Abstract
Enabling formulation based on hydroxypropyl-β-cyclodextrins (HPβCD), micellar preparation and liposomes have been designed to deliver the racemic mixture of a lipophilic CB2 agonist, MDA7. The antiallodynic effects of MDA7 formulated in these three different systems were compared after intravenous administration in rats. Stoichiometry of the inclusion complex formed by MDA7 in HPβCD was determined by continuous variation plot, ESI-MS analysis, phase solubility and NMR studies and indicate formation of exclusively 1:1 adduct. Morphology and particle sizes determined by DLS and TEM show the presence of a homogenous population of closed round-shaped oligolamellar MDA7 containing-liposomes, with average size of 117nm [polydispersity index (PDI) <0.1]. Monodisperse micelles exhibited average size of 15 nm (PDI 0.1). HPβCD based formulation administrated in vivo was composed of two discrete particles populations with a narrow size distribution of 3 nm (PDI<0.1) and 510 nm (PDI<0.1). HPβCD based formulation dramatically improved antiallodynic effect of MDA7 in comparison to the liposomes preparation. Through inclusion complexation and possibly formation of aggregates, HPβCD can enhance the aqueous solubility of lipophilic drugs thereby improving their bioavailability for i.v. administration.
Keywords: Chirality, Complexation, Cyclodextrins, Micelle, Bioavailability, CNS, Liposomes
Introduction
Following our ongoing project to develop novel cannabinoid type 2 (CB2) receptor agonist for the prevention and treatment of pain1,2, we recently reported the synthesis of MDA7, a CB2 selective agonist3. However, the racemic mixture of MDA7 showed poor water solubility restricting its evaluation in vivo4. Different approaches have been described to improve water solubility for lipophilic molecules. Formation of inclusion complexes with functionalized cyclodextrins, micellar systems and liposomes are the most commonly used. Cyclodextrins (CDs) are cyclic cone shaped α-1,4-linked d-glucopyranose units (Fig. 1). The interior of the cavity formed by the glucopyranose units are hydrophobic while the outer surface remains hydrophilic. CDs are one of the most used drug-carriers owing to their ability to form inclusion complexes with a wide variety of small lipophilic molecules. Inclusion complexes are water soluble and furthermore complexation can increase the stability of the guest molecules. Among chemically modified cyclodextrins, FDA-approved HPβCD have attracted growing interest based on their low toxicity, water solubility and complexation potential. However, complexation of cyclodextrins with racemic mixtures forms diastereomeric inclusion complexes which can lead to enantiodifferentiation. Due to this property, cyclodextrins have been extensively used as chiral stationary phases in HPLC and capillary electrophoresis. The eutomer of MDA7 has been identified and enantiodifferentiation can be induced by different complexation mode which could impact the solubility of both enantiomers and thereby the in vivo activity of MDA7. Nevertheless, considering the advantages of HPβCD, we prepared MDA7 inclusion complexes to enhance its aqueous solubility. MDA7:HPβCD inclusion complexes were studied to identify a potential enantiodifferenciation of racemic MDA7. For this purpose, MDA7:HPβCD inclusion complexes were characterized by phase-solubility diagram, Job's plots and differential scanning calorimetry. Association mode in solution was determined by NMR spectroscopy and ESI-mass spectrometry. HPβCD aggregates were characterized by Transmission Electron Microscopy (TEM) and Dynamic Light Scattering (DLS). Even though enantiodifferentiation between the two diastereomeric complexes formed by HPβCD and racemic MDA7 were detected by NMR studies and in silico modeling of the binding mode, no difference in term of solubility was detected for the two enantiomers. In order to ensure accurate and reproducible in vivo exposure, micellar solution and liposomes have been characterized. Finally, MDA7:HPβCD inclusion complex, MDA7 containing-liposomes and MDA7 micellar solution were administrated intravenously (i.v.) and compared using a rat model of neuropathic pain, the spinal nerve ligation model. To the best of our knowledge, this constitutes the first characterization of diastereomeric inclusion complexes of a chiral CB2 agonist complexed by HPβCD, followed by an in vivo comparison with other drug delivery systems.
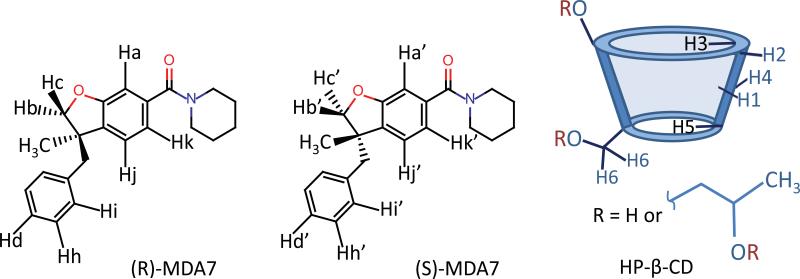
Figure 1.
Numbered chemical structures of the two enantiomers of MDA7 and of HPβCD.
Material and methods
MDA7 was synthesized according to the method previously described.4,5 2-Hydroxypropyl-β-cyclodextrin (HPβCD) with an average degree of molar substitution of 4.4 was purchased from CTD Holdings Inc (Alachua, FL, USA). Ethyl Alcohol, 190 Proof, 95%, ACS/USP was purchased from PHARMCO-AAPER (Shelbyville, KY, USA). Dimyristoylphosphatidylcholine (DMPC) was purchased from Avanti Polar Lipids (Alabaster, AL, USA). Dimethyl sulfoxide was purchased from SIGMA-ALDRICH (St. Louis, MO, USA). Phosphate Buffer Saline (PBS) was purchased from VWR. Deuterium oxide (D2O) and Methanol-d4 (MeOH-d4) were purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA, USA). Acrodisc® Syringe Filters with Nylon Membrane were purchased from PALL Corporation (Port Washington, NY, USA). Other reagents and chemicals were obtained as gift samples, N-methyl-2-pyrrolidone (NMP or Pharmasolve™) from ISP, propylene glycol USP and Cremophor™ ELP from BASF (Ludwigshafen, Germany), Super refined™ Polysorbate 20 from CRODA International Inc. (Edison, NJ, USA). All chemicals were used as received. All experiments were carried out using ultrapure water.
Preparation of complexes
MDA7 and HPβCD were mixed in appropriate ratios and solvents (ultrapure water or D2O or D2O/MeOH-d4 or PBS according to the study) for six days under constant magnetic stirring (200 rpm) at room temperature and protected from light.
Preparation of liposomes
MDA7 was dissolved in DMSO to afford a solution at 40 mg/mL and mixed to a solution of dimyristoylphosphatidylcholine (DMPC) at 50 mg/mL in tert-butanol in a molar ratio of 1:10 drug/lipids.6 Polysorbate 20 was added to the preparation in an amount corresponding to 5% w/w of the total amount of MDA7+lipids. 95 mL of tert-butanol was then added for each 5 mL of MDA7/lipids mixture previously obtained. The resulting solution was freeze-dried (Labconco-freeze dry system/Freezone® 1) for 48H, providing a white and dried powder containing drug, lipids and surfactant. Before administration to animals, the dry drug/lipids powder was hydrated with PBS to obtain the desired volume of administration, and stirred at 100 rpm for 20 min at 30°C, a temperature above the gel-liquid crystal transition temperature of DMPC. This process resulted in homogeneous milky-like oligolamellar vesicles (OLVs) liposomal suspensions.
Preparation of the micellar system
MDA7 (6 mg/mL) was stirred at 40°C in NMP (30 % v/v) until dissolution. Propylene glycol (30 % v/v), ethanol (10 % v/v) and Cremophor™ ELP (10 % w/v) were added dropwise at 40°C. The micellar system was obtained after dropwise addition of PBS or saline solution (qs 100 %v/v).
NMR spectroscopy
One-dimensional 1H NMR spectra were recorded at room temperature on Bruker Avance III™ spectrometer at 400 MHz using a 5 mm probe and a simple pulse-acquire sequence. Acquisition parameters consisted of spectral width 4000 Hz with an acquisition time 3.98 s, number of scans of 128, and relaxation delay of 1 s. Rotating-frame overhauser effect spectroscopy (ROESY) experiments were acquired in the phase sensitive mode with the same spectrometer and Bruker standard parameters (pulse program roesyphpr). For ROESY spectra, the time domain data was zero filled to 1024 points in F2 and 256 points in F1. The ROESY data was acquired with a spin-lock mixing time of 200 ms, number of scans 8, and a relaxation delay of 2 s. Complexes were prepared as described above in D2O.
Phase solubility studies
The phase solubility method was carried out according to Higuchi and Connors7 method to study the effect of complexation on MDA7 solubility in water. Complexes were prepared as described above in pure water. Excess amount of MDA7 (10 mg, 29.8 μmol) were mixed with increasing concentrations of HPβCD (0.5, 1, 2, 3, 4 molar equivalent). After equilibration, undissolved MDA7 was removed from the suspension by filtration using 0.45 μm membrane filter (Acrodisc® Syringe Filters). Quantities of MDA7 dissolved were assessed using NMR quantification, with maleic acid in D2O as external standard. Molar concentration of solubilized MDA7 ([MDA7]) were plotted versus molar concentration of HPβCD ([HPβCD]). From the slope of the linear fit, the apparent association constant was determined for MDA7 and HPβCD at the linear increase of MDA7 solubility according to equation 1. Complex stoichiometry is assumed to be 1:1 for determining the apparent association constant K1:1.
The complexation efficiency (CE) was calculated as a product or the intrinsic solubility (S0) and the association constant K1:1.
Continuous variation method (Job's plots)
Stoichiometry of inclusion complex was determined using continuous variation method. The 1H NMR spectrum of MDA7 in D2O could not be determined due to its very low aqueous solubility. Therefore, plot was determined from NMR chemical shift measurement in D2O/Methanol-d4 (1:1, v/v). The chemical shifts were referenced relative to the residual peak of MeOH-d4 at 3.310 ppm. The total molar concentration of MDA7 and HPβCD was kept constant (6 mM) and their mole fraction (r) varied from 0.1 to 0.9. In order to calculate the stoichiometry, the product of molar ratio (r) by chemical shift variations (Δδ) of the H1, H5 and CH3 HPβCD protons of HPβCD and Ha, Hb and Hc and CH3 protons of MDA7 were plotted versus r.
ESI-MS analysis
Stoichiometry of the inclusion complex prepared as described above in ultrapure water was confirmed by ESIMS analysis using acetonitrile/water as mobile phase. ESI-MS Experiments were performed on a Waters Micromass LCT Premier™ that utilizes a high-resolution time-of-flight (TOF) analyzer equipped with a W-Optics and a regular and nanoelectrospray ionization sources. The sample solutions were directly infused into the ESI source with a flow rate of 0.4 ml.min−1, capillary voltage 2900 V, sample cone voltage 95 V, desolvation temperature 150°C, source temperature 100°C in the positive ion mode.
Differential scanning calorimetry (DSC)
Complexes were prepared by dissolving MDA7 (15 mg, 44.7 μmol) and HPβCD as described above in 1.35 ml of ultrapure water at 1:1, 1:4, 1:6.7 molar ratio. After equilibration undissolved MDA7 was removed from the suspension with one equivalent of HPβCD by filtration using 0.45 μm membrane filter (Acrodisc® Syringe Filter). MDA7 was soluble at MDA7:HPβCD molar ratio of 1:4 and 1:6.7. Solutions were freeze-dried (Labconco-freeze dry system/Freezone® 1) for 24 hours under vacuum. The resulting solid inclusion complexes were collected and analyzed. A physical mixture was prepared by thoroughly mixing MDA7 and HPβCD at 1:4 molar ratio. An excess of HPβCD was employed to reflect the molar ratio used for in vivo studies. The samples were placed in hermetically sealed aluminum pans and the experiments run in a calorimeter (Mettler Toledo DSC1 STARe system) at 10°C/min heating rate over 25-400°C. The instrument was calibrated with indium. Thermograms for inclusion complexes were compared to physical mixture.
Transmission electron microscopy (TEM)
MDA7, HPβCD physical mixture and MDA7:HPβCD solid complexes were prepared as described above in ultrapure water. 1 μl aliquot of sample was placed in 4 μl of demineralized water on a formvar-coated 400 mesh copper grid. The sample was kept on the grid for 5 minutes, the liquid was wicked off with filter paper and the grid was air dried. The grid was stained by placing it on a drop of 2% Uranyl acetate for 1 minute. The stain was rinsed off by dipping the grid in demineralized water, the water removed by wicking, and the grid air dried. The grid was imaged in a Hitachi H-7100 TEM at 75kV. Liposomal preparation samples were placed on single slot formvar coated copper grids treated with poly-l-lysine for 1 hour. Excess samples were blotted with filter paper, and then stained with filtered 1% uranyl acetate for 1 min. Stain was blotted dry from the grids with filter paper and samples were allowed to dry. Samples were then examined in a JEM 1010 transmission electron microscope (JEOL, USA, Inc., Peabody, MA) at an accelerating voltage of 80 Kv. Digital images were obtained using the AMT Imaging System (Advanced Microscopy Techniques Corp., Danvers, MA, USA).
Dynamic light scattering (DLS)
Particle sizes of complexes prepared as described above in ultrapure water were determined using DLS measurements. The DLS experiments were performed on 132 mM of pure HPβCD solution, MDA7-HPβCD solutions with concentration of 33 mM MDA7 and 33 mM of HPβCD, 33 mM MDA7 and 132 mM of HPβCD, 33 mM MDA7 and 221 mM of HPβCD (1:1, 1:4, 1:6.7 molar ratio respectively). The samples were run at 25°C using a Zetasizer nano ZS (Malvern Instruments Ltd., Malvern, UK) DLS instrument. Before each DLS scan, the samples were stabilized for 10 min.
Molecular Modeling
The 2-hydroxypropyl cyclodextrin structure was based on the cyclodextrin crystal structure provided by the Structural Data Base System of the Cambridge crystallographic Data Center.8 Two 2-hydroxypropyl groups were randomly added at O-6 positions and two at O-2, O-3 positions to the crystal structure of cyclodextrin based on the expected 4 degree of substitution of 2-hydroxypropyl cyclodextrin. The 2-hydroxypropyl groups were added and MDA7 structure was built using ChemDraw 3D. The HPβCD and MDA7 structures were individually optimized by energy minimization using a MM2 force field with a convergence criterion of 0.05 kcal/mol. The AutoDock 4.0 software was used to obtain a MDA7/ HPβCD complex. Lamarckian genetic algorithm was employed to generate a total population of 100 complexes. An initial population of 100 individuals with a maximal number of energy evaluations of 25,000,000 and a maximal number of generations of 50,000 were the parameters used for the global search. A grid box of 70 × 70 × 70 points was used to calculate the energy maps. The points were separated from each other by 0.375 Å.
Drug administration
All dosing solutions were prepared within 1 h prior to injection and stored at room temperature until use. MDA7 (10 mg/Kg) dosing solution was administrated as a single bolus injection into the tail vein of the animal.
Assessment of mechanical withdrawal thresholds
As described in our previous study,3 six adult male Sprague-Dawley rats (Harlan Sprague Dawley, Indianapolis, IN, USA) weighing 120-150 g and were used in experimental procedures approved by the Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center. Animals were housed 3 per cage on a 12/12-h light/dark cycle with water and food pellets available ad libitum.
Rats were placed in a compartment with a wire mesh bottom and allowed to acclimate for a minimum of 30 min before testing. Sensory thresholds for the development of allodynia to mechanical stimuli were assessed. Mechanical sensitivity was assessed by using a series of Von Frey filaments with logarithmic incremental stiffness (Stoelting Co., Wood Dale, IL, USA), as previously described,9 and 50% probability paw withdrawal thresholds were calculated with the up-down method.10 In brief, filaments were applied to the plantar surface of a hind paw for about 6 s in an ascending or descending order after a negative or positive withdrawal response, respectively. Six consecutive responses after the first change in response were used to calculate the paw withdrawal threshold (in grams). In rats, when response thresholds occurred outside the range of detection, the paw withdrawal threshold was assigned at 15.00 g for continuous negative responses and at 0.25 g for continuous positive responses.11 Behavioral assessment was performed by one investigator who was blinded to group allocation.
Results
Differential scanning calorimetry
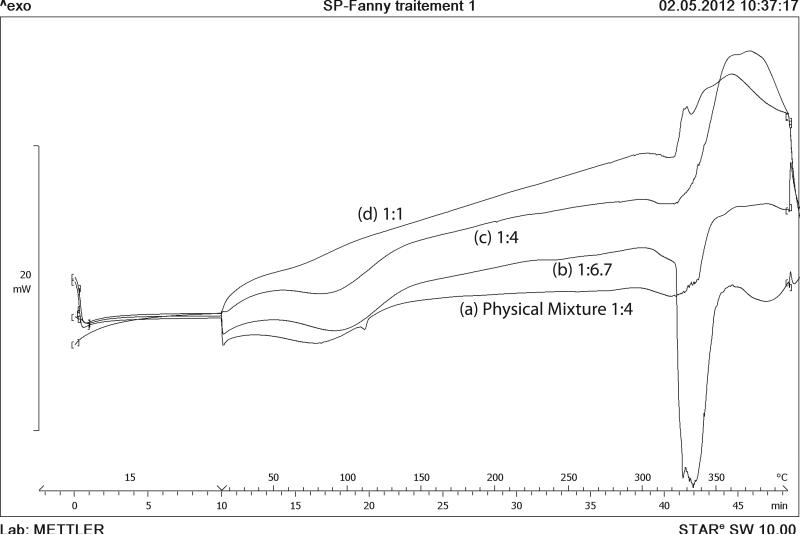
DSC curves of the inclusion complexes were compared with physical mixture (Fig. 2). MDA7 showed an endothermic peak at 108°C corresponding to melting point.4 HPβCD exhibited two broad endothermic peaks in the range of 50-150°C and 300-350°C.12 The first endothermic event assigned to loss of water and the second one to thermal decomposition.13 The DSC curve of MDA7:HPβCD physical mixture showed the endotherms corresponding to HPβCD (50-150°C and 300-350°C) and the melting peak of MDA7 at 108°C (see Supplementary Data for DSC of freeze-dried MDA7). The absence of MDA7 melting peak in the inclusion complexes with MDA7:HPβCD molar ratio of 1:1, 1:4 and 1:6.7 is a strong indication of inclusion complex formation.14 The broad endotherm at 50-150°C due to loss of water molecules absorbed and bonded to HPβCD is dramatically decreased with 1:1 molar ratio and increased with MDA7:HPβCD molar ratio. This effect reflects that higher active substance content of the complex means less water content. This complexation effect correlated with the loss of water indicates that inclusion complex formation results essentially in displacement of water molecules.
Figure 2.
DSC thermograms of (a) HPβCD:MDA7 physical mixture molar ratio of 1:4; (b) HPβCD:MDA7 inclusion complex molar ratio 1:6.7; (c) HPβCD:MDA7 inclusion complex molar ratio 1:4 and (d) HPβCD:MDA7 inclusion complex molar ratio 1:1.
ESI-MS analysis
Electrospray mass spectrometry (ESI-MS) has been used to investigate noncovalent complexes of different classes of compounds.15 ESI-MS allows detecting non-covalent interactions of supramolecular inclusion host-guest complexes in the gas phase. Electrospray provides a faithful image of the composition of the complex formed in solution and subsequently its stoichiometry in the range of concentrations used for in vivo studies,16 provided that all species are kinetically stable enough to reach the detector intact.15 However it has been suggested that to avoid non-specific aggregation occurring upon droplet evaporation, analyzed sample should be diluted and electrospray flow rate decrease.15,17,18
The ESI mass spectra were observed for mixtures of uncomplexed HPβCD and the supramolecular complexes with MDA7 (see Supplementary Data S1). HPβCD is a mixture of several hydroxylpropylated CD with a degree of substitution ranging from 1 to 8, which correspond to the m/z peak distribution from 1193 to 1600. The ions ranging from m/z 1645 to 1820 correspond to [HPβCDs+MDA7+H]+ with a degree of hydroxylpropyl substitution ranging from 3 to 6: 1309+335+1, 1368+335+1, 1426+335+1, 1484+335+1. Figure S1 shows the presence of MDA7:HPβCD complex at a ratio of 1:1. No signal of higher order drug:HPβCD have been detected in the range of m/z studied (1900-5000).
Continuous variation method (Job's plots)
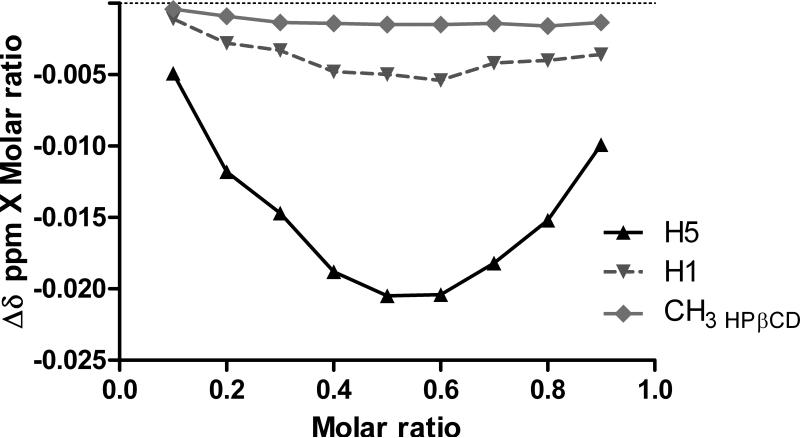
Stoichiometry of the inclusion complex was determined by continuous variation plot (Job's plot) studying 1H NMR spectra. Continuous variation curves were obtained by monitoring chemical shifts of the outer proton H1, the internal cavity proton H5 and CH3 HPβCD protons located in the hydroxypropyl moieties of the host (HPβCD) which were well separated from those of MDA7. HPβCD is not a single compound but rather a mixture of randomly hydroxypropylated molecules of βCD resulting in complexe NMR spectra. No new peaks were observed in NMR spectra indicating that free and complexed forms of MDA7 were in a rapid exchange relative to NMR timescale. The plots obtained for H5 reach a minimum at an r value of 0.5 on the abscissa axis indicating formation of exclusively 1:1 adduct (Fig. 3). As expected largest chemical shift variation was obtained for H5 compared to H1 and CH3 HPβCD of HPβCD with an up-field of Δδ 0.041 ppm. Since H5 is located in the cavity of HPβCD the chemical shift observed clearly suggests the presence of MDA7 in the cavity. The upfield shielding effect probably results from a diamagnetic anisotropy effect due to the inclusion of π-electrons belonging to MDA7 into the hydrophobic central cavity of the HPβCD.19,20 In addition, the effects of HPβCD on the 1H NMR chemical shift of MDA7 were investigated (see Supplementary Data S2). Continuous variation curves were obtained by recording chemical shifts of protons located in different regions of the guest, MDA7: Ha,a’; Hb,b’; Hc,c’ and CH3. These protons were selected because no peaks overlaps were observed. 1H NMR chemical shifts of MDA7 protons upon complexation by HPβCD are summarized in table 1. Proton Ha,a’; Hb,b’ are probably located close to an oxygen of HPβCD since a downfield displacement of the host protons indicates that they are close to an electronegative atom such as oxygen. In contrast, protons Hc,c’ and from the CH3 are probably located close to an hydrogen atom as indicated by an upfield displacement due to changes in the local polarity due to the inclusion of the drug into the lipophilic central cavity or Van der Waals interactions between MDA7 and hydrogen atoms of HPβCD cavity.21 Magnitude of the shift difference for protons of the guest molecule is dependent on the relative strength of the two types of interaction with oxygen or hydrogen atoms with the HPβCD cavity. Shielding effects for Ha,a’; Hb,b’ were the most intense compared to Hc,c’ and CH3 indicating that the phenyl ring of the benzofuran moiety is deeply located in the HPβCD cavity .
Figure 3.
Continuous variation plot corresponding to the chemical shift induced displacement of HPβCD protons, H5, H1 and CH3 HPβCD in an equimolar solution of D2O and methanol-d4 at 300 K.
Table 1.
1H NMR chemical shifts (δ in ppm) and changes (Δδ in ppm) of HPβCD protons upon complexation of MDA7 at 0.5 molar ratio in an equimolar solution of D2O and methanol-d4 at 300 K.
| HPβCD protons | δ free (ppm) | δ complex (ppm) | Δδ (ppm) | MDA7 protons | δ free (ppm) | δ complex (ppm) | Δδ (ppm) |
|---|---|---|---|---|---|---|---|
| H1 | 5.024 | 5.014 | −0.01 | Ha | 6.5745 | 6.6370 | 0.0625 |
| H5 | 3.846 | 3.805 | −0.041 | Ha’ | 6.5745 | 6.6540 | 0.0795 |
| CH3 HPβCD | 1.144 | 1.141 | −0.003 | Hc | 4.1700 | 4.1270 | −0.0430 |
| Hc’ | 4.1700 | 4.1060 | −0.0640 | ||||
| Hb | 4.6175 | 4.6285 | 0.0110 | ||||
| Hb’ | 4.6175 | 4.6230 | 0.0055 | ||||
| CH3 | 1.439 | 1.430 | −0.009 |
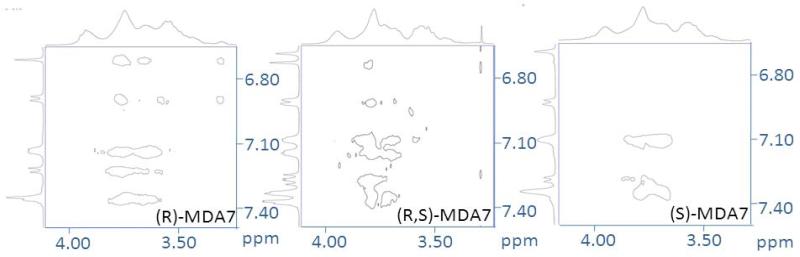
Enantiomeric recognition
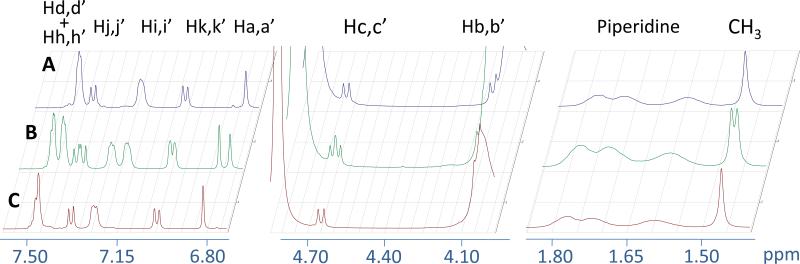
We decided to formulate and test the racemic mixture in vivo even though the eutomer of MDA7 has been isolated and identified as being (S)-MDA74. It was reasonable to expect that HPβCD will form diastereomeric inclusion complexes with racemic mixture of MDA7. 1H NMR spectra of enantiomerically pure (R)-MDA7 and (S)-MDA7 were compared to MDA7 racemic mixture to infer the 1H NMR chemical shifts for each enantiomer. Enantiomeric recognition was observed in equimolar solution of D2O and methanol-d4 at the low concentrations which were used for Job's plots studies (Supplementary Data S4) and in the system used for in vivo studies in D2O (Fig.4 and Supplementary Data S3). The Job plots showed a marked difference between the two enantiomers. Chemical shift differences do not vary in the same manner for each enantiomer (e.g. Ha,a’; Hb,b’ and Hc,c’). (S)-MDA7:HPβCD complex appears to have larger Δδ for Ha’ and Hc’ than that of (R)-MDA7:HPβCD complex (Ha and Hc). On the other hand, Δδ for Hb from (R)-MDA7: HPβCD complex is larger than that of Hb’ from (S)-MDA7:HPβCD complex. It is difficult to conclude which enantiomer has the higher affinity for HPβCD since the positioning of the two enantiomers in HPβCD is different according to chemical shift differences but the 1H NMR study is possible only for protons well resolved and not overlapping. As expected, chiral recognition was reduced when the concentration of HPβCD was reduced in methanol-d4 and D2O. Chiral recognition for the aromatic and furanic protons decreases when 0.5 to 4 equivalents of HPβCD were used in D2O while no discrimination was observed for 4 and 5 equivalents. Surprisingly, enantiomeric recognition was observed for aromatic protons when 6.7 equivalent of HPβCD were used. On the other hand, Chiral recognition was observed for the methyl moiety of the benzofuran scaffold at 0.5 to 6.7 equivalents of HPβCD were used in D2O.
Figure 4.
Portions of NMR spectra of an equimolar mixture of (A) (R)-MDA7, of (B) (R,S)-MDA7 and of (C) (S)-MDA7 upon complexation by HPβCD at a concentration of 33 mM in D2O at 300 K.
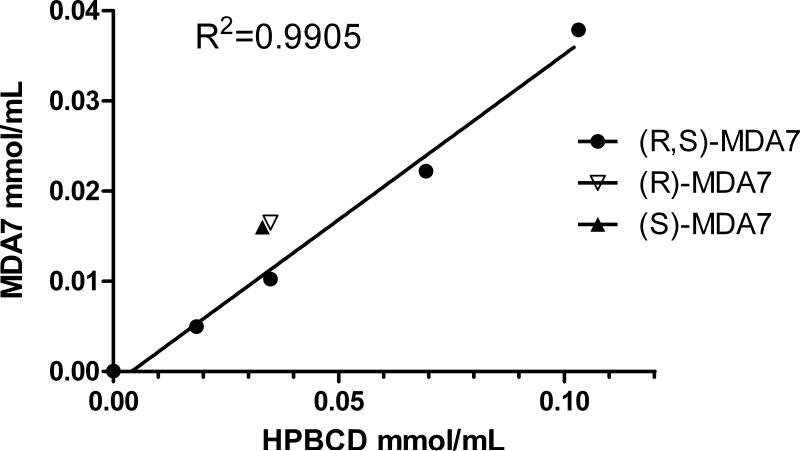
Phase solubility studies
MDA7 is a lipophilic compound showing very limited water solubility (at pH=7: 5.69 μg/mL) with a clog P of 4.54 +/− 0.37 (calculated in silico using ACD PhysChem software, Canada).
The determination of K1:1 between MDA7 and HPβCD was based on the quantitation of the concentration of soluble MDA7 determined by NMR upon inclusion. The increase on MDA7 solubility occurred as a linear function of HPβCD concentration (Fig. 5) corresponding to an AL-type profile defined by Higuchi and Connors. This relationship confirmed a first order kinetics on the complex formation between MDA7 and HPβCD and confirmed the formation of a 1:1 complex.
Figure 5.
Phase-solubility diagram for (R,S)-MDA7 at increasing HPβCD concentrations and (R)-MDA7 and (S)-MDA7 solubility values at a MDA7:HPβCD molar ratio of 1:1.
A stability constant (K1:1) of 18 200 M−1 was calculated from the slope and the intrinsic solubility (S0) of the drug in the aqueous complexation medium according to Eq. 1 with intrinsic water solubility of MDA7 = 31.8 μM and slope = 0.367.
| (1) |
Where [MDA7:HPβCD] is the concentration of dissolved complex, [HPβCD] is the concentration of dissolved free HPβCD assuming that [MDA7] is the intrinsic solubility of MDA7 and Slope is the slope of the phase-solubility plot.
Intrinsic solubility (S0) of a drug is expected to be determined by the intercept with Y axe. In the case of MDA7 the Y-intercept is −0.001475 mol/mL. However, an intrinsic solubility of 31.8 μM was previously determined using the shake flask method.4 In this case, a more accurate method to determine the solubilizing effect of HPβCD is to determine the complexation efficiency (CE). CE is the concentration ratio between free HPβCD and HPβCD complexing MDA7. CE is independent of both S0 and the intercept and is more reliable for poorly soluble guest. A CE of 0.58 was determined according to Eq. 2.23,24
| (2) |
The drug:cyclodextrin molar ratio in water saturated with MDA7 can be determined according to Eq. 3.
| (3) |
MDA7:HPβCD ratio has a value of 1:2.7 indicating that, on a 1:1 MDA7:HPβCD complexation, just one out of approximately 3 HPβCD molecules is forming an inclusion complex with MDA7. Since HPβCD is a mixture of randomly hydroxypropylated CDs with different molecular weights, MDA7:HPβCD ratio is similar to the experimental MDA7:HPβCD ratio determined to solubilized MDA7 with a value between 3 and 4. Both (R)-MDA7 and (S)-MDA7 enantiomers exhibited a comparable water solubility value with the racemic mixture after complexation with HPβCD at a molar ratio of 1:1 (Fig. 5).
2D NMR spectroscopy
Two-dimensional rotating-frame NOE spectroscopy (ROESY) has been used widely for the characterization of CD inclusion complex (Fig. 6). NOE cross-peaks resulting from dipolar coupling after complexation indicate a distance smaller than 5 Å between two protons of the host and the guest molecules. As mentioned above HPβCD is a mixture of randomly hydroxypropylated molecules of βCD and in addition, just one out of 3 HPβCD molecules is forming an inclusion complex with MDA7. These parameters are adding complexity on the interpretation of ROESY experiments. 2D ROESY spectra of the complex MDA7:HPβCD were acquired at different molar ratio in pure D2O. Aromatic protons Hi,i’, Hd,d’ and Hh,h’ of MDA7 at 1:1 molar ratio exhibited intermolecular cross-peaks with the NMR signals including H5 and H6 of HPβCD. These observations suggest that the benzyl moiety of MDA7 is positioned deeply inside the cavity. In this case, cross-peak between MDA7 and H3 should confirm the hypothesis. It was difficult to conclude about possible interactions between CH3 and the benzylic CH2 of MDA7 with H3 of HPβCD since H3 is overlapping with Hb,b’ protons. (R)-MDA7 protons Ha, Hj and Hk had additional cross-peaks with NMR region including H5 and H6 of HPβCD compared to (S)-MDA7. A dynamic equilibrium between two different complexes in the NMR timescale can explained these additional interactions. Benzofuran moiety of (R)-MDA7 could interact with HPβCD in addition to the benzyl moiety. These two complexes could coexist in solution. (R,S)-MDA7 ROESY studies at 1:1 molar ratio confirm the data obtained with pure enantiomers. (R,S)-MDA7 ROESY studies at 1:4 molar ratio did not show significant differences compared to 1:1 ratio. Hk,k’ cross-peaks were weak in both spectra. Interaction between NMR region including H5 and H6 of HPβCD and Ha,a’ indicates that at higher concentration of HPβCD, (R)-MDA7 is forming the 2 complexes.
Figure 6.
Partial contour plots of 2D ROESY spectrum of MDA7:HPβCD inclusion complexes in D2O at 300 K upon complexation by HPβCD at a concentration of 33 mM in D2O at 300 K. For the proton labeling see Figure 1.
Morphology and size
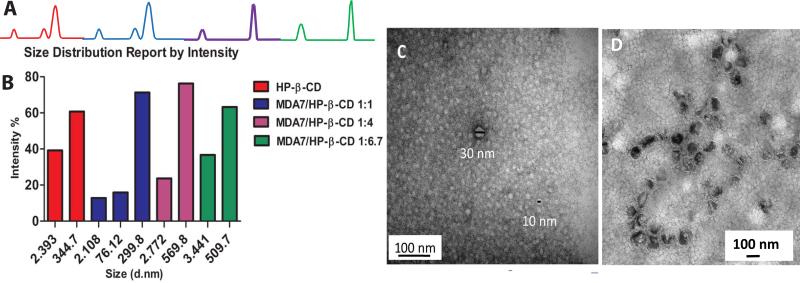
Morphology and size of MDA7 liposomal preparation was investigated by TEM 24H after manufacturing (Fig.7D) and showed the presence of an homogenous population of closed round-shaped oligolamellar (OLVs) MDA7 containing-liposomes, with an average size of approximately 100 nm. No MDA7 crystallization occurred in the liposomal powder. In DLS studies, MDA7 liposomal powder hydrated in PBS yielded to a monodisperse distribution of OLVs with an average mean diameter of 117 nm [polydispersity index (PDI)<0.1] corroborating the TEM results. Monodisperse micelles exhibited an average size of 15nm(PDI 0.1). Hydrodynamic radius of samples containing HPβCD at 130 mM, and mixture of MDA7 and HPβCD at 1:1, 1:4 and 1:6.7 molar ratio with a concentration of respectively 30 mM, 130 mM and 230 mM were determined by DLS (Fig. 7A and Fig. 7B). The Z-average and the PDI results given by the single mode cumulant couldn't be exploited since different populations of particles are present in the dispersion. However pure HPβCD solution is composed of a mixture of two discrete populations with an average diameter of 2 nm (PDI<0.1) and 345nm (PDI<0.1). A hydrodynamic radius of approximately 2 nm is in reasonable agreement with the size expected for a single HPβCD25.
Figure 7.
A, DLS results showing size distribution by intensity of the aqueous solutions of HPβCD and MDA7:HPβCD at 1:1, 1:4 and 1:6.7 molar ratio; B, Flow charts corresponding to the size distribution by intensity; C, TEM images of aggregated 230 mM 1:6.7 MDA7:HPβCD; D, TEM images of MDA7 in liposomal dispersions.
This population was found in all the samples containing MDA7 and HPβCD at different molar ratio. Complexation of HPβCD with MDA7 is not expected to dramatically change the hydrodynamic radius of single MDA7:HPβCD complex compared to a single HPβCD. The second population was composed by particles with a mean hydrodynamic radius of 345 nm corresponding probably of self-aggregated HPβCD forming nanoparticles. We observed the same bimodal distribution for the samples containing MDA7 and HPβCD at 1:4 and 1:6.7 molar ratios. However, the mean hydrodynamic radii corresponding to self-aggregated HPβCD were larger for samples containing MDA7 and HPβCD than in the sample containing pure HPβCD. MDA7 can affect the size of the self-aggregated HPβCD inducing formation of larger aggregates.24
Surprisingly, a third population with a mean hydrodynamic radius of 76nm (PDI<0.1) was identified in the sample with MDA7:HPβCD at a molar ratio of 1:1, furthermore the mean diameter of the larger population was similar than for pure HPβCD. The lower concentration of HPβCD in the former sample compared to the sample with pure HPβCD could explain the differences in term of distribution and diameter. The aggregation and the size of the aggregates increase with increasing cyclodextrin concentration.24 The morphology and the size of self-aggregates of HPβCD were also analyzed by TEM. Two populations of spherical aggregates of HPβCD with diameter of 5-10 nm and 30 nm were observed (Fig. 7C). These results were not in agreement with DLS experiments but sample preparation can affect the size of the aggregates observed by TEM. Nevertheless, TEM results confirm that spherical aggregates of different sizes are present in aqueous MDA7:HPβCD solutions as previously shown with other host molecules.26
Molecular modeling
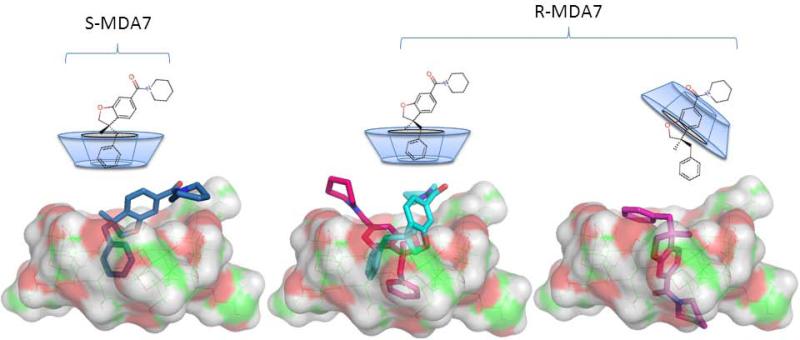
To further explore the mode of complexation of MDA7 by HPβCD we performed molecular modeling studies. We observed that the complexes with the highest score were with the benzyl ring inserted into the HPβCD cavity (ES-MDA7 = −537.0 kcal mol−1, ER-MDA7 pink = −627.0 kcal mol−1, ER-MDA7 cyan = −579.0 kcal mol−1) which is in agreement with the results obtained by NMR spectroscopy. In addition the complex with the benzofuran ring into the HPβCD cavity for (R)-MDA7 was among the complexes with the highest score (ER-MDA7 purple = −614.0 kcal mol−1), confirming that the two modes of complexation can coexist (Fig. 11).
In vivo characterization of MDA7 formulated
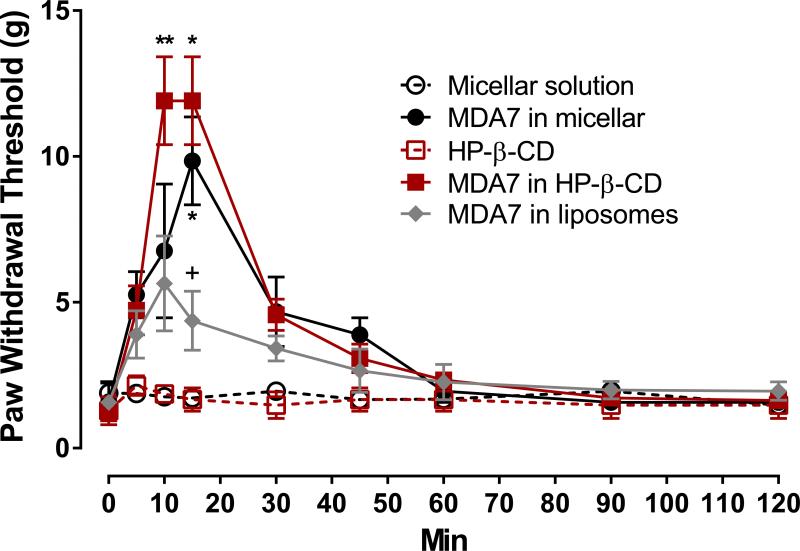
Antinociceptive effect of MDA7 evaluated in vivo in three Drug Delivery System (DDS)
In vivo experiments were carried out to study the influence of three different DDS, HPβCD, liposomes and micellar preparation of the CB2 agonist MDA7, to modify its efficacy in vivo at therapeutic dose. Formulation concepts based on colloidal systems such as micellar solution, liposomes and inclusion complexes were selected because of their common use in intravenous formulations and their high solubilizing ability for lipophilic drugs such as MDA7 thereby improving drug biodistribution and therapeutic efficacy. In the spinal nerve ligation neuropathic pain model, MDA7 (i.v.) in the three different DDS attenuated tactile allodynia (Fig. 9). MDA7 formulated in HPβCD produced a significantly (P<0.01) greater antiallodynic effect than that noted with liposomes or with the vehicle. A similar or slightly reduced antiallodynic effect was obtained with MDA7 formulated in micellar preparation. Liposomes used for this study were not coated with hydrophilic polymers and elimination by immune system was expected. This effect can explain the reduced effect of MDA7 prepared in liposomes compared to the two other DDS. A combination of different factors such as the hydrophobic-core-hydrophilic-shell structure of cyclodextrin and micelles -based DDS as well as their smaller size distribution (respectively ≃ 3nm and ≃15 nm) compared to liposomes (>100 nm) may have contributed in preventing recognition by the mononuclear phagocyte system (MPS) and subsequent preliminary elimination of the drug.
Figure 9.
Effect of MDA7 formulated in HPβCD, liposomes and micellar preparation administered by i.v. injection on tactile allodynia in a spinal nerve ligation neuropathic pain model at a dose of 10 mg/kg. MDA7 effects were greatest in HPβCD ≥ micellar> liposomes preparations at increasing the paw withdrawal threshold of the nerve-injured paw. **P<0.01, *P<0.05 versus corresponding vehicle. +P<0.05 versus MDA7 in HPβCD or in micellar preparations (two-way analysis of variance followed by Student-Newman-Keuls multiple comparisons test).
Discussion
Significant solubility enhancement was obtained for MDA7 owing to suitable solubilizing properties of the three DDS developed for a dose of 10 mg/Kg administered intravenously. Although the solubilizing system should not influence the intrinsic pharmacokinetics (PK) of the drug, evidence indicates that there is an impact of DDS on drugs PK profile by affecting transporters, metabolic enzymes, proteins binding, possibly cytochrome-mediated metabolism, and biodistribution.27,28,29 MDA7 is a lipophilic molecule with a Log P value of 4.54, poorly water soluble (31.8 μM) and moderately protein bound (82.8%).4 Excipients modulate protein binding and cellular partitioning of compounds in blood which is expected to impact the overall drug disposition. Half time life of MDA7 in the plasma is short (0.18 h)2 and effect of drug-DDS interactions on unbound fraction may result in better antinociceptive effect. Surfactant–cosolvent mixtures yielded to suitable in vivo activity, liposomes resulted in significantly reduced efficacy whereas HPβCD dramatically improved MDA7 in vivo performance. MDA7 exhibited a high affinity constant (K1:1= 18 200 M−1) for HPβCD. After parental administration and dilution in the biological fluids, drugs appear to be rapidly and quantitatively released from CDs inclusion complexes, particularly for K value < 105 M–1. This is a predominant mechanism involved in drug release from the drug:HPβCD complex that continuously forms and dissociates with lifetimes in the range of milliseconds or less.30 An excess of HPβCD (6.7 mole equivalents) was used to deliver MDA7. We demonstrated that at this molar ratio HPβCD form aggregates which are expected to dissociate with an increased dilution in vivo. Complexation by HPβCD and aggregates formation can decrease MDA7 metabolism and renal excretion of the complexes, thereby increasing MDA7 plasma concentration.
Micelles represent versatile, easy to prepare and characterize DDS for enabling formulations of lipophilic cannabinoids but may suffer from significant toxicity after parenteral administration with high solvents concentration, when chronic administration is required. However Cremophor™ELP has been shown to reduce protein binding in plasma, thereby increasing the free drug fraction available to exert its efficacy.
Lipid excipients such as phosphatydilcholine used to prepare the liposomes are generally considered as safe and have proven to be minimally toxic even in applications where they have been employed at very high concentrations. Liposomal DDS are usually more stable to dilution in contrast to cosolvent-micelles approaches and have a good potential for preventing in vivo drug precipitation. We prepared non sterically stabilized liposomes which are expected to be cleared quite rapidly31 and may not have been circulating long enough for the drug to exert its therapeutic effect. Steric stabilization of conventional liposomes using inert hydrophilic polymers (often referred as “stealth” liposomes) has been reported to increase their colloidal stability as well as circumventing mononuclear phagocyte system uptake and clearance. However, some authors have reported that conventional liposomes could be more efficient than the pegylated ones32 or cyclodextrin 33 and address the issue of accelerated clearance of sterically stabilized liposomes upon repeated injection.34
Conclusions
The FDA approved and chiral HPβCD appears to be a well-adapted enabling formulation approach for the in vivo delivery of lipophilic and chiral molecule such as MDA7. A full characterization of the diasteriomeric MDA7:HPβCD inclusion complex allowed an accurate and reproducible dosing, a prerequisite for performing pharmacology pre-clinical studies. Our study has shown that using inclusion complexation and possibly formation of aggregates, HPβCD can enhance the aqueous solubility of lipophilic drugs without compromising their intrinsic ability to permeate biological membranes. We also demonstrated HPβCD could be successfully used for the delivery of chiral drugs such as MDA7 since the same proportion of each enantiomer was solubilized by the formation of a 1:1 complex and no enantiomeric discrimination in term of solubility was observed. Differences in chemical shifts observed for the diastereomeric complexes formed by HPβCD and racemic MDA7 may result from differences in equilibrium constants or modes of complexation of the various possible complexes as demonstrated by our molecular modeling study. Rapid exchange occurs between free and complexed forms of MDA7 relative to NMR timescale, minimizing an enantiodifferentiation in term of water solubility for both enantiomers. Enabling formulation using HPβCD improved antiallodynic effect of the CB2 agonist MDA7 in comparison to incorporation in liposomes. The use of micellar system may be a suitable strategy when the lipophilic compound has inappropriate geometry, size or physicochemical characteristics to form host/guest complex formation, if chronic administration is not required.
Overall, HPβCD present many advantages related to their high safety of use in pre-clinical and clinical studies for racemic mixtures, their good performance in vivo, easy characterization of the complexes formed and their versatility, forming easy-to obtain water soluble inclusion complexes with lipophilic drugs.
In addition, HPβCD are well adapted DDS for oral administration, a more appropriate route of administration for the treatment of chronic disease. Since Biopharmaceutics Classification System (BCS) Class II (poor solubility, high intestinal permeability) preclinical drug candidates to treat neuropathic pain such as MDA7 could behave as BCS Class I (high solubility, high instestinal permeability) drug candidates by using HPβCD - based DDS as an early formulation strategy, their use may result in an improved oral bioavailability and an increased in vivo efficacy.
Supplementary Material
Figure 8.
Schematic structures and lowest energy inclusion complexes of (S)-MDA7 and (R)-MDA7 in HPβCD. In Blue (S)-MDA7:HPβCD complex with the benzyl ring inserted into the cavity of HPβCD, in cyan and pink two conformations of (R)-MDA7:HPβCD complexes with the benzyl ring inserted into the cavity of HPβC and, in purple (R)-MDA7:HPβCD complex with benzofuran ring inserted into the cavity of HPβCD.
Acknowledgements
FAD, SWM and PhD acknowledge National Institutes of Health (NIH) grant P30-NS055022. Images, electron microscopy services and resources for liposome were provided by the High Resolution Electron Microscopy Facility at MD Anderson Cancer Center, Institutional Core Grant #CA16672. Images, electron microscopy services and resources for HPβCD formulations were provided by the Electron Microscopy Facility, Division of Biological Sciences, University of Montana, Missoula, MT. The EM Facility is supported, in part, by National Center for Research Resources (5P20RR016455-11) and the National Institute of General Medical Sciences (8 P20 GM103474-11). P.D. and M.N are named on 2 patent applications submitted by the University of Texas MD Anderson Cancer Center.
References
- 1.Xu JJ, Diaz P, Astruc-Diaz F, Craig S, Munoz E, Naguib M. Pharmacological Characterization of a Novel Cannabinoid Ligand, MDA19, for Treatment of Neuropathic Pain. Anest Analg. 2010;111(1):99–109. doi: 10.1213/ANE.0b013e3181e0cdaf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naguib M, Xu JJ, Diaz P, Brown DL, Cogdell D, Bie B, Hu J, Craig S, Hittelman WN. Prevention of Paclitaxel-Induced Neuropathy Through Activation of the Central Cannabinoid Type 2 Receptor System. Anesth Analg. 2012;114(5):1104–1120. doi: 10.1213/ANE.0b013e31824b0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naguib M, Diaz P, Xu JJ, Astruc-Diaz F, Craig S, Vivas-Mejia P, Brown DL. MDA7: a novel selective agonist for CB2 receptors that prevents allodynia in rat neuropathic pain models. Br J Pharmacol. 2008;155(7):1104–1116. doi: 10.1038/bjp.2008.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz P, Phatak SS, Xu J, Fronczek FR, Astruc-Diaz F, Thompson CM, Cavasotto CN, Naguib M. 2,3-Dihydro-1-Benzofuran Derivatives as a Series of Potent Selective Cannabinoid Receptor 2 Agonists: Design, Synthesis, and Binding Mode Prediction through Ligand-Steered Modeling. ChemMedChem. 2009;4(10):1615–1629. doi: 10.1002/cmdc.200900226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo Z, Naguib M. A synthetic approach for (S)-(3-benzyl-3-methyl-2,3-dihydro-benzofuran-6-yl)-piperidin-1-yl-methanone, a selective CB2 receptor agonist. Tetrahedron Lett. 2012 (0) [Google Scholar]
- 6.Lopez-Berestein G, Tari AM, Lim S-J. Method to incorporate N-(4-hydroxyphenyl) retinamide in liposomes. TUo; Texas: 2002. [Google Scholar]
- 7.Higuchi T, Connors KA. Advances in Analytical Chemistry and Instrumentation. Phase Solubility Studies; 1965. pp. 117–212. Chapter 4. [Google Scholar]
- 8.Aree T, Schulz B, Reck G. Crystal Structures of β-Cyclodextrin Complexes with Formic Acid and Acetic Acid J of Inclusion Phenom Macrocyclic Chem. 2003;47(1-2):39–45. [Google Scholar]
- 9.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53(1):55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 10.Dixon W. The up-and-down method for small samples. J Am Stat Assoc. 1965;60:967–978. [Google Scholar]
- 11.Brainin-Mattos J, Smith ND, Malkmus S, Rew Y, Goodman M, Taulane J, Yaksh TL. Cancer-related bone pain is attenuated by a systemically available δ-opioid receptor agonist. Pain. 2006;122(1-2):174–181. doi: 10.1016/j.pain.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 12.Veiga MD, Merino M, Fernández D, Lozano R. Characterization of some cyclodextrin derivatives by thermal analysis. J Therm Anal Calorim. 2002;68:511–516. [Google Scholar]
- 13.de Araújo VG, Mr, Vieira EKB, Lázaro GS, de Souza Conegero L, Ferreira OP, Almeida LE, Barreto LS, da Costa NB, Jr, Gimenez IF. Inclusion complexes of pyrimethamine in 2-hydroxypropyl-β-cyclodextrin: Characterization, phase solubility and molecular modelling. Bioorg Med Chem Lett. 2007;15(17):5752–5759. doi: 10.1016/j.bmc.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Orgoványi J, Pöppl L, H-Otta K, Lovas GA. Characterization of some cyclodextrin derivatives by thermal analysis. Journal of Thermal Analysis and Calorimetry. 2005;81:261–266. [Google Scholar]
- 15.Gabelica V, Galic N, De Pauw E. On the specificity of cyclodextrin complexes detected by electrospray mass spectrometry. J Am Soc Mass Spectrom. 2002;13(8):946–953. doi: 10.1016/S1044-0305(02)00416-6. [DOI] [PubMed] [Google Scholar]
- 16.Smith RD, Light-Wahl KJ. The observation of non-covalent interactions in solution by electrospray ionization mass spectrometry: Promise, pitfalls and prognosis. Biological Mass Spectrometry. 1993;22(9):493–501. [Google Scholar]
- 17.Smith RD, Light-Wahl KJ. The observation of non-covalent interactions in solution by electrospray ionization mass spectrometry: Promise, pitfalls and prognosis. Biol Mass Spectrom. 1993;22(9):493–501. [Google Scholar]
- 18.Schalley CA. Supramolecular chemistry goes gas phase: the mass spectrometric examination of noncovalent interactions in host-guest chemistry and molecular recognition. Int J Mass Spectrom. 2000;194(1):11–39. [Google Scholar]
- 19.Djedaïni F, Lin SZ, Perly B, Wouessidjewe D. High-field nuclear magnetic resonance techniques for the investigation of a β-cyclodextrin:indomethacin inclusion complex. J Pharm Sci. 1990;79(7):643–646. doi: 10.1002/jps.2600790721. [DOI] [PubMed] [Google Scholar]
- 20.Loftsson T, Olafsdottir BJ, Frioriksdottir H, Jonsdottir S. Cyclodextrin complexation of NSAIDSs: physicochemical characteristics. Eur J Pharm Sci. 1993;1(2):95–101. [Google Scholar]
- 21.Zornoza A, Martín C, Sánchez M, Vélaz I, Piquer A. Inclusion complexation of glisentide with α-, β- and γ-cyclodextrins. Int J Pharm. 1998;169(2):239–244. [Google Scholar]
- 22.Loftsson T, Brewster ME. Pharmaceutical applications of cyclodextrins: basic science and product development. Journal of Pharmacy and Pharmacology. 2010;62(11):1607–1621. doi: 10.1111/j.2042-7158.2010.01030.x. [DOI] [PubMed] [Google Scholar]
- 23.Loftsson T, Brewster ME. Pharmaceutical applications of cyclodextrins: basic science and product development. Journal of Pharmacy and Pharmacology. 62(11):1607–1621. doi: 10.1111/j.2042-7158.2010.01030.x. [DOI] [PubMed] [Google Scholar]
- 24.Loftsson T, Brewster ME. Cyclodextrins as functional excipients: Methods to enhance complexation efficiency. J Pharm Sci. 2012;101(9):3019–3032. doi: 10.1002/jps.23077. [DOI] [PubMed] [Google Scholar]
- 25.He Y, Fu P, Shen X, Gao H. Cyclodextrin-based aggregates and characterization by microscopy. Micron. 2008;39(5):495–516. doi: 10.1016/j.micron.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Messner M, Kurkov SV, Brewster ME, Jansook P, Loftsson T. Self-assembly of cyclodextrin complexes: Aggregation of hydrocortisone/cyclodextrin complexes. Int J Pharm. 2011;407(1-2):174–183. doi: 10.1016/j.ijpharm.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Bittner B, Mountfield RJ. Intravenous administration of poorly soluble new drug entities in early drug discovery: the potential impact of formulation on pharmacokinetic parameters. Curr Opin Drug Discov Devel. 2002;5(1):59–71. [PubMed] [Google Scholar]
- 28.Egger-Heigold B. The effect of excipients on pharmacokinetic parameters of parenteral drugs. Basel University; Basel: 2005. [Google Scholar]
- 29.Dhanikula AB, Singh DR, Panchagnula R. In vivo pharmacokinetic and tissue distribution studies in mice of alternative formulations for local and systemic delivery of Paclitaxel: gel, film, prodrug, liposomes and micelles. Curr Drug Deliv. 2005;2(1):35–44. doi: 10.2174/1567201052772852. [DOI] [PubMed] [Google Scholar]
- 30.Stella VJ, Rao VM, Zannou EA, Zia V. Mechanisms of drug release from cyclodextrin complexes. Adv Drug Delivery Rev. 1999;36(1):3–16. doi: 10.1016/s0169-409x(98)00052-0. [DOI] [PubMed] [Google Scholar]
- 31.Drummond DC, Meyer O, Hong K, Kirpotin DB, Papahadjopoulos D. Optimizing liposomes for delivery of chemotherapeutic agents to solid tumors. Pharm Rev. 1999;51(4):691–743. [PubMed] [Google Scholar]
- 32.Rouf MA. In: Development of Rapamycin drug delivery systems, their characterization and investigation of the antiproliferative effect on cell cultures. Health Sciences Institute, Pharmaceutical Technology, editor. Hacettepe University; Ankara: 2007. [Google Scholar]
- 33.Bruzell EM, Morisbak E, Tonnesen HH. Studies on curcumin and curcuminoids. XXIX. Photoinduced cytotoxicity of curcumin in selected aqueous preparations. Photochem Photobiol Sci. 2005;4(7):523–530. doi: 10.1039/b503397g. [DOI] [PubMed] [Google Scholar]
- 34.Ishida T, Harashima H, Kiwada H. Liposome clearance. Biosci Rep. 2002;22(2):197–224. doi: 10.1023/a:1020134521778. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.