Abstract
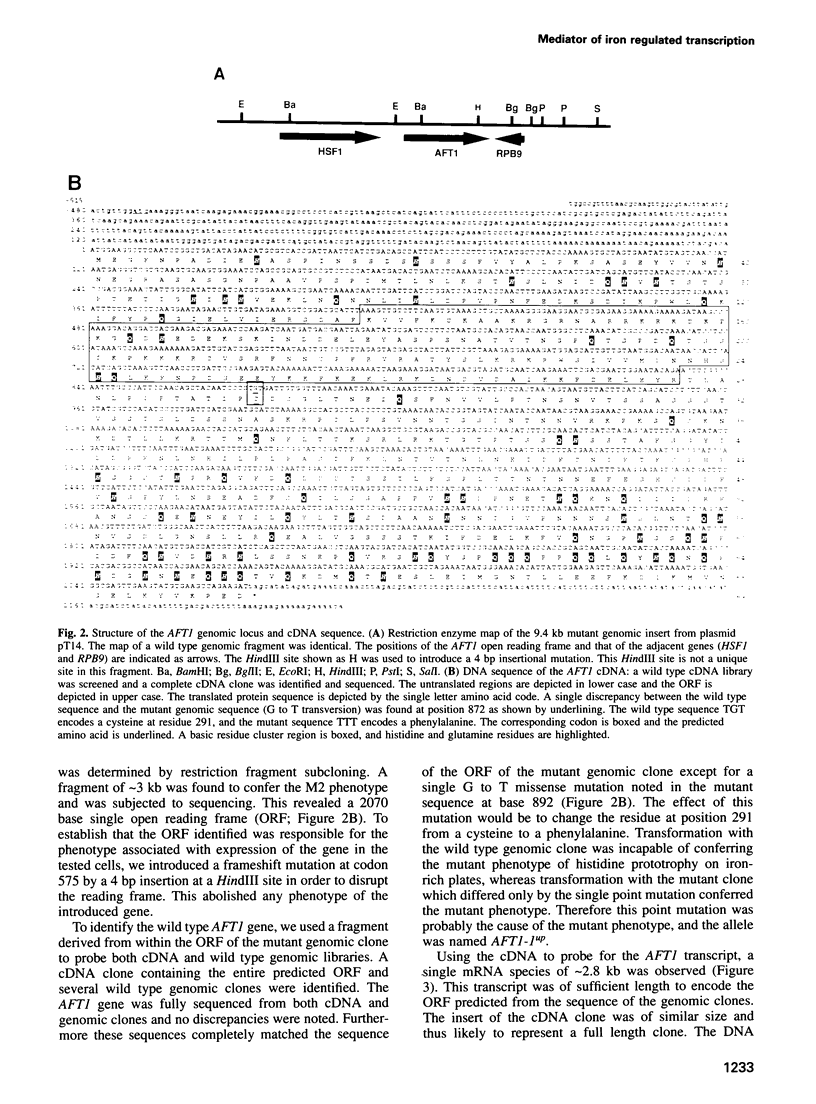
Using a scheme for selecting mutants of Saccharomyces cerevisiae with abnormalities of iron metabolism, we have identified a gene, AFT1, that mediates the control of iron uptake. AFT1 encodes a 78 kDa protein with a highly basic amino terminal domain and a glutamine-rich C-terminal domain, reminiscent of transcriptional activators. The protein also contains an amino terminal and a C-terminal region with 10% His residues. A dominant mutant allele of this gene, termed AFT1-1up, results in high levels of ferric reductase and ferrous iron uptake that are not repressed by exogenous iron. The increased iron uptake is associated with enhanced susceptibility to iron toxicity. These effects may be explained by the failure of iron to repress transcription of FRE1, FRE2 and FET3. FRE1 and FRE2 encode plasma membrane ferric reductases, obligatory for ferric iron assimilation, and FET3 encodes a copper-dependent membrane-associated oxidase required for ferrous iron uptake. Conversely, a strain with interruption of the AFT1 gene manifests low ferric reductase and ferrous iron uptake and is susceptible to iron deprivation, because of deficient expression of FRE1 and negligible expression of FRE2 and FET3. Thus, AFT1 functions to activate transcription of target genes in response to iron deprivation and thereby plays a central role in iron homeostasis.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold F. H., Haymore B. L. Engineered metal-binding proteins: purification to protein folding. Science. 1991 Jun 28;252(5014):1796–1797. doi: 10.1126/science.1648261. [DOI] [PubMed] [Google Scholar]
- Askwith C., Eide D., Van Ho A., Bernard P. S., Li L., Davis-Kaplan S., Sipe D. M., Kaplan J. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell. 1994 Jan 28;76(2):403–410. doi: 10.1016/0092-8674(94)90346-8. [DOI] [PubMed] [Google Scholar]
- Bagg A., Neilands J. B. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987 Aug 25;26(17):5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- Bagg A., Neilands J. B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987 Dec;51(4):509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P. F., Chen Y., Potts W. E., Chaney R. L., Angle J. S. A reevaluation of the Fe(II), Ca(II), Zn(II), and proton formation constants of 4,7-diphenyl-1,10-phenanthrolinedisulfonate. Biol Trace Elem Res. 1991 Aug;30(2):125–144. doi: 10.1007/BF02990349. [DOI] [PubMed] [Google Scholar]
- Chaney R. L., Brown J. C., Tiffin L. O. Obligatory reduction of ferric chelates in iron uptake by soybeans. Plant Physiol. 1972 Aug;50(2):208–213. doi: 10.1104/pp.50.2.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancis A., Haile D., Yuan D. S., Klausner R. D. The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J Biol Chem. 1994 Oct 14;269(41):25660–25667. [PubMed] [Google Scholar]
- Dancis A., Klausner R. D., Hinnebusch A. G., Barriocanal J. G. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2294–2301. doi: 10.1128/mcb.10.5.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancis A., Roman D. G., Anderson G. J., Hinnebusch A. G., Klausner R. D. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3869–3873. doi: 10.1073/pnas.89.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancis A., Yuan D. S., Haile D., Askwith C., Eide D., Moehle C., Kaplan J., Klausner R. D. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell. 1994 Jan 28;76(2):393–402. doi: 10.1016/0092-8674(94)90345-x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D., Davis-Kaplan S., Jordan I., Sipe D., Kaplan J. Regulation of iron uptake in Saccharomyces cerevisiae. The ferrireductase and Fe(II) transporter are regulated independently. J Biol Chem. 1992 Oct 15;267(29):20774–20781. [PubMed] [Google Scholar]
- Frankel A. D., Kim P. S. Modular structure of transcription factors: implications for gene regulation. Cell. 1991 May 31;65(5):717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- Georgatsou E., Alexandraki D. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1994 May;14(5):3065–3073. doi: 10.1128/mcb.14.5.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R., Zueco J., Sentandreu R., Herrero E. RCS1, a gene involved in controlling cell size in Saccharomyces cerevisiae. Yeast. 1991 Jan;7(1):1–14. doi: 10.1002/yea.320070102. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Haile D. J., Rouault T. A., Harford J. B., Kennedy M. C., Blondin G. A., Beinert H., Klausner R. D. Cellular regulation of the iron-responsive element binding protein: disassembly of the cubane iron-sulfur cluster results in high-affinity RNA binding. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11735–11739. doi: 10.1073/pnas.89.24.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J., Reins H. A., Lee J., Romeo A., Hassett R., Kosman D., Jentsch S. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 1993 Dec 15;12(13):5051–5056. doi: 10.1002/j.1460-2075.1993.tb06198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M. C., Mende-Mueller L., Blondin G. A., Beinert H. Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11730–11734. doi: 10.1073/pnas.89.24.11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner R. D., Rouault T. A., Harford J. B. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993 Jan 15;72(1):19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- Köhrer K., Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Lesuisse E., Raguzzi F., Crichton R. R. Iron uptake by the yeast Saccharomyces cerevisiae: involvement of a reduction step. J Gen Microbiol. 1987 Nov;133(11):3229–3236. doi: 10.1099/00221287-133-11-3229. [DOI] [PubMed] [Google Scholar]
- Liu H., Krizek J., Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992 Nov;132(3):665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei B., Budde A. D., Leong S. A. sid1, a gene initiating siderophore biosynthesis in Ustilago maydis: molecular characterization, regulation by iron, and role in phytopathogenicity. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):903–907. doi: 10.1073/pnas.90.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Philpott C. C., Klausner R. D., Rouault T. A. The bifunctional iron-responsive element binding protein/cytosolic aconitase: the role of active-site residues in ligand binding and regulation. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):7321–7325. doi: 10.1073/pnas.91.15.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja K. B., Simpson R. J., Peters T. J. Investigation of a role for reduction in ferric iron uptake by mouse duodenum. Biochim Biophys Acta. 1992 Jun 10;1135(2):141–146. doi: 10.1016/0167-4889(92)90129-y. [DOI] [PubMed] [Google Scholar]
- Roman D. G., Dancis A., Anderson G. J., Klausner R. D. The fission yeast ferric reductase gene frp1+ is required for ferric iron uptake and encodes a protein that is homologous to the gp91-phox subunit of the human NADPH phagocyte oxidoreductase. Mol Cell Biol. 1993 Jul;13(7):4342–4350. doi: 10.1128/mcb.13.7.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Schäffer S., Hantke K., Braun V. Nucleotide sequence of the iron regulatory gene fur. Mol Gen Genet. 1985;200(1):110–113. doi: 10.1007/BF00383321. [DOI] [PubMed] [Google Scholar]
- Segal A. W., Abo A. The biochemical basis of the NADPH oxidase of phagocytes. Trends Biochem Sci. 1993 Feb;18(2):43–47. doi: 10.1016/0968-0004(93)90051-n. [DOI] [PubMed] [Google Scholar]
- Tsai S. F., Martin D. I., Zon L. I., D'Andrea A. D., Wong G. G., Orkin S. H. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989 Jun 8;339(6224):446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- Voisard C., Wang J., McEvoy J. L., Xu P., Leong S. A. urbs1, a gene regulating siderophore biosynthesis in Ustilago maydis, encodes a protein similar to the erythroid transcription factor GATA-1. Mol Cell Biol. 1993 Nov;13(11):7091–7100. doi: 10.1128/mcb.13.11.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederrecht G., Seto D., Parker C. S. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell. 1988 Sep 9;54(6):841–853. doi: 10.1016/s0092-8674(88)91197-x. [DOI] [PubMed] [Google Scholar]
- Woychik N. A., Lane W. S., Young R. A. Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. J Biol Chem. 1991 Oct 5;266(28):19053–19055. [PubMed] [Google Scholar]
- Zhou P. B., Thiele D. J. Isolation of a metal-activated transcription factor gene from Candida glabrata by complementation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6112–6116. doi: 10.1073/pnas.88.14.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]







