Abstract
Heat shock protein 70 (HSP70) is one of the most abundant and best characterized heat shock protein family that consists of highly conserved stress proteins, expressed in response to stress, and plays crucial roles in environmental stress tolerance and adaptation. The present study was conducted to identify major types of genes under the HSP70 family and to quantify their expression pattern in heat- and cold-adapted Indian goats (Capra hircus) with respect to different seasons. Five HSP70 gene homologues to HSPA8, HSPA6, HSPA1A, HSPA1L, and HSPA2 were identified by gene-specific primers. The cDNA sequences showed high similarity to other mammals, and proteins have an estimated molecular weight of around 70 kDa. The expression of HSP70 genes was observed during summer and winter. During summer, the higher expression of HSPA8, HSPA6, and HSPA1A was observed, whereas the expression levels of HSPA1L and HSPA2 were found to be lower. It was also observed that the expression of HSPA1A and HSPA8 was higher during winter in both heat- and cold-adapted goats but downregulates in case of other HSPs. Therefore, both heat and cold stress induced the overexpression of HSP70 genes. An interesting finding that emerged from the study is the higher expression of HSP70 genes in cold-adapted goats during summer and in heat-adapted goats during winter. Altogether, the results indicate that the expression pattern of HSP70 genes is species- and breed-specific, most likely due to variations in thermal tolerance and adaptation to different climatic conditions.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-013-0469-0) contains supplementary material, which is available to authorized users.
Keywords: Goat, Adaptation, Heat stress, Cold stress, Heat shock protein (HSP) 70, Gene expression
Introduction
Ambient temperature plays a crucial role in determining the distribution and performance of animals. Despite having well-developed mechanisms of thermoregulation, ruminants do not maintain strict homeothermy under heat stress (Lu 1989). Heat stress is the major constraint on animal productivity in tropical climatic conditions. Growth, production, and reproduction are impaired as a result of the drastic changes in biological functions caused by heat stress.
The ability to survive and adapt to thermal stress is a fundamental requirement of cellular life. Thermal stresses trigger a complex program of gene expression and biochemical adaptive responses (Fujita 1999; Lindquist 1986). Heat shock proteins (HSPs), a large protein family, allow cells to adapt to gradual environment changes and are considered to play crucial roles in environmental stress tolerance and thermal adaptation (Feder and Hofmann 1999; Frydenberg et al. 2003; Hoffmann et al. 2003; Sørensen et al. 2003). These proteins are highly conserved across evolutionary lines (Lindquist 1986; Parsell and Lindquist 1993) and represent between 2–15 % of total cellular proteins expressed by all living organisms (Morimoto et al. 1994). Their expression follows both constitutively expressed and inducible patterns. Classified according to their molecular weight (Kregel 2002), one of the most abundant and the best characterized HSP is the 70-kDa family (HSP70), which has both inducible (HSP72) and cognate (HSP73) forms (Kiang and Tsokos 1998). Changes in gene expression in response to heat shock that are associated with acclimation include elevation of the constitutive HSP system and reduced threshold for systemic heat shock responses (Horowitz 2001). The mechanism by which the HSPs confer stress tolerance is not completely understood but may relate to the important role of HSPs in the processing of stress-denatured proteins (Mizzen and Welch 1988).
Goat, a small ruminant, is popularly known as the “poor man's cow” because of their immense economic importance (Mac Hugh and Bradley 2001). These animals are found in almost every corner of the world as they are well adapted under different geographical and environmental conditions including extreme and harsh climates. In India and other developing countries, goat is mainly reared by the small and marginal farmers, including landless agricultural laborers (Rekib 1998). Goats adapted to a harsh environment perform better than other domesticated ruminants (Devendra 1990; King 1983; Shkolnik and Silanikove 1981). These animals have developed adaptive mechanisms that allow their survival at very high temperatures (45 to 50 °C) as well as cold temperatures (−20 to −40 °C). However, despite their extreme tolerance (Al-Tamimi 2007), the productivity of these animals often declines due to thermal stress. However, a large number of information is available on physiological changes in heat and cold stress, but information in cellular and genetic levels are still very few. Elucidating the structure and expression profiling of major HSP70 genes might provide some insight into the mechanisms underlying the heat and cold adaptation in goats. The information generated will be of immense value for understanding the adaptive response of goats to heat and cold shock and also for the development of therapeutics, since adaptation to one stress often leads to cross protection. Therefore, the present study was undertaken to identify the major HSP70 genes and their expression pattern in heat- and cold-adapted goats (Capra hircus).
Materials and methods
Location, animals, and experimental protocol
All the experiments were carried out at the National Dairy Research Institute, Karnal, India. Two heat-adapted (Sirohi and Barbari) and two cold-adapted breeds (Gaddi and Chegu) were selected for the study. Sirohi and Barbari goat breeds are well adapted to the dry hot climatic conditions of Indian states of Rajasthan, Uttar Pradesh, and Gujarat, whereas Gaddi and Chegu breeds are found at high altitude and cold climatic conditions of Himachal Pradesh state. Sampling from the Sirohi and Barbari goat breeds was conducted at the Central Institute for Research on Goats (Mathura, India; 27°10′N, 70°02′E) and from Gaddi and Chegu breeds at Himachal Pradesh Krishi Viswavidyalaya (Palampur, India; 32.12°N, 76.53°E).
Twenty nonlactating and nongestating females (doe) from each breed were randomly selected for this study. The goats were 1.5 to 2 years old and had an average body wt of 25–30 kg. All the animals were clinically healthy and free from any physical or anatomical abnormalities. The experiments were carried out during three distinct phases coinciding with three seasons of the year, viz. winter (mid-December–mid-February), spring (mid-February–mid-April), and summer (mid-April–mid-June). All the animals were closely monitored and were provided similar managemental inputs during experimental period. The data on meteorological variables (ambient temperature, relative humidity) were recorded by weather station installed at CIRG, Mathura and HPKVV, Palampur. Temperature humidity index (THI) was calculated from dry and wet bulb temperature by using the following formula (Johnson et al. 1963).
 |
where Dbt is the dry bulb temperature in degrees Celsius, and Wbt is the wet bulb temperature in degrees Celsius.
Blood collection and RNA extraction
About 10 ml of blood samples was collected from each animal in lithium heparin-coated vacutainer tubes (BD Biosciences, USA) under aseptic conditions and was immediately transported to laboratory under refrigeration. A total 240 blood samples (20 animals from four breeds in three seasons) were collected. Within a season (i.e., summer, winter, or spring), blood sample collection from all the animals in a breed was not possible in a single day, so samples were collected in three consecutive days in each breed in a season, and the environmental conditions in each date of collection were recorded and considered during data analysis. Total RNA from blood was extracted using Trizol method. The concentrations were measured in NanoDrop (Thermo Scientific, USA), and purity was analyzed by agarose (1.5 %) gel electrophoresis. To exclude any genomic DNA contamination, total RNA was treated with RNase-free DNAse I (Kappa Biosystem, USA).
cDNA synthesis, cloning, and sequencing of HSP70 genes
The primers for amplification of full-length cDNA of HSP70 genes (HSPA8, HSPA6, HSPA1A, HSPA1L, and HSPA2) were designed on the basis of prior sequence information available at National Center for Biotechnology Information (NCBI) on other mammalian species (Table S1, Supplementary data). First-strand cDNA was prepared using SuperScript III Kit (Life Technology, USA). Gene-specific PCR amplifications were carried out as per standard cycling parameters [95 °C for 2 min, (95 °C for 30 s, 56–58 °C for 30 s, and 72 °C for 1 min) × 35 cycles, and 72 °C for 10 min], and the products were analyzed by agarose (1.5 %) gel electrophoresis (Fig. 1).
Fig. 1.
cDNA-amplified products of HSP70 genes on agarose gel (1.5 %). 1(a) cDNA-amplified product of HSPA8. 1(b) cDNA-amplified product of HSPA6. 1(c) cDNA-amplified product of HSPA1A. 1(d) cDNA-amplified product of HSPA1L. 1(a) cDNA-amplified product of HSPA2
Purified PCR products were inserted into the pGEM-T Easy plasmids (Promega) and amplified by culturing in XL1-Blue super competent cells. Plasmid DNA was then isolated and sequenced using forward, reverse, and internal primers in automated ABI 377 Sequencer (PerkinElmer Applied Biosystem, Foster City, CA, USA) by Xcelris Privet Limited (Ahmadabad, India). The predicted protein was done by ExPASy Translate Tool and BioEdit. The predicted molecular weight, isoelectric points (pI), and charges of the translated HSP70 proteins were estimated using Protein Calculator v3.3.
Phylogenetic analysis
For phylogenetic analysis, available sequences of the five HSPs (HSPA8, HSPA6, HSPA1A, HSPA1L, and HSPA2) of different mammalian species were obtained from the NCBI protein database. Phylogenetic analyses were conducted in MEGA5.1 (Tamura et al. 2011). The evolutionary history was inferred using the maximum parsimony method (Swofford 1998). The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein 1985). A test of the homogeneity of substitution patterns between sequences was performed by disparity index test (Kumar and Gadagkar 2001). A Monte Carlo test (1,000 replicates) was used to estimate the P values (North et al. 2002), and values smaller than 0.05 were considered significant.
Real-time PCR (qPCR)
Relative expression level of mRNA transcripts of HSP70 genes was measured by quantitative real-time PCR (qPCR) in Roche LC480 using the SYBR Green I (Roche Diagnostics, Switzerland) chemistry. The primers for qPCR were designed on the basis of prior sequence information from other animals (Table S2, Supplementary data). Along with the target genes (HSPA8, HSPA6, HSPA1A, HSPA1L, and HSPA2), two housekeeping (glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and β-actin) genes were amplified for relative expression measurements (Fig. 2). Each sample had triple replicates, and in all cases, samples of total RNA were used as negative control. Two housekeeping genes, GAPDH and β-actin, were used in this study. Proper validation was done to determine that its expression is unaffected by the experimental treatments (heat and cold). The results indicated that, for our study, β-actin was better than GAPDH. So expression data of β-actin were used for analysis of relative expression data. Relative quantification of a target gene was done by comparing the expression levels of reference gene (β-actin), as per the method of Livak and Schmittgen (2001).
Fig. 2.
Real-time PCR-amplified products of HSP70 genes (HSPA8, HSPA6, HSPA1A, HSPA2, and HSPA1L) and housekeeping genes (GAPDH and β-actin) on agarose gel (3 %)
Statistical analysis
All the experiments were replicated three times. Results were expressed as the means ± SEM. A difference with value P < 0.05 was considered statistically significant. Data were analyzed by analysis of variance, and statistical differences between the various treatment group means were determined by Duncan's multiple range test (DMRT) using the statistical Product and Service Solutions, Version 17.0.1 software (SPSS Inc., Chicago, IL, USA).
Results
Climatic conditions
The average climatic parameters recorded during the experimental periods are presented in Table 1. In both the places during spring, the THI was below 72, reflecting that the animals were in thermoneutral condition. During summer, THI was above 72, representing heat stress, whereas during winter, THI was nearly 50 in both places, indicating cold stress.
Table 1.
Environmental condition during the experimental period
| Place | Season | Average temperature (°C) | Relative humidity (%) | THI |
|---|---|---|---|---|
| Mathura | Winter | 11.98 | 63.5 | 51.38 |
| Spring | 22.8 | 59.4 | 69.78 | |
| Summer | 42.10 | 45.0 | 93.81 | |
| Palampur | Winter | 9.84 | 59.0 | 48.87 |
| Spring | 19.6 | 52.2 | 64.91 | |
| Summer | 32.10 | 60.5 | 83.13 |
Winter (mid-December to mid-February), spring (mid-February to mid-April), summer (mid-April to mid-June)
Identification of HSP70 gene homologues in goat
Five HSP70 cDNAs (HSPA8, HSPA6, HSPA1A, HSPA1L, and HSPA2) were cloned and sequenced. HSPA8 cDNA was 1,953 bp long and encoding 650 amino acids. The cDNA sequence exhibited the highest identity and similarity to heat shock 70-kDa protein 8 (HSPA8) from cattle (97 %), followed by pig (92 %), horse (91 %), and human (90 %) as well as substantial homology with 70-kDa heat shock proteins from many other species (Table S3, Supplementary data). HSPA6 cDNA was 1,912 bp long, encoded for 637 amino acids and showed substantial identity and homology to 70-kDa heat shock protein (HSPA6) from many different organisms, e.g., cattle (97 %), camel (92 %), horse (91), pig (90 %), human (90 %), etc. (Table S3, Supplementary data). Similarly, other three genes, i.e., HSPA1A, HSPA1L, and HSPA2 cDNA, were 1,926, 1,926, and 1,911 bp long, encoded for 641, 641, and 636 amino acids, respectively. The amino acid translation of these fragments exhibited the highest identity and similarity to heat shock 70-kDa protein 1A (HSPA1A), heat shock 70-kDa protein 1-like (HSPA1L), and heat shock 70-kDa protein 2 (HSPA2), respectively, from the cattle, horse, pig, and human as well as substantial homology with 70-kDa heat shock proteins from many other animals (Table S3, Supplementary data). The sequences of HSP70 genes (HSPA8, HSPA6, HSPA1A, HSPA1L, and HSPA2) in goat were submitted in NCBI GenBank with accession nos. JN616380, JN616381, JN656104, JN656105, and JN656106, respectively. The predicted molecular weights of HSP70 proteins estimated from amino acid sequences by using Protein Calculator v3.3 showed that the proteins have an estimated molecular weight around 70 kDa and with pI around 6.0 (Table S4, Supplementary data).
Phylogenetic analysis of mammalian HSPs (Fig. 3) showed three different clusters with HSPA2 and HSPA8 in one group, and HSP1A and HSPA6 in another. HSPIL formed a separate cluster. All goat HSPs were found to be grouped with other ruminants and porcine species, except HSPA6, where ruminants and porcine species were grouped differently. Monte Carlo analysis for the test in homogeneity of substitution patterns revealed that the HSPA6, after its divergence from other HSPs, has evolved independently in mammalians (Table S5, Supplementary data). A similar trend was also found for sheep and goat HSP1L that was found to evolve differently than cattle (Bos taurus). The substitution patterns of amino acids of other HSPs among different mammals were found to be nonsignificant.
Fig. 3.
Phylogenetic relationship of five HSP70 (HSPA8, HSPA6, HSPA1A, HSPA1L, and HSPA2) amino acid sequences from different mammalian species obtained from the NCBI protein database using MEGA5.1
Relative expression profile of HSP70 genes
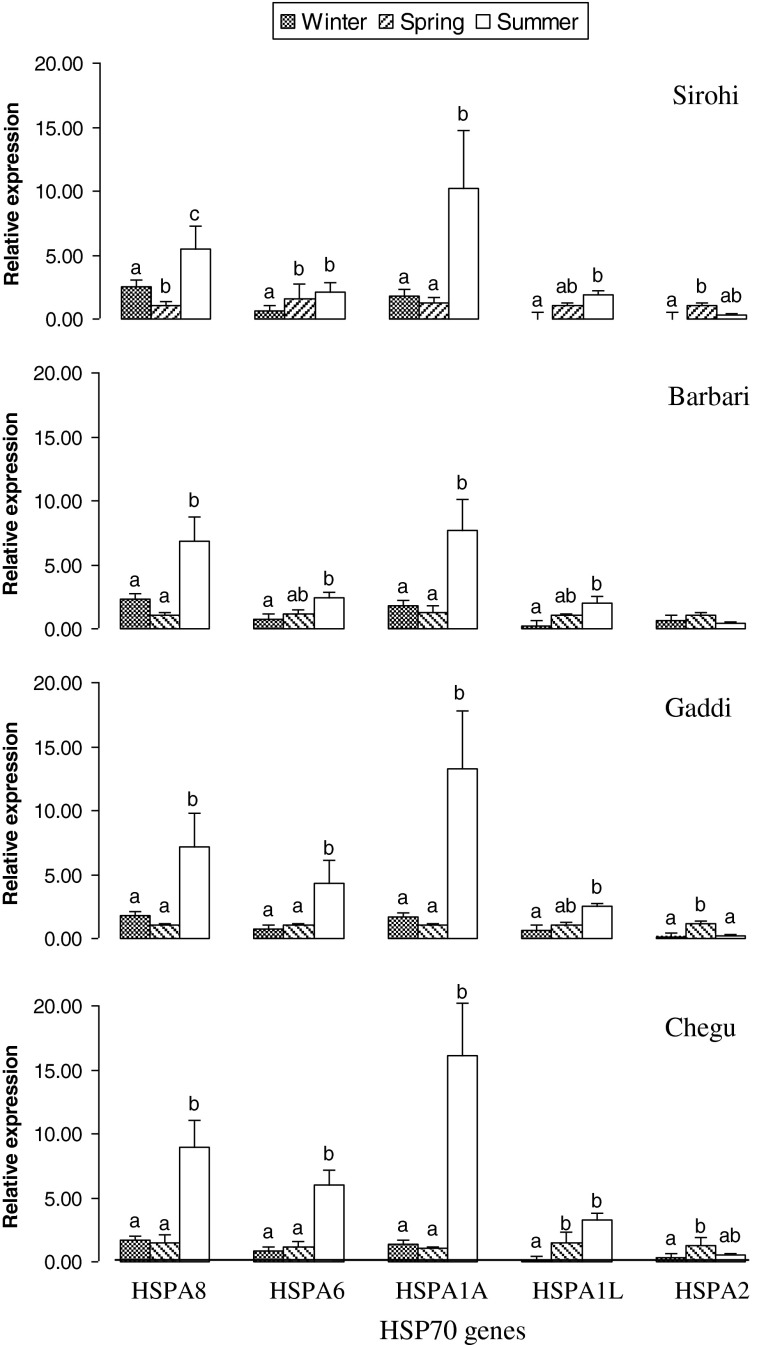
The expression of HSP70 genes (HSPA8, HSPA6, HSPA1A, HSPA1L, and HSPA2) showed temperature sensitivity and seasonal variation. Relative expression of HSP70 genes varied markedly among the heat- and cold-adapted goat breeds with a moderate variation between breeds and showed a good response to increased or decreased ambient temperature (Fig. 4). Statistical analysis revealed a significant variation between different seasons (P < 0.01) for all HSP70 gene expression. The expression of HSPA8, HSPA6, and HSPA1A was higher, but the relative mRNA expression of HSPA1L and HSPA2 was very low. Results indicated that the expression level of HSPA8 and HSPA1A was higher during both winter and summer. The expression level of HSPA6 and HSPA1L was higher only during summer. HSPA2 was observed to be downregulated during the summer and winter seasons. During summer, the relative expressions of all the HSP70 genes were higher in cold-adapted breeds (Gaddi and Chegu) than the heat-adapted breeds (Sirohi and Barbari) (Fig. 4). In Chegu, i.e., a cold-adapted breed, the expression of all the genes was higher as compared to the other cold-adapted breed of goat, Gaddi, during summer. The HSPA8 and HSPA1A expression during winter in heat-adapted breeds was observed to be higher than cold-adapted breeds.
Fig. 4.
Relative expression profile of HSP70 genes by real-time PCR during winter and summer in comparison to spring in heat-adapted (Sirohi and Barbari) and cold-adapted (Gaddi and Chegu) goats. Relative quantities of mRNA were normalized to beta actin. Values are the means ± SEM of three experiments performed. Columns with different superscript letters (a, b, c) indicate significant difference (P < 0.05). Statistical differences between various treatment group means were determined by Duncan's multiple range test
Discussion
The cloning and sequencing of five full-length cDNAs encoding the HSP70 family genes (HSPA8, HSPA6, HSPA1A, HSPA1L, and HSPA2) in goat were carried out for the first time. The BLAST results indicated that the goat HSP70 gene sequences were highly conversed and shared a high similarity (up to 91 %) with heat shock proteins of other mammals. The highest nucleotide sequence identities of goat HSP70 genes were observed with cow and pig followed by horse, camel, and human, indicating a close evolutionary relationship among HSP70 family genes. Similarly, high homology with other HSP70 genes has also been studied with the HSP70 gene from goat (Gade et al. 2010), where maximum identity with cow HSP70 (97.8 %) was observed.
The phylogenetic analysis of mammalian HSPs indicated that goat HSPs have close relationship with other ruminants including cattle (Bos indicus and B. taurus), buffalo, and sheep. They share homology with pig, camel, and horse, but are fairly distant from HSPs of human, chimpanzee, orangutan, dog, and mouse. The amino acid substitution pattern of different HSPs indicated that HSPA6 and HSP1L have evolved independently in mammalians compared to other HSPs. It will be interesting to find the biological significance of this divergence of HSP homologs in different mammalian species.
During summer and winter, animals were exposed to ambient temperature beyond the comfort zone, so they were under thermal stress in both the seasons. The findings of the present study indicated that HSP70 genes were expressed in both heat- and cold-adapted goats during summer as well as winter. The expression level of HSP70 genes, except HSPA2, was observed to be higher during summer in both heat- and cold-adapted breeds than that of winter. Our findings are in accordance with those of previous studies where heat stress-induced HSP70 expression was observed in bovine lymphocytes (Lacetera et al. 2006; Patir and Upadhyay 2007, 2010). Among all HSP70 genes studied, a significant increase was observed with respect to HSPA1A, HSPA6, and HSPA8. Similarly, heat stress-induced upregulation of HSPA8, HSPA6, and HSPA1A gene expression was observed in human blood (Sonna et al. 2002, 2004). In Brown Swiss cattle, increased HSP72 or HSPA1A mRNA levels in peripheral blood mononuclear cells due to heat stress have also been reported by Lacetera et al. (2006). Our results were also supported by earlier studies in camel showing that temperature elevation increases the level of constitutively expressed HSP70 in camel (Garbuz et al. 2011; Ulmasov et al. 1993).
A nonsignificant upregulation of HSPA1A and HSPA8 was also observed in response to cold exposure during winter. It is not surprising to see the HSP70 upregulation in response to cold as the cold-induced denaturation of proteins is a general phenomenon (Kostal and Tollarova -Borovanská 2009; Privalov 1990; Tsai et al. 2002), and in turn, partially denatured or misfolded proteins are potent triggers of rapid HSP70 accumulation (Feder and Hofmann 1999; Welch 1993). An earlier study on Drosophila suggested that the induction of HSPs following cold exposure (0 °C) was due to the shift from 0 °C back to the control temperature of 25 °C, i.e., due to heat shock, rather than to the cold treatment itself (Burton et al. 1988). Other studies on mammalian species observed that the induction of HSP70 gene expression after cold treatment (4 °C) occurred upon recovery at control temperature (37 °C). The magnitude and the kinetics of the response were, however, related to the duration of cold stress (Liu et al. 1994). However, HSPs have been implicated in cold survival, but this link has not been established conclusively (Michaud and Denlinger 2004). Therefore, further studies are highly required to differentiate between the cold exposure and the subsequent recovery phase in order to establish the role of HSPs during cold stress in goats. Low temperature affects many different structures and processes simultaneously, and therefore, cold tolerance is a highly complex adaptation and must be explained as a combination of different mechanisms (Danks 1996; Lee 1991; Murray et al. 2007). From the findings of this study, we suggest that the HSP70 is closely related to the cold tolerance in animals.
Increase of HSP70 gene expression during summer was observed to be higher in cold-adapted goat breeds (Gaddi and Chegu) as they experience higher stress during summer than heat-adapted goats (Sirohi and Barbari). In contrast, during winter, the expression of HSP70 genes was higher in heat-adapted goats than the cold-adapted goats. In different species of Drosophila, decreased HSP70 expression after acute heat stress seems to be the evolutionary consequence if the lines are exposed to chronically stressful high-temperature conditions or originate from natural populations that inhabit warm environments (Sørensen et al. 2003). The heat-adapted goats are adapted to dry hot environment in Rajasthan and parts of the U.P. region in India, while the cold-tolerant breeds are native to the relatively cold and hilly regions (Jammu-Kashmir and Himachal Pradesh) of India. Stress-induced HSP accumulation has been observed to be associated with thermotolerance and the ability to survive otherwise lethal heat stress. The difference observed in relative expression of HSP70 genes in cold-tolerant and heat-tolerant goats could be attributed to breed difference and their adaptation to different environmental conditions. The ability of the HSPs to confer thermotolerance in both cultured cells and in animals is well documented (Li 1985; Weshler et al. 1984). Stress tolerance is a complex characteristic, and the mechanism of stress tolerance by HSPs is not yet completely understood, but according to Mizzen and Welch (1988), this may be related to the important role of HSPs in the processing of stress-denatured proteins.
Among the two cold-adapted breeds, Chegu is more susceptible to heat stress than Gaddi breed because Chegu is adapted to higher altitude and low temperature. Both heat-adapted breeds, Sirohi and Barbari, showed almost similar pattern of expression, but the expression level in Sirohi goat was observed to be relatively lower. There is a growing body of literature supporting the role for HSPs in the whole organism's adaptation to heat other than through thermotolerance. A survey of lizard species inhabiting a variety of environments, including highlands, forests, and deserts, demonstrated a remarkable diversity of constitutive HSP70 levels that were correlated with the lizard's environmental temperatures (Ulmasov et al. 1992). The present study demonstrates that though HSP70 genes are highly conserved across evolutionary lines, the HSP70 gene expression is species- and breed-specific. The species-specific differences in HSP70 isoforms are most likely due to variations in thermal tolerance (Yamashita et al. 2004), and isoform expression may vary with regard to thermotolerance (Hightower et al. 1999). The role of HSP70 in thermal tolerance and adaptation to heat and cold should be uncovered in the future by exploring various cellular and animal models, enabling to directly investigate the interactions between these vitally important genomic loci.
Conclusions
The sequences of HSP70 genes in goat are well conserved and have similarities with other mammalian species. The major findings that emerged from this study include the higher expression of HSPA1A and HSPA8 and lower expression of HSPA1L and HSPA2 during both winter and summer seasons, whereas the expression of HSPA6 was higher only during summer. The expression level of HSP70 genes was higher in cold-adapted goats, and expression during winter was higher in heat-adapted goats. Regardless of reasonably high evolutionary conservation, the expression of HSP70 isoforms is species-specific, most likely due to variations in thermal tolerance. HSP70 gene expression can be used as a marker for thermal adaptation in different species, and information generated will have significant implications in the development of strategy to cope up with the challenges of climate change.
Electronic supplementary material
(DOC 34 kb)
(DOC 39 kb)
(DOC 64 kb)
(DOC 30 kb)
(XLS 41 kb)
Acknowledgments
This work was supported by World Bank-funded National Agricultural Innovation Project (NAIP-1900, code C4/C30033). The authors are thankful to the director of the National Dairy Research Institute, Karnal, India. The authors are also thankful to the director of the Central Institute for Research on Goat, Mathura, India, and to the head of the division of Veterinary Physiology, Himachal Pradesh Krishi Viswavidyalaya, Palampur, India, for providing goat blood samples.
References
- Al-Tamimi HJ. Thermoregulatory response of goat kids subjected to heat stress. Small Rumin Res. 2007;71:280–285. doi: 10.1016/j.smallrumres.2006.04.013. [DOI] [Google Scholar]
- Burton V, Mitchell HK, Young P, Petersen NS. Heat shock protection against cold stress of Drosophila melanogaster. Mol. Cell Biol. 1988;8:3550–3552. doi: 10.1128/mcb.8.8.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danks HV. The wider integration of studies on insect cold-hardiness. Eur J Entomol. 1996;93:383–403. [Google Scholar]
- Devendra C (1990) Comparative aspects of digestive physiology and nutrition in goats and sheep. In: Devendra, C., I mazumi, E. (Ed.), Ruminant nutrition and physiology in Asia. Japan Society of Zootechnical Science Ed., Sendai, pp. 45–60
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;6:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Frydenberg J, Hoffmann AA, Loeschcke V. DNA sequence variation and latitudinal associations in hsp23, hsp26 and hsp27 from natural populations of Drosophila melanogaster. Mol Ecol. 2003;12:2025–2032. doi: 10.1046/j.1365-294X.2002.01882.x. [DOI] [PubMed] [Google Scholar]
- Fujita J. Cold shock response in mammalian cells. J Mol Microbiol Biotechnol. 1999;1:243–255. [PubMed] [Google Scholar]
- Gade N, Mahapatra RK, Sonawane A, Singh VK, Doreswamy R, Saini M. Molecular characterization of heat shock protein 70–1 gene of goat (Capra hircus) Mol. Biol. Int. 2010 doi: 10.4061/2010/108429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbuz DG, Astakhova LN, Zatsepina OG, Arkhipova IR, Nudler E, Evgen'ev MB. Functional organization of hsp70 cluster in camel (Camelus dromedarius) and other mammals. PLoS ONE. 2011;6:e27205. doi: 10.1371/journal.pone.0027205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower LE, Norris CE, di Iorio PJ, Fielding E. Heat shock responses of closely related species of tropical and desert fish. Integr Comp Biol. 1999;39:877–888. doi: 10.1093/icb/39.6.877. [DOI] [Google Scholar]
- Hoffmann AA, Sorensen JG, Loeschcke V. Adaptation of Drosophila to temperature extremes: bringing together quantitative and molecular approaches. J Therm Biol. 2003;28:175–216. doi: 10.1016/S0306-4565(02)00057-8. [DOI] [Google Scholar]
- Horowitz M. Heat acclimation: phenotypic plasticity and cues to the underlying molecular mechanisms. J Therm Biol. 2001;26:357–363. doi: 10.1016/S0306-4565(01)00044-4. [DOI] [Google Scholar]
- Johnson HD, Ragsdale A C, Berry IL, Shanklin MD (1963) Temperature–humidity effects including influence of acclimation in feed and water consumption of Holstein cattle. University of Missouri Res Bull no. 846
- Kiang JG, Tsokos GC. Heat shock protein 70 kDa: molecular biology, biochemistry and physiology. Pharmacol Ther. 1998;80:183–201. doi: 10.1016/S0163-7258(98)00028-X. [DOI] [PubMed] [Google Scholar]
- King JM (1983) Livestock water needs in pastoral Africa in relation to climate and forage. Res report no. 7. Int. Lives Center Africa (ILCA), Addis Ababa, Ethiopia
- Kostal V, Tollarova -Borovanská M. The 70 kDa heat shock protein assists during the repair of chilling injury in the insect, Pyrrhocoris apterus. PLoS ONE. 2009;4:e4546. doi: 10.1371/journal.pone.0004546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kregel KC. Molecular biology of thermoregulation Invited review: heat shock proteins modifying factors in physiological stress responses and acquired thermotolerence. J Appl Physiol. 2002;92:2177–2186. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- Kumar S, Gadagkar SR. Disparity index: a simple statistic to measure and test the homogeneity of substitution patterns between molecular sequences. Genetics. 2001;158:1321–1327. doi: 10.1093/genetics/158.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacetera N, Bernabucci U, Scalia D, Basirico L, Morera P, Nardone A. Heat stress elicits different responses in peripheral blood mononuclear cells from Brown-Swiss and Holstein cows. J Dairy Sci. 2006;89:4606–4612. doi: 10.3168/jds.S0022-0302(06)72510-3. [DOI] [PubMed] [Google Scholar]
- Lee RE., Jr . Principles of insect cold tolerance. In: Lee RE Jr, Denlinger DL, editors. Insects at low temperature. New York: Chapman and Hall; 1991. pp. 17–46. [Google Scholar]
- Li GC. Elevated levels of 70,000 dalton heat shock protein in transiently thermotolerant Chinese hamster fibroblasts and in their stable heat resistant variants. Int J Radiat Oncol Biol Phys. 1985;11:165–177. doi: 10.1016/0360-3016(85)90376-1. [DOI] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Liu AYC, Bian H, Huang LE, Lee YK. Transient cold shock induces the heat shock response upon recovery at 37°C in human cells. J Biol Chem. 1994;269:14768–14775. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu CD. Effects of heat stress on goat production. Small Rumin Res. 1989;2:151–162. doi: 10.1016/0921-4488(89)90040-0. [DOI] [Google Scholar]
- Mac Hugh DE, Bradley DG. Livestock genetic origins: goats buck the trend. Proc Natl Acad Sci USA. 2001;98:5382–5384. doi: 10.1073/pnas.111163198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud MR, Denlinger DL. Molecular modalities of insect cold survival: current understanding and future trends. Int Congress Ser. 2004;1275:32–46. doi: 10.1016/j.ics.2004.08.059. [DOI] [Google Scholar]
- Mizzen L, Welch W. Effects on protein synthesis activity and the regulation of heat shock protein 70 expression. J Cell Biol. 1988;106:1105–1116. doi: 10.1083/jcb.106.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI, Tissieres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1994. pp. 1–593. [Google Scholar]
- Murray P, Hayward SAL, Govan GG, Gracey AY, Cossins AR. An explicit test of the phospholipid saturation hypothesis of acquired cold tolerance in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:5489–5494. doi: 10.1073/pnas.0609590104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BV, Curtis D, Sham PC. A note on the calculation of empirical P values from Monte Carlo procedures. Am J Hum Genet. 2002;71:439–441. doi: 10.1086/341527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Patir H, Upadhyay RC. Interrelationship between heat shock protein 70 (HSP70) and lymphocyte proliferation in thermal exposed buffalo heifers. Italian Journal of Animal Science. 2007;6:1344–1346. [Google Scholar]
- Patir H, Upadhyay RC. Purification, characterization and expression kinetics of heat shock protein 70 from Bubalus bubalis. Research in Veterinary Science. 2010;88:258–262. doi: 10.1016/j.rvsc.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Privalov PL. Cold denaturation of proteins. Crit Rev Biochem Mol Biol. 1990;25:281–305. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- Rekib A. Grazing resources and livestock productivity with special reference to goat production. Ind J Anim Sci. 1998;68:846–848. [Google Scholar]
- Shkolnik A, Silanikove N. Water economy, energy metabolism and productivity in desert ruminants. In: Morand-Fehr P, Borbouse A, De Simiane M, editors. Nutrition and systems of goat feeding. France: Tours; 1981. pp. 236–246. [Google Scholar]
- Sonna LA, Gaffin SL, Pratt RE, Cullivan ML, Angel KC, Lilly CM. Effect of acute heat shock on gene expression by human peripheral blood mononuclear cells. J Appl Physiol. 2002;92:2208–2220. doi: 10.1152/japplphysiol.01002.2001. [DOI] [PubMed] [Google Scholar]
- Sonna LA, Wenger CB, Flinn S, Sheldon HK, Sawka MN, Lilly CM. Exertional heat injury and gene expression changes: a DNA microarray analysis study. J Appl Physiol. 2004;96:1943–1953. doi: 10.1152/japplphysiol.00886.2003. [DOI] [PubMed] [Google Scholar]
- Sørensen JG, Kristensen TN, Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol Lett. 2003;6:1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x. [DOI] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (and other methods) Sunderland: Sinauer Associates; 1998. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Maizel JV, Nussinov R. The hydrophobic effect: a new insight from cold denaturation and two-state water structure. Crit Rev Biochem Mol Biol. 2002;37:55–69. doi: 10.1080/10409230290771456. [DOI] [PubMed] [Google Scholar]
- Ulmasov HA, Karaev KK, Lyashko VN, Evgen'ev MB. Heat-shock response in camel (Camelus dromedarius) blood cells and adaptation to hyperthermia. Comp Biochem Physiol. 1993;106:867–872. doi: 10.1016/0305-0491(93)90043-5. [DOI] [PubMed] [Google Scholar]
- Ulmasov KA, Shammakov S, Karaev K, Evgen'Ev MB. Heat shock proteins and thermoresistance in lizards. Proc Natl Acad Sci USA. 1992;89:1666–1670. doi: 10.1073/pnas.89.5.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch WJ. How cells respond to stress. Sci Am May. 1993;1993:34–41. doi: 10.1038/scientificamerican0593-56. [DOI] [PubMed] [Google Scholar]
- Weshler Z, Kapp DS, Lord PF, Hayes T. Development and decay of systemic thermotolerance in rats. Cancer Res. 1984;44:1347–1351. [PubMed] [Google Scholar]
- Yamashita M, Hirayoshi K, Nagata K. Characterization of multiple members of the HSP70 family in platyfish culture cells: molecular evolution of stress protein HSP70 in vertebrates. Gene. 2004;336:207–218. doi: 10.1016/j.gene.2004.04.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 34 kb)
(DOC 39 kb)
(DOC 64 kb)
(DOC 30 kb)
(XLS 41 kb)