Abstract
Rhomboid domain containing 2 (RHBDD2) was previously observed overexpressed and amplified in breast cancer samples. In order to identify biological pathways modulated by RHBDD2, gene expression profiles of RHBDD2 silenced breast cancer cells were analyzed using whole genome human microarray. Among the statistically significant overrepresented biological processes, we found protein metabolism—with the associated ontological terms folding, ubiquitination, and proteosomal degradation—cell death, cell cycle, and oxidative phosphorylation. In addition, we performed an in silico analysis searching for RHBDD2 co-expressed genes in several human tissues. Interestingly, the functional analysis of these genes showed similar results to those obtained with the microarray data, with negative regulation of protein metabolism and oxidative phosphorylation as the most enriched gene ontology terms. These data led us to hypothesize that RHBDD2 might be involved in endoplasmic reticulum (ER) stress response. Thus, we specifically analyzed the unfolding protein response (UPR) of the ER stress process. We used a lentivirus-based approach for stable silencing of RHBDD2 mRNA in the T47D breast cancer cell line, and we examined the transcriptional consequences on UPR genes as well as the phenotypic effects on migration and proliferation processes. By employing dithiothreitol as an UPR inducer, we observed that cells with silenced RHBDD2 showed increased expression of ATF6, IRE1, PERK, CRT, BiP, ATF4, and CHOP (p < 0.01). We also observed that RHBDD2 silencing inhibited colony formation and decreased cell migration. Based on these studies, we hypothesize that RHBDD2 overexpression in breast cancer could represent an adaptive phenotype to the stressful tumor microenvironment by modulating the ER stress response.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-013-0466-3) contains supplementary material, which is available to authorized users.
Keywords: RHBDD2, Breast cancer, ER stress, UPR
Introduction
The Rhomboid gene family constitutes a heterogeneous group of proteases that are conserved throughout evolution (Freeman 2008). Rhomboid 1, from Drosophila melanogaster, was the first member described in this family (Sturtevant et al. 1993). Since then, many genes have been classified as rhomboids based on sequence similarities despite lacking a catalytic domain. These members therefore have been denominated rhomboid pseudoproteases. So far, 14 human rhomboid family member proteins have been identified: five rhomboid proteases (RHBDL1/2/3/4 and PARL) and nine pseudoproteases (iRhom1/2, Derlin1/2/3, RHBDD2/3, UBAC2, and TMEM115) (Bergbold and Lemberg 2013).
Rhomboids have been implicated in numerous processes, ranging from cell signaling and apoptosis to parasitic shedding and endoplasmic reticulum-associated degradation (ERAD), a cellular pathway associated to the unfolded protein response (UPR) (McQuibban et al. 2003; Urban and Freeman 2003; Liu and Kaufman 2003; Cipolat et al. 2006; Zou et al. 2009; Adrain et al. 2011; Fleig et al. 2012). In addition, various rhomboid protein family members have been associated with diseases such as neurodegenerative disorders and cancer (Whitworth et al. 2008; Zou et al. 2009; Abba et al. 2009; Blaydon et al. 2012; Lacunza et al. 2012; Etheridge et al. 2013). The specific molecular function of several of the proteolytically inactive rhomboids in particular remains unknown.
In previous studies, we identified rhomboid domain containing 2 (RHBDD2), a pseudoprotease family member, to be markedly overexpressed in primary invasive breast carcinomas from patients that developed recurrent disease (Abba et al. 2007; Abba et al. 2009). More recently, we also observed significant RHBDD2 mRNA and protein overexpression in the advanced stages of colorectal cancer (Lacunza et al. 2012), suggesting that RHBDD2 up-modulation might be associated with breast and colon cancer malignant progression. We also determined that RHBDD2 protein expression is upregulated in colon cancer cells treated with the chemotherapeutic agent 5-fluourouracil (5FU) (Lacunza et al. 2012), which has recently been shown to be an inducer of ER stress in hepatocellular carcinoma cells (Yadunandam et al. 2012).
With the aim of identifying the biological processes modulated by RHBDD2 and to elucidate its role in breast cancer progression, we report here the transcriptomic profiling of two breast cancer cell lines transiently silenced for RHBDD2 expression. Results obtained and previous evidence led us to hypothesize that RHBDD2 might be involved in the UPR pathway; therefore, we evaluated the transcriptional effect of RHBDD2 stably silenced T47D breast cancer cells on the UPR pathway genes, as well as the phenotypic consequences on proliferation and cell migration processes.
Material and methods
Cell culture and RHBDD2 gene expression analysis
Estrogen-dependent breast cancer cell lines MCF7 and T47D were cultured in RPMI medium (Gibco, Gaithersburg, MD) and Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Gaithersburg, MD), respectively, supplemented with 10 % fetal bovine serum (FBS) (Bioser, Argentina), 10 U/mL penicillin, and 10 μg/mL streptomycin.
RT-qPCR analysis of RHBDD2 mRNA was evaluated for different breast cancer cell lines. Expression of each sample was normalized with mRNA from 18S rRNA as housekeeping gene. Total RNA was isolated using TRIzol (Invitrogen, USA) and cDNAs were synthesized using High Capacity Reverse Transcription Kit (Applied Biosystems, USA). The following primers were designed and used: forward 5′-GGTGTTTGGCATGGTTGTG-3′ and reverse 5′-CGATGGAATAGCAGTAGGTGAG-3′. The thermal profile was 94 °C for 2 minute and then 40 cycles of 94 °C for 40 s, 57 °C for 45 s, and 72 °C for 40 s.
Gene expression profiling of RHBDD2 silencing cells
MCF7 and T47D cell lines were cultured on 12-well plates at 40 % of confluence in Opti-MEM I Reduced Serum Medium and were transiently transfected with 40 pmol/μL of siRNA mixed with Lipofectamine according to the manufacturer’s protocol (Invitrogen, USA). We used a siRNA of 19-mer against RHBDD2 mRNA (RHBDD2-siRNA, 5′-CUGUGUUGGGUACUUUGAUdTdT-3′) as was previously described (Abba et al. 2009). In addition, the AccuTargetTM biotin-labeled negative control siRNA (NegCt-siRNA, 5′-CCUACGCCACCAAUUUCGUdTdT-3′) (Bioneer Inc., South Korea) that exhibits no homology to any human genome sequence was used as a non-silencing reference. Cells were incubated during 72 h.
In order to analyze the differential gene expression profiling of RHBDD2 silencing and control cells, total RNA was isolated from duplicate experiments using TRIzol reagent and purified using the TRIreagent and NucleoSpin RNA Clean-up Kit (Macherey-Nagel). RNA concentration and integrity were measured on an Agilent Bioanalyzer RNA 6000 Nanochip (Agilent Technologies). Briefly, aminoallyl-amplified RNA (aRNA) was synthesized from 1 μg of total RNA with the Amino Allyl MessageAmp™ II aRNA Amplification Kit (Ambion) and subsequently labeled with Cy5 Mono-ReactiveDyePack (GE Healthcare Bioscience). One microgram of labeled aRNA was probed using the whole genome Toray 3D-Gene™ Human Oligo Chip 25k V2.1 (GPL13915). Target labeling and hybridization to Chips were carried out in the Genomics Core Facility at Toray Inc. Raw datasets have been submitted to NCBI GEO database with accession number GSE43015.
Bioinformatics and statistical analysis
To compare the control siRNA vs. RHBDD2-siRNA treatments in each breast cancer cell line models, we employed the Rank Products’ test (Breitling et al. 2004). Statistical analysis, heatmap visualization, and comparison of overlapping differentially expressed genes between MCF7 and T47D cell lines were done with the MultiExperiment Viewer software (MeV 4.8) (Saeed et al. 2003). The number and identity of genes commonly affected in both models were determined. We used the normal approximation to the binomial distribution as previously described (Smid et al. 2003) to calculate the number of matching genes derived from each pairwise comparison at the p < 0.05 statistical significance level.
For automated functional annotation and gene enrichment analysis, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID, http://david.niaid.nih.gov/david) (Huang et al. 2008). The DAVID resource calculates overrepresentation of specific biological themes/pathways with respect to the total number of genes assayed and annotated. REViGO resource (http://revigo.irb.hr) was employed to summarize and visualize the enriched gene ontology (GO) terms in an interactive graph based on the p values obtained by DAVID. This allows one to identify biological themes/pathways within a specific list of differentially expressed genes.
To further analyze possible pathways associated with RHBDD2, we employed the “guilt by association” principle, which states that gene co-expression might indicate shared regulatory mechanisms and roles in related biological processes. Briefly, RHBDD2 co-expressed genes in different tissue localizations (adrenal gland, brain, breast, colon, head and neck, lung, ovary, small intestine, and stomach) were obtained by using the web-based bioinformatics tool Multiexperiment Matrix (http://biit.cs.ut.ee/mem/; Adler et al. 2009). For each tissue, we selected the 200 best positive correlated genes (p < 0.0001), which were compiled into one Excel pivot table for comparison of overlapping genes between the tissues, selecting those genes that were present in at least four localizations. Finally, we built a matrix with the selected genes (rows) across the nine tissues (columns) and performed a hierarchical clustering (HCL) by using the MeV 4.8. The DAVID and REViGO tools were employed for the functional enrichment analysis.
Silencing of RHBDD2 gene on T47D cells
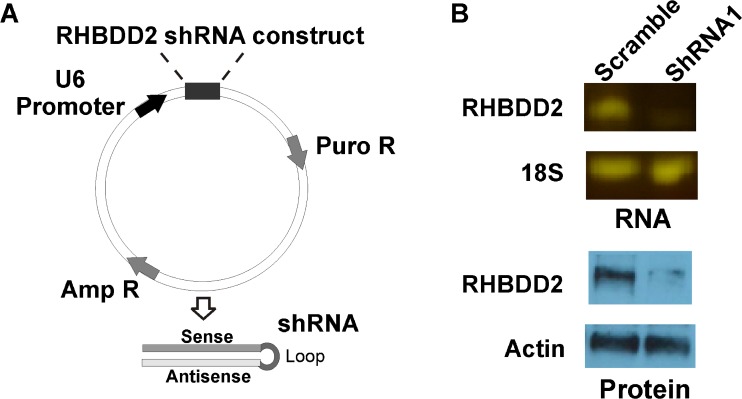
The pLKO.1–TRC Cloning Vector (Addgene) was employed for silencing RHBDD2 gene on the T47D cell line. pLKO.1 is a replication-incompetent lentiviral vector that can be used for the production of infective lentiviral particles containing the shRNA of interest using the HEK293T helper cell line. Following manufacturer’s instructions, a target sequence of 21 base pair oligonucleotides was designed to obtain RHBDD2 siRNA for cloning into plKO.1: forward ccggaaGTCTACGAGAATCCCATCTctcgagAGATGGGATTCTCGTAGACtttttttg and reverse aattcaaaaaaaGTCTACGAGAATCCCATCTctcgaGAGATGGGATTCTCGTAGACTT. Capital letters indicate sense and antisense target sequences. These sequences are flanked by sequences that are compatible with the sticky ends of EcoRI and AgeI restriction enzymes. After digestion, forward and reverse oligos were annealed and ligated into the pLKO.1 vector, producing a final plasmid that expresses the RHBDD2 shRNA. Plasmid was transformed into 25 μL competent DH5 alpha cells and plated on lysogeny broth (LB) agar plates containing 100 μg/mL ampicillin. Colonies were grown in LB/100 μg/mL ampicillin and DNA was purified by using a Miniprep Kit (Qiagen, USA). Positive clones were verified by conducting a sequencing reaction. To produce lentiviral particles, 1 μg of plKO1-shRNA RHBDD2 and plKO1-shRNA scramble (provided with the kit) were mixed in separate tubes with 750 ng of psPAX2 packaging plasmid and 250 ng of pMD2.G envelope plasmid. Cocktail was transfected into HEK293T cells by using FUGENE6 transfection reagent (Roche, USA). Twenty-four hours after transfection, media containing lentiviral particles were harvested, and lentiviral concentration was estimated with the lentiviral titer kit, Lenti-X GoStix (Clontech, USA) Lentiviral infection was performed by adding 1 mL of lentiviral particle solution on 70 % confluent T47D cells grown in 6 cm plates with fresh media containing 8 μg/mL polybrene. Infected cells were selected 24 h after infection with 1 μg/mL of puromycin. Three days post-infection, cells were harvested to the extraction of RNA and proteins. Efficiency of the knockdown of the gene was assayed by real-time PCR and Western blot.
Real-time quantitative PCR analysis of genes associated with the UPR signaling pathway
To activate the UPR pathway, T47D scramble and T47D silenced cells (shRNA1) were incubated in 10-cm plates, by triplicate, with dithiothreitol (DTT, 2 mM) during a time series of 4, 6, 7, and 8 h. Total RNA was isolated from controls and treated cells using TRI Reagent (MRC, USA). Approximately, 5 μg of the total RNA was reverse transcribed using RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, Canada). About 5 μL of cDNA was amplified for seven UPR pathway genes (CRT, BiP, PRKR-like endoplasmic reticulum kinase (PERK), inositol-requiring enzyme 1 (IRE1), activating transcription factor 6 (ATF6), activating transcription factor 4 (ATF4), and CHOP) and the data normalized to RPL19 endogenous control gene. Primer sequences were obtained from Balakhrishnan et al. (2013). PCR reaction was performed in a 25-μL volume, using Taq DNA polymerase and EVA green dye. PCR condition was set at an initial denaturation at 94 °C for 2 min, 40 cycles at 94 °C for 40 s, 55 °C for 40 s, and 72 °C for 40 s. For IRE1 and PERK, different annealing conditions were employed, setting 52 °C for 40 s. Data was captured and analyzed using the Stratagene MxPro-Mx3000P system 3.2 Software. The relative gene expression between T47D Scramble Control cells and Scramble and shRNA1 DTT treated cells was measured by the comparative threshold cycle (2–∆∆CT) method, and values >2-fold were considered as differentially regulated between groups at the p < 0.05 statistical significance level. Fold changes were always referred to the scramble control cells.
Soft agar growth and wound healing assays
For soft agar assay, a base layer of 1.5 mL of the corresponding culture media containing 0.5 % agarose and 10 % FBS was added to 35-mm plates. After the base layer was solidified, 5,000 cells were resuspended in 1.5 mL of culture media containing 0.35 % agarose and 10 % FBS and added to the plates. Plates were incubated at 37 °C in a humidified incubator for 10 to 20 days. Colonies were stained with 0.005 % Crystal Violet and counted using an inverted microscope.
For the wound healing assay, T47D (scramble and shRNA1) cells were grown to confluence on 10-mm plates and wounded six times in the cell monolayer with a 200-μL standard pipette tip. Cells were then washed twice with PBS to remove cell debris and incubated with DMEM medium as was previously described. The area of cell-free wound was captured as images at 0, 24, 48, and 72 h using an inverted microscope (Olympus, IX71) equipped with a digital camera (Olympus) under ×100 magnification. To quantify the migration rate of the cells, the width of the wound was measured at ten points for each time and replicate, and the mean and standard deviation were calculated.
Results
Gene expression profiling of RHBDD2 silenced cells showed an enrichment in ontological terms associated to ER stress
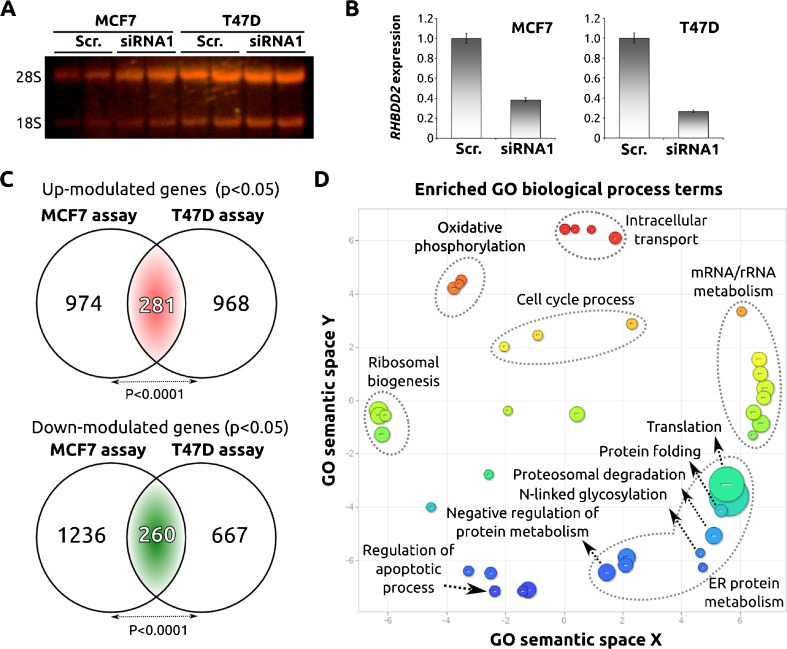
In order to identify RHBDD2-modulated pathways, we analyzed the transcriptomic profiles of breast cancer cell lines MCF7 and T47D transiently silenced for RHBDD2 expression (Fig. 1a, b). Statistical analysis of the gene expression profiling data identified 541 commonly differentially expressed genes (281 up-modulated and 260 down-modulated transcripts) in association to the RHBDD2 knockdown in both breast cancer cell lines (Fig. 1c and Supplementary File 1). We used the DAVID resource for automated annotation and functional classification of the differentially expressed genes. Among the top biofunctions, we found protein metabolism, ribosomal biogenesis, oxidative phosphorylation, regulation of cell cycle, and apoptosis (Fig. 1d). Protein metabolism was the ontological term most significantly enriched. This GO cluster includes terms associated to ER stress biology such as protein folding, proteosomal degradation, ubiquitination, and translation (Fig. 1d). The GO molecular functions most represented categories include terms associated with unfolded protein binding, structural constituents of ribosomes, oxidoreductase activity, and posttranslational protein modification.
Fig. 1.
Transcriptional effect of RHBDD2-siRNA silencing in breast cancer cell lines. RNA obtained from the different cell lines was run in a denaturing agarose gel to determine the quality samples, indicated by the presence of the ribosomal RNAs 28s and 18s. (a) The effect of the siRNA was then validated by real-time PCR as it is shown with the bar chart. (b) Venn diagram showing the overlap between transcripts modulated in association to RHBDD2 silencing in both breast cancer cell lines. Statistical analysis showed a significant number of overlapping genes between cell lines (p < 0.001). (c) Scatter plot graph of the 541 commonly deregulated genes (281 up-modulated and 260 down-modulated genes) showing the representative clusters, after redundancy reduction of the statistical significant GO terms (p < 0.01) enriched in the deregulated gene list, in a two-dimensional space related to GO terms’ semantic similarities. Bubble color indicates the p value of GO terms (expressed as log10 p value) and bubble size indicates the frequency of the GO term in the underlying GOA database (bubbles of more general terms are larger) (d)
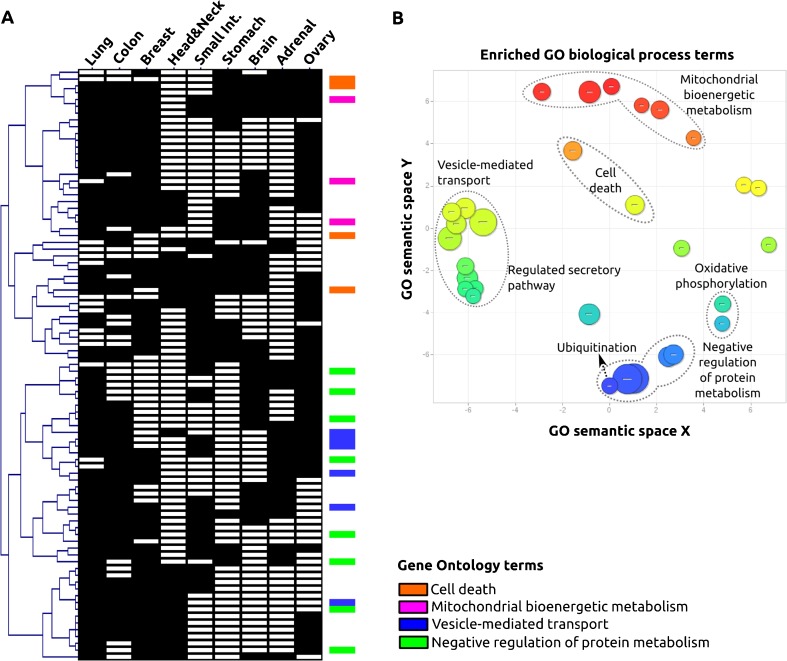
To further explore RHBDD2-associated biological pathways from another perspective, we performed an in silico analysis based on the hypothesis that genes that are co-expressed in different locations could be associated with common functional mechanisms. We searched for RHBDD2 co-expressed genes in different tissues and selected the 200 best correlated genes (p < 0.0001) for each tissue. Finally, we extracted genes overlapping in at least four different tissues (>40 %), reducing the list of genes to 87 (Fig. 2a and Supplementary File 2). The functional classification indicated enrichment in the following biological terms: negative regulation of protein metabolism, vesicle-mediated transport, regulation of cell death, and mitochondrial metabolism (Fig. 2b). These four main categories segregated in two main nodes of the HCL dendogram, determining two sets of functional related genes (Fig. 2a). Interestingly, the GO terms are very similar to those found overrepresented in the breast cancer cell line study, reinforcing the observation that RHBDD2 might be involved in the ER stress–protein folding-related pathway.
Fig. 2.
Gene ontology classification of genes co-expressed with RHBDD2 in multiple human tissues. Hierarchical clustering of the resulting array of RHBDD2 co-expressed genes (N = 87) and different tissue localizations (N = 9). (a) Scatter plot graph of the 87 RHBDD2 co-expressed genes showing the representative functional clusters according GO terms with a statistical significance of p < 0.01 (b)
RHBDD2 stable silencing affects the expression of genes associated to the UPR pathway
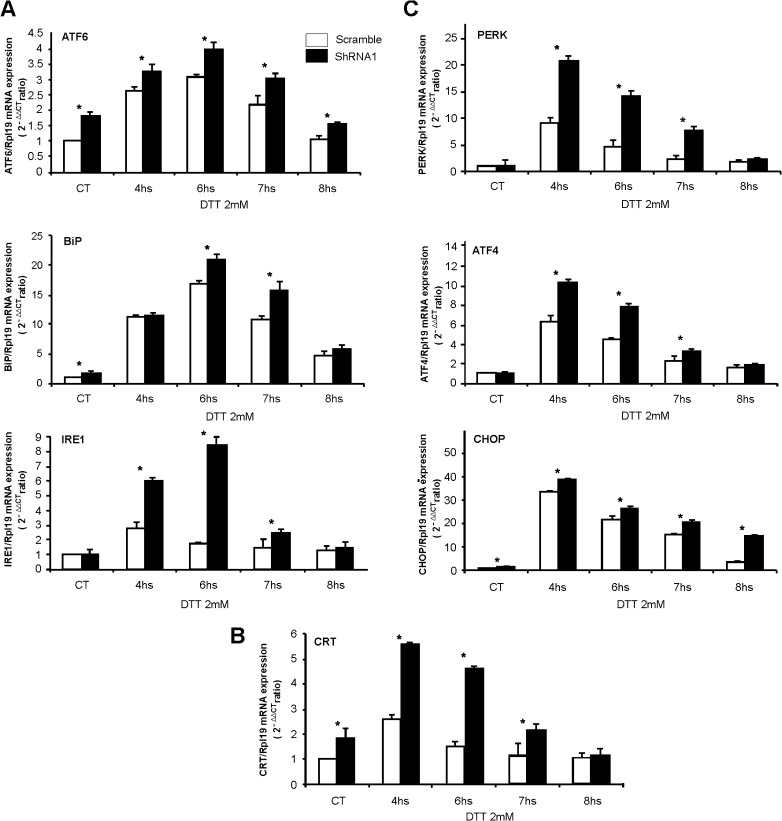
Gene expression profiling information led us to evaluate the effect of RHBDD2 stable silencing in the T47D cell line on the UPR signaling pathway under stressful ER conditions. After verifying the knockdown of RHBDD2 at mRNA and protein levels (Fig. 3), we evaluated seven genes falling in the three major axes of the UPR pathway, in which the sensor molecules are PERK, IRE1, and ATF6. The ER stress was induced by DTT treatment in a time series of four points (4, 6, 7, and 8 h). Interestingly, all the genes responded transiently to DTT, reaching a peak at 4 or 6 h with a consequent significant decrease after 7 and 8 h of exposure (Fig. 4). Importantly, cells silenced for RHBDD2 expression showed a more dramatic upregulation of all the responsive genes upon exposure to the ER stressor DTT when compared with scrambled controls. For instance, the expression of chaperones CRT and BiP increased 4.8- and 23-fold, respectively, in RHBDD2 silenced cells at 6 h of treatment, compared with 1.5- and 17-fold, respectively, in scrambled control cells (p < 0.001; Fig. 4a, b). Remarkably, a slight increase in the basal expression of both genes was also observed in control silenced cells as compared to the control scramble cells. The UPR sensor molecule ATF6 increased 4-fold in expression in RHBDD2 silenced cells vs. 3.1-fold in scramble cells (p < 0.01; Fig. 4a). Furthermore, the baseline level expression of ATF6 in RHBDD2 silenced cells unexposed to DTT was also higher than scrambled control cells (p < 0.01; Fig. 4a). A significant increase in DTT response of silenced cells was also observed in the other two sensor molecules, IRE1 and PERK (Fig. 4a, c), being the latter the sensor molecule that showed the highest response. Moreover, the downstream target of PERK, ATF4, behaved similarly to the previous genes, with a peak of 4.5-fold of expression above the scramble cells after 4 h of exposition to DTT (p < 0.01; Fig. 4c). Interestingly, CHOP, a downstream target of ATF4 and ATF6, also showed a higher increase of expression after DTT exposition in silenced cells (40-fold) compared with scramble cells (32-fold), with the peak at 4 h time point (Fig. 4c). In this sense, the results illustrate two types of kinetics of response to DTT: one with a peak at 4 h of treatment that includes the genes PERK, ATF4, CHOP, and CRT and the other with a peak at 6 h, represented by ATF6, BiP, and IRE1.
Fig. 3.
RHBDD2 stable silencing in T47D cells. Schematic representation of the plKo system employed for stable silencing of RHBDD2 and its validation at RNA and protein level. After inserting RHBDD2 siRNA, this is transcribed in the form of shRNA by the strong U6 promoter. (a) Silencing efficiency of lentiviral particles obtained from the plkO system was evidenced at mRNA and protein levels (b)
Fig. 4.
Transcriptional effect of RHBDD2 stable silencing on UPR genes under ER stress activation with DTT. Bar charts indicate the level of mRNA expression of each gene under time-dependent ER stress conditions in both scramble and RHBDD2 silenced cells (shRNA1) compared to their respective controls. A significant greater increase of the molecular sensors ATF6 and IRE1 and the chaperone BiP in response to DTT is observed in RHBDD2 silenced cells compared with scramble cells, with the peak of response at 6 h of treatment; additionally, these genes showed high levels of their RNAs in the untreated silenced cells compared with the controls (a). This situation was also evident for the chaperone CRT, but in this case, the peak of response is observed at 4 h (b), as well as observed for PERK, ATF4, and CHOP, which also showed a greater response in silenced cells compared with the scramble (c). Asterisk: Statistically significant differences (p < 0.01)
RHBDD2 stable silencing reduces anchorage-independent growth and cell migration in vitro
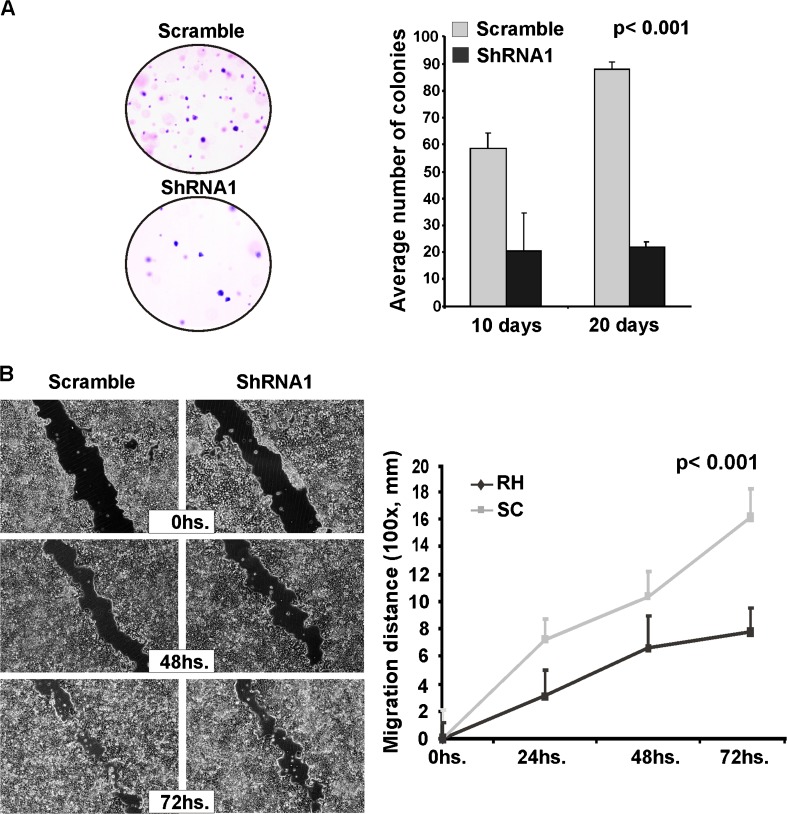
In order to evaluate the phenotypic effects of RHBDD2 stable silencing, the effect on anchorage-independent growth by the soft agar colony formation assay was evaluated. Significantly lower number of colonies was formed by the RHBDD2 silenced cells after 10 days of culture. Whereas control cells showed an average of 60 colonies per optical field, in silenced cells, an average of 30 colonies was observed (p < 0.01). Furthermore, in control cells, the number of colonies continued to increase even after 20 days of culture, while RHBDD2 silenced cells showed approximately the same number of colonies as the 10-day time point (Fig. 5a).
Fig. 5.
Phenotypic effects of RHBDD2 stable silencing in the T47D cells on colony formation and cell monolayer migration. Knocking down RHBDD2 expression in T47D cells decreased the mean number of colonies grown in agar. (a) Microphotographs of T47D cell line monolayer (×100), in which the wound is observed, are shown at three time points. A statistical significant decrease in cell migration in RHBDD2 silenced cells was observed (b)
We also analyzed directional cell migration in vitro by means of the standard wound healing assay. Similar to the result obtained in colony formation, the migration rate was significantly lower in RHBDD2 silenced cells (p < 0.001) when compared with controls, which indicates that RHBDD2 abrogation would also inhibit cell migration (Fig. 5b).
Discussion
In previous studies, we demonstrated that RHBDD2 is overexpressed at mRNA and protein levels in advanced stages of breast and colorectal carcinomas, suggesting that RHBDD2 up-modulation might be associated with tumor progression (Abba et al. 2009: Lacunza et al. 2012). In the present study, with the aim of identifying molecular pathways associated to RHBDD2, by the analysis of gene expression microarray data and subsequent experimental approaches by quantitative real-time PCR, we determined that RHBDD2 abrogation in T47D breast cancer cells disrupts the UPR signaling pathway, affecting cellular proliferation and migration in vitro.
The UPR is mainly controlled by the sensor molecules IRE1, ATF6, and PERK. In the absence of stress, these sensor molecules are maintained inactive via their association with BiP (immunoglobulin heavy chain-binding protein, also known as GRP78). Upstream in the ER, the quality control chaperone CRT (calreticulin) interacts with misfolded and unfolded proteins to achieve a native conformation; if this is not achieved, BiP dissociates from the molecular sensors (thus activating them) and binds to the unfolded proteins. As a consequence, ATF4 is activated by PERK, IRE1 is activated by homo-oligomerization followed by autophosphorylation which enables both its kinase and endoribonuclease activities to transduce stress response signals, and ATF6 is translocated to the Golgi apparatus and cleaved. The activated ATF4, the cleaved ATF6, and the products of IRE1 signaling finally translocate to the nucleus to bind UPR elements and induce transcription of several stress genes including BiP and CHOP (CCAAT/enhancer-binding protein homologous protein) (Yoshida et al. 2001; Liu and Kaufman 2003; Chakrabarti et al. 2011; Balakrishnan et al. 2013).
ER stress induction over T47D cells by using DTT showed a transient response of all the UPR genes analyzed, reaching a peak of expression at 4 and 6 h of treatment. Interestingly, in all cases, the kinetics of response was higher in RHBDD2 silenced cells than scramble cells. Remarkably, results also indicated that RHBDD2 silenced cells show an endogenous overexpression of the UPR genes BiP, CRT, ATF6, and CHOP, which would suggest that RHBDD2 abrogation might be a stressful effect in itself, partially disrupting the UPR signaling pathway. Additionally, in the balance of responses of the three sensor molecules, PERK showed the highest difference between the silenced and scramble cells, which would be consequent with the also high response of the PERK downstream genes ATF4 and CHOP. Moreover, all these genes reached the peak of response at 4 h of treatment compared to the delayed response shown by the other genes. It has been established that PERK and ATF6 would be activated before IRE1 and that this ordering would be consistent with the signals that each branch—governed by a sensor molecule—transduces (Szegezdi et al. 2006). While PERK and ATF6 pathways promote ER adaptation to misfolding, IRE1 has a dual role, transmitting survival signals and pro-apoptotic signals. According to the results shown in this study, the branch constituted by PERK, ATF4, and CHOP would be the more affected by RHBDD2 silencing.
Interestingly, CHOP (also known as GADD153) that was originally identified as a member of the DNA damage response pathway has a role in the induction of cell death by promoting protein synthesis and oxidation in the stressed ER (Di Sano et al. 2006). Therefore, the high expression of CHOP in RHBDD2 silenced cells would explain at least in part the observed decrease in cell migration and colony formation, which would be in line with the observation that RHBDD2 transient abrogation decreased MCF7 breast cancer cell viability as well (Abba et al. 2009).
The fact that RHBDD2 would be involved in the ER stress response according to the results obtained in this study is also supported by previous findings with RHBDD2 and with other members of the rhomboid family. In this sense, we recently demonstrated that the chemotherapy agent 5FU treatment on colorectal cancer cells significantly increases RHBDD2 mRNA and protein expression, compared with the untreated cells, suggesting that RHBDD2 up-modulation might be induced as an adaptive cellular response to the 5FU cytotoxic effects (Lacunza et al. 2012). Interestingly, a recent study showed that 5FU promotes ER stress, activating the mitochondrial apoptotic cell death process (Yadunandam et al. 2012). An association of different rhomboid member’s genes and the ER quality control process has also been established. It was determined that Derlins 1, 2, and 3, which are rhomboid pseudoproteases phylogenetically associated to RHBDD2, have been involved in the ERAD, a necessary process of the UPR before the ubiquitin-proteasome system (Oda et al. 2006; Greenblatt et al. 2011). Similarly, the RHBDL4 protein has been recently found upregulated upon ER stress and participates actively in the classical ERAD pathway, cleaving membrane proteins with atypical positively charged transmembrane domains (Fleig et al. 2012). Moreover, UBAC2, another rhomboid pseudoprotease, is upregulated under ER stress conditions suggesting a vital role in the control of ER protein homeostasis (Olzmann et al. 2013).
Cancer cells are often exposed to ER stress conditions, which are originated in the tumor microenvironment such as nutrient starvation, hypoxia, redox environment, and the activation of cellular processes that demand a high rate of protein biosynthesis (e.g., cell proliferation). These ER stress stimuli lead to the activation of the UPR pathway that can provide either pro-survival signals to reestablish cellular homeostasis or cell death signals if the stress is prolonged and unresolved (Scriven et al. 2009; Li et al. 2011).
Several studies have also established that the interplay between protein synthesis, ubiquitin-mediated degradation, processing, and modification regulates a broad array of basic cellular processes such as cell proliferation and migration (Adams 2003; Guerriero and Brodsky 2012). Previous reports have demonstrated that the UPR and the overexpression of UPR genes influence on cell migration by weakening cell adhesion to substrate promoting the rearrangement of cytoskeleton components, such as actins and microtubules (Rongjian et al. 2003; Kviecinski et al 2012). In this sense, Rongjian et al. who analyzed UPR on cell migration in organogenesis in vitro inferred that UPR would be an adaptive mechanism for stress cells to win more time for repairing their functions in the process of migration during organogenesis. We found that RHBDD2 abrogation directly affects cell migration and colony formation independently of substrate anchorage, which might be in agreement with the idea that stressful conditions would impact on cell fate.
In this study, we demonstrated that RHBDD2 abrogation not only deregulates UPR gene expression, but also directly affects cell migration and proliferation as well as the anchorage-independent growth. In conclusion, our findings raise the possibility that RHBDD2 overexpression could be important for tumor cells to thrive under stressful conditions by modulating their ER stress response; however, more studies will be necessary to confirm this hypothesis.
Electronic supplementary material
(XLS 166 kb)
(XLS 274 kb)
Acknowledgments
This work was supported by CONICET (PIP no. 2131) and FONCYT (PICT 0275) grants (Abba MC).
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abba MC, Sun H, Hawkins KA, Drake JA, Hu Y, Nunez MI, Gaddis S, Shi T, Horvath S, Sahin A, Aldaz CM. Breast cancer molecular signatures as determined by SAGE: correlation with lymph node status. Mol Cancer Res. 2007;5:881–890. doi: 10.1158/1541-7786.MCR-07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abba MC, Lacunza E, Nunez MI, Colussi A, Isla-larrain M, Sahin A, Segal-Eiras A, Croce MV, Aldaz CM. Rhomboid domain containing 2 (RHBDD2): a novel cancer-related gene amplified and overexpressed in invasive breast carcinomas. Biochim Biophys Acta: Mol Basis Dis. 2009;1792:988–997. doi: 10.1016/j.bbadis.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J. The proteasome: structure, function, and role in the cell. Cancer Treat Rev. 2003;29:3–9. doi: 10.1016/S0305-7372(03)00081-1. [DOI] [PubMed] [Google Scholar]
- Adler P, Kolde R, Kull M, Tkachenko A, Peterson H, Reimand J, Vilo J (2009) Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol 10(12):R139. doi:10.1186/gb-2009-10-12-r139 [DOI] [PMC free article] [PubMed]
- Adrain C, Strisovsky K, Zettl M, Hu L, Lemberg MK, Freeman M. Mammalian EGF receptor activation by the rhomboid protease RHBDL2. EMBO Rep. 2011;12:421–427. doi: 10.1038/embor.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan B, Sen D, Hareendran S, Roshini V, David S, Srivastava A, Jayandharan GR. Activation of the cellular unfolded protein response by recombinant adeno-associated virus vectors. PLoS One. 2013;8(1):e53845. doi: 10.1371/journal.pone.0053845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergbold MK, Lemberg N. Emerging role of rhomboid family proteins in mammalian biology and disease. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbamem.2013.03.025. [DOI] [PubMed] [Google Scholar]
- Blaydon DC, Etheridge SL, Risk JM, Hennies HC, Gay LJ, Carroll R, Plagnol V, McRonald FE, Stevens HP, Spurr NK, Bishop DT, Ellis A, Jankowski J, Field JK, Leigh IM, South AP, Kelsell DP. RHBDF2 mutations are associated with tylosis, a familial esophageal cancer syndrome. Am J Hum Genet. 2012;90(2):340–346. doi: 10.1016/j.ajhg.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1–3):83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Chakrabarti A, Chen AW, Varner JD. A review of the mammalian unfolded protein response. Biotechnol Bioeng. 2011;108(12):2777–2793. doi: 10.1002/bit.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D’Adamio L, Derks C, Dejaegere T, Pellegrini L, D’Hooge R, Scorrano L, De Strooper B. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Di Sano F, Ferraro E, Tufi R, Achsel T, Piacentini M, Cecconi F. Endoplasmic reticulum stress induces apoptosis by an apoptosome-dependent but caspase 12-independent mechanism. J Biol Chem. 2006;281(5):2693–2700. doi: 10.1074/jbc.M509110200. [DOI] [PubMed] [Google Scholar]
- Etheridge SL, Brooke MA, Kelsell DP, Blaydon DC. Rhomboid proteins: a role in keratinocyte proliferation and cancer. Cell Tissue Res. 2013;351(2):301–307. doi: 10.1007/s00441-012-1542-1. [DOI] [PubMed] [Google Scholar]
- Fleig L, Bergbold N, Sahasrabudhe P, Geiger B, Kaltak L, Lemberg MK. Ubiquitin-dependent intramembrane rhomboid protease promotes ERAD of membrane proteins. Mol Cell. 2012;47(4):558–569. doi: 10.1016/j.molcel.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Freeman M. Rhomboid proteases and their biological functions. Annu Rev Genet. 2008;42:191–210. doi: 10.1146/annurev.genet.42.110807.091628. [DOI] [PubMed] [Google Scholar]
- Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant alpha-1 antitrypsin from the endoplasmic reticulum. Nature Struct Mol Biol. 2011;18:1147–1153. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero CJ, Brodsky JL. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev. 2012;92(2):537–576. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kviecinski MR, Pedrosa RC, Felipe KB, Farias MS, Glorieux C, Valenzuela M, Sid B, Benites J, Valderrama JA, Verrax J, Buc Calderon P (2012) Inhibition of cell proliferation and migration by oxidative stress from ascorbate-driven juglone redox cycling in human bladder-derived T24 cells. Biochem Biophys Res Commun 421:268–273 [DOI] [PubMed]
- Lacunza E, Canzoneri R, Rabassa ME, Zwenger A, Segal-Eiras A, Croce MV, Abba MC. RHBDD2: a 5-fluorouracil responsive gene overexpressed in the advanced stages of colorectal cancer. Tumor Biol. 2012;33(6):2393–2399. doi: 10.1007/s13277-012-0503-3. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang K, Li Z. Unfolded protein response in cancer: the physician’s perspective. J Hematol Oncol. 2011;4:8. doi: 10.1186/1756-8722-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Kaufman RJ. The unfolded protein response. J Cell Sci. 2003;116:1861–1862. doi: 10.1242/jcs.00408. [DOI] [PubMed] [Google Scholar]
- McQuibban GA, Saurya S, Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423(6939):537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol. 2006;172(3):383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Richter CM, Kopito RR. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc Natl Acad Sci U S A. 2013;110(4):1345–1350. doi: 10.1073/pnas.1213738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongjian S, Yuhua C, Jindan S (2003) The effects of the unfolded protein response (UPR) on cell migration in organogenesis in vitro. Chinese Journal of Cell Biology 25(6):380–384
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Scriven P, Coulson S, Haines R, Balasubramanian S, Cross S, Wyld L. Activation and clinical significance of the unfolded protein response in breast cancer. Br J Cancer. 2009;101(10):1692–1698. doi: 10.1038/sj.bjc.6605365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smid M, Dorssers LCJ, Jenster G. Venn mapping: clustering of heterologous microarray data based on the number of co-occurring differentially expressed genes. Biogeosciences. 2003;19:2065–2071. doi: 10.1093/bioinformatics/btg282. [DOI] [PubMed] [Google Scholar]
- Sturtevant MA, Roark M, Bier E (1993) The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev 7(6):961–73 [DOI] [PubMed]
- Szegezdi E, Logue SE, Gorman AM, Samali A (2006) Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep 7(9):880–885 [DOI] [PMC free article] [PubMed]
- Urban S, Freeman M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol Cell. 2003;11(6):1425–1434. doi: 10.1016/S1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- Whitworth AJ, Lee JR, Ho VM, Flick R, Chowdhury R, McQuibban GA. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson’s disease factors Pink1 and Parkin. Dis Model Mech. 2008;1(2–3):168–174. doi: 10.1242/dmm.000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadunandam AK, Yoon JS, Seong YA, Oh CW, Kim GD. Prospective impact of 5-FU in the induction of endoplasmic reticulum stress, modulation of GRP78 expression and autophagy in Sk-Hep1 cells. Int J Oncol. 2012;41(3):1036–1042. doi: 10.3892/ijo.2012.1506. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zou H, Thomas SM, Yan Z, Grandis JR, Vogt A, Li L. Human rhomboid family-1 gene RHBDF1 participates in GPCR-mediated transactivation of EGFR growth signals in head and neck squamous cancer cells. FASEB J. 2009;23:425–432. doi: 10.1096/fj.08-112771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS 166 kb)
(XLS 274 kb)