–607. doi: 10.1093/nar/gkg157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abtin A, Eckhart L, Mildner M, et al. Degradation by stratum corneum proteases prevents endogenous RNase inhibitor from blocking antimicrobial activities of RNase 5 and RNase 7. The Journal of investigative dermatology. 2009;129:2193–2201. doi: 10.1038/jid.2009.35. [DOI] [PubMed] [Google Scholar]
- 17.Blackburn P, Wilson G, Moore S. Ribonuclease inhibitor from human placenta. Purification and properties The Journal of biological chemistry. 1977;252:5904–5910. [PubMed] [Google Scholar]
- 18.Futami J, Tsushima Y, Murato Y, et al. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA and cell biology. 1997;16:413–419. doi: 10.1089/dna.1997.16.413. [DOI] [PubMed] [Google Scholar]
- 19.Kobe B, Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 20.Hofsteenge J, Kieffer B, Matthies R, et al. Amino acid sequence of the ribonuclease inhibitor from porcine liver reveals the presence of leucine-rich repeats. Biochemistry. 1988;27:8537–8544. doi: 10.1021/bi00423a006. [DOI] [PubMed] [Google Scholar]
- 21.Lee FS, Vallee BL. Structure and action of mammalian ribonuclease (angiogenin) inhibitor. Progress in nucleic acid research and molecular biology. 1993;44:1–30. doi: 10.1016/s0079-6603(08)60215-9. [DOI] [PubMed] [Google Scholar]
- 22.Dickson KA, Haigis MC, Raines RT. Ribonuclease inhibitor: structure and function. Progress in nucleic acid research and molecular biology. 2005;80:349–374. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liau DF, Yin NX, Ryan SF. Isolation of human polymorphonuclear leukocyte elastase by chromatography on immobilized benzamidine. Preparative biochemistry. 1993;23:439–447. doi: 10.1080/10826069308544568. [DOI] [PubMed] [Google Scholar]
- 24.Porter E, Yang H, Yavagal S, et al. Distinct defensin profiles in Neisseria gonorrhoeae and Chlamydia trachomatis urethritis reveal novel epithelial cell-neutrophil interactions. Infection and immunity. 2005;73:4823–4833. doi: 10.1128/IAI.73.8.4823-4833.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulvey MA, Schilling JD, Martinez JJ, et al. Bad bugs and beleaguered bladders: interplay between uropathogenic Escherichia coli and innate host defenses. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:8829–8835. doi: 10.1073/pnas.97.16.8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mobley HL, Jarvis KG, Elwood JP, et al. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1–4) beta Gal binding in virulence of a wild-type strain. Mol Microbiol. 1993;10:143–155. doi: 10.1111/j.1365-2958.1993.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 27.Torrent M, Badia M, Moussaoui M, et al. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. The FEBS journal. 277:1713–1725. doi: 10.1111/j.1742-4658.2010.07595.x. [DOI] [PubMed] [Google Scholar]
- 28.Torrent M, Nogues MV, Boix E. Eosinophil cationic protein (ECP) can bind heparin and other glycosaminoglycans through its RNase active site. J Mol Recognit. doi: 10.1002/jmr.1027. [DOI] [PubMed] [Google Scholar]
- 29.Wood SJ, Miller KA, David SA. Anti-endotoxin agents. 1. Development of a fluorescent probe displacement method optimized for the rapid identification of lipopolysaccharide-binding agents. Comb Chem High Throughput Screen. 2004;7:239–249. doi: 10.2174/1386207043328832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurutas EB, Ciragil P, Gul M, et al. The effects of oxidative stress in urinary tract infection. Mediators of inflammation. 2005;2005:242–244. doi: 10.1155/MI.2005.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Celik S, Gorur S, Aslantas O, et al. Caffeic acid phenethyl ester suppresses oxidative stress in Escherichia coli-induced pyelonephritis in rats. Molecular and cellular biochemistry. 2007;297:131–138. doi: 10.1007/s11010-006-9337-x. [DOI] [PubMed] [Google Scholar]
- 32.Blazquez M, Fominaya JM, Hofsteenge J. Oxidation of sulfhydryl groups of ribonuclease inhibitor in epithelial cells is sufficient for its intracellular degradation. The Journal of biological chemistry. 1996;271:18638–18642. doi: 10.1074/jbc.271.31.18638. [DOI] [PubMed] [Google Scholar]
- 33.Moenner M, Vosoghi M, Ryazantsev S, et al. Ribonuclease inhibitor protein of human erythrocytes: characterization, loss of activity in response to oxidative stress, and association with Heinz bodies. Blood cells, molecules & diseases. 1998;24:149–164. doi: 10.1006/bcmd.1998.0182. [DOI] [PubMed] [Google Scholar]
- 34.Hofsteenge J, Servis C, Stone SR. Studies on the interaction of ribonuclease inhibitor with pancreatic ribonuclease involving differential labeling of cysteinyl residues. The Journal of biological chemistry. 1991;266:24198–24204. [PubMed] [Google Scholar]
- 35.Fominaya JM, Hofsteenge J. Inactivation of ribonuclease inhibitor by thiol-disulfide exchange. The Journal of biological chemistry. 1992;267:24655–24660. [PubMed] [Google Scholar]
- 36.Yamasaki K, Schauber J, Coda A, et al. Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. FASEB J. 2006;20:2068–2080. doi: 10.1096/fj.06-6075com. [DOI] [PubMed] [Google Scholar]
- 37.Panyutich A, Shi J, Boutz PL, et al. Porcine polymorphonuclear leukocytes generate extracellular microbicidal activity by elastase-mediated activation of secreted proprotegrins. Infection and immunity. 1997;65:978–985. doi: 10.1128/iai.65.3.978-985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scocchi M, Skerlavaj B, Romeo D, et al. Proteolytic cleavage by neutrophil elastase converts inactive storage proforms to antibacterial bactenecins. Eur J Biochem. 1992;209:589–595. doi: 10.1111/j.1432-1033.1992.tb17324.x. [DOI] [PubMed] [Google Scholar]
- 39.Wolfe AJ, Toh E, Shibata N, et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol. 2012;50:1376–1383. doi: 10.1128/JCM.05852-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinovitch MR, Sreebny LM, Smuckler EA. Ribonuclease and ribonuclease inhibitor of the rat parotid gland and its secretion. The Journal of biological chemistry. 1968;243:3441–3446. [PubMed] [Google Scholar]
- 41.Nadano D, Yasuda T, Takeshita H, et al. Ribonuclease inhibitors in human blood: comparative studies on the inhibitors detected in erythrocytes, platelets, mononuclear leukocytes and granulocytes. The international journal of biochemistry & cell biology. 1995;27:971–979. doi: 10.1016/1357-2725(95)00063-u. [DOI] [PubMed] [Google Scholar]
- 42.Arnold U, Leich F, Neumann P, et al. Crystal structure of RNase A tandem enzymes and their interaction with the cytosolic ribonuclease inhibitor. The FEBS journal. 2011;278:331–340. doi: 10.1111/j.1742-4658.2010.07957.x. [DOI] [PubMed] [Google Scholar]
- 43.Johnson RJ, McCoy JG, Bingman CA, et al. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. Journal of molecular biology. 2007;368:434–449. doi: 10.1016/j.jmb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LiVolsi VA, Clausen KP, Grizzle W, et al. The Cooperative Human Tissue Network. An update Cancer. 1993;71:1391–1394. doi: 10.1002/1097-0142(19930215)71:4<1391::aid-cncr2820710434>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 45.Roberts KB. Urinary tract infection: clinical practice guideline for the diagnosis and management of the initial UTI in febrile infants and children 2 to 24 months. Pediatrics. 2011;128:595–610. doi: 10.1542/peds.2011-1330. [DOI] [PubMed] [Google Scholar]
- 46.Li D, Liu B, Chen M, et al. A multiplex PCR method to detect 14 Escherichia coli serogroups associated with urinary tract infections. Journal of microbiological methods. 2010;82:71–77. doi: 10.1016/j.mimet.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. The Journal of infectious diseases. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 48.Spencer JD, Hains DS, Porter E, et al. Human alpha defensin 5 expression in the human kidney and urinary tract. PloS one. 2012;7:e31712. doi: 10.1371/journal.pone.0031712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torrent M, Navarro S, Moussaoui M, et al. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry. 2008;47:3544–3555. doi: 10.1021/bi702065b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.