Abstract
Surgical resection remains a mainstay of treatment and is highly effective for localized colorectal cancer. However, ~30-40% of patients develop recurrence following surgery and 40-50% of recurrences are apparent within the first few years after initial surgical resection. Several variables factor into the ultimate outcome of these patients, including the extent of disease, tumor biology, and patient co-morbidities. Additionally, the time from initial treatment to the development of recurrence is strongly associated with overall survival, particularly in patients who recur within one year of their surgical resection. Current post-resection surveillance strategies involve physical examination, laboratory, endoscopic and imaging studies utilizing various high and low-intensity protocols. Ultimately, the goal is to detect recurrence as early as possible, and ideally in the asymptomatic localized phase, to allow initiation of treatment that may still result in cure. While current strategies have been effective, several efforts are evolving to improve our ability to identify recurrent disease at its earliest phase. Our aim with this article is to briefly review the options available and, more importantly, examine emerging and future options to assist in the early detection of colon and rectal cancer recurrence.
Keywords: Colorectal cancer, recurrence, biomarkers.
Introduction
More than 140,000 people in the United States will be diagnosed with colorectal cancer (CRC) this year.1 Of those diagnosed and undergo a potentially curable surgical resection, approximately 25-40% will develop a tumor recurrence, many of whom will die from their disease.2 Confounding the situation, the optimal strategy to accurately detect recurrences at the earliest possible time is a highly debated concept in the current colorectal cancer literature. It is well known that most recurrences occur within 5 years. The current recommendations for follow-up include routine visits with laboratory evaluation every 3-4 months for the first 2 years (then every 6 months for 2 years then annually), CT scan every 6 months for the first few years, and a colonoscopy within 12 months of the resection. Patients should be offered another colonoscopy after 3 years if the examination is normal. Further surveillance depends on the results of the examination, imaging, labs and endoscopy; however, if that examination is normal, the patients may undergo routine examination annually and endoscopic surveillance every 5 years.3 It is in this post-surgical window where efforts at both predicting and identifying recurrent disease need to be expanded. More importantly, detection of these early recurrences (prior to widespread metastases) can be effectively improved through predictive nomograms, biomarkers, and/or imaging studies, and allows for potentially more effective re-intervention, and a longer overall survival. The aim of this article is to inform readers of novel and evolving concepts regarding early detection of tumor recurrence.
Predicting Tumor Recurrence
Regardless of the adequacy of resection and the use of adjuvant therapy, recurrence remains a major problem. While debate regarding the ideal surveillance strategy (i.e., intensive versus standard) exists, the focus remains on identifying recurrent disease at the earliest possible instance, when it is potentially still curable with additional treatment. One of the ways this can be achieved is by improving the ability to predict those that will recur, which could lead to more focused or intensive follow-up. To aid in this process, several patient and disease-related factors have been identified that can help better predict the risk of recurrence, including depth of invasion, number of nodes involved, site of disease, obstruction, perforation, resection margins, and pathologic grade.4,5 Based on these factors, patients are typically stratified into cohorts with a similar prognosis to aid with predicting outcomes (i.e., disease-free and overall survival) following resection. Select patients (i.e., Stage III and high-risk Stage II) will be offered modern adjuvant therapy, which has been shown to lead to a reduction in recurrence and an overall improved prognosis.6, 7 However, a key factor remains the ability to identify those patients who will truly benefit from such regimens and not expose those that will not to undue harm. The difficulty with many of the current strategies is that patients are often “lumped” together without considering other factors such as tumor biology or individual characteristics that may lead to better delineation of risk categories.8 To combat this, various predictive models are becoming more available, validated, and widely used by the practicing surgeon and medical oncologist.
Predictive Nomograms
Nomograms are prediction stemming from hundreds of patients who present with the same condition. Using the characteristics of these patients, the individual can determine their relative risk. They are typically easier to understand for the average patient due to a risk estimation on a 0-100% scale. In regards to colon cancer, nomograms can reliably predict the probability of a patient's overall survival from colon cancer five years after surgical removal of all cancerous tissue.
Adjuvant! Online
Adjuvant! Online provides a model to predict the risk of recurrence as well as the potential benefit of adjuvant chemotherapy.9 This model was originally created using the SEER database for patients with breast cancer and has subsequently been updated for patients with colon cancer. It is based upon several clinical parameters such as tumor size and number of positive nodes. The program then adds the individual's comorbidities to estimate an overall survival.9 The model is extremely useful and patient friendly, as it provides a visual bar-graph representation of the benefit of adjuvant therapy in terms of disease recurrence and overall survival, as well as likelihood of death based on co-morbidities alone (http://www.adjuvantonline.com).10
MSKCC
Another popular and useful nomogram has been created by the Memorial Sloan Kettering Cancer Center (MSKCC) for colorectal cancer.11 The model is based on an institutional database of 1,320 patients, of whom 243 had developed disease recurrence at last follow-up. The nomogram was developed, validated, and compared with the American Joint Committee on Cancer fifth and sixth editions with a concordance rate of 0.77, with regards to identifying those most at risk for recurrence at 10 years.12 This decision aid takes into account site of disease, preoperative carcinoembryonic antigen, lymphovascular invasion, and perineural invasion (http://nomograms.mskcc.org).12,13
Bayesian Belief Network
A Bayesian Belief Network (BBN) is a hierarchical network of associations amongst clinical factors in a registry data set that provides multivariate mapping of complex data.8 This modeling process uses mathematical computations based on a conditional dependence between several different variables to provide a probability estimate of a given outcome of interest. Unique to this system, the model goes beyond simple TNM staging by combining interactions amongst known variables (i.e., patient demographics, tumor biology, histopathologic characteristics) to provide individualized estimates of case-specific risks for a given patient (i.e., overall and disease-specific survival). BBNs have been shown to improve prognostic estimates and to guide clinical decision-making and counseling for appropriate treatment and follow-up surveillance testing for several different tumors including colorectal cancer.8, 14, 15
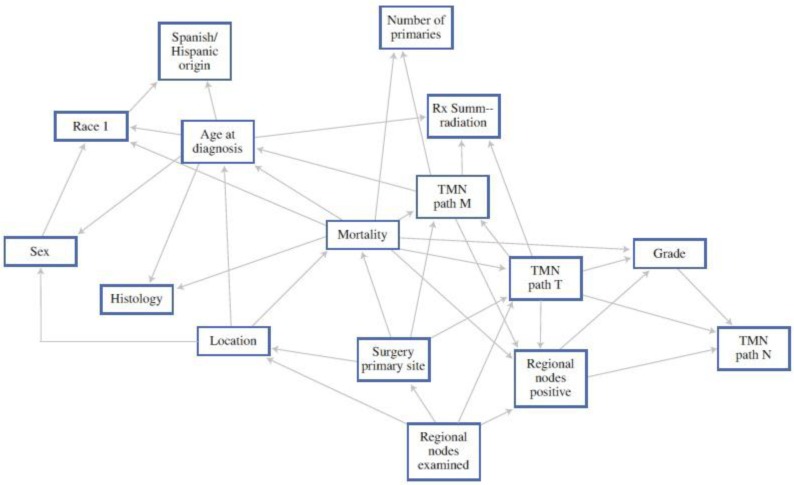
As an example, Stojadinovic et al. reported on a BBN specifically looking at colorectal cancer. When compared with the AJCC staging system alone, they showed superior sensitivity and specificity in estimating mortality.8 These inferences can then be used to assist patients and providers to estimate outcomes much more specific to the individual patient (Figure 1). While not currently available, if the above models could be placed into computer or smart-phone “app” formats, they may assist the provider with real-time clinical decision-making in terms of use of (or avoidance of) adjuvant treatment.
Figure 1.
A 12-month Bayesian Belief Network. Each box in the figure represents a feature within the study data set, the edges represent patterns of dependence between features. This allows an estimate of mortality based on the different combinations. 8 (With Permission, Springer Publishing)
Genetic Components of Recurrence
Markers that can reliably predict the likelihood of recurrence or responsiveness to chemotherapy would be particularly helpful in Stage II or node-negative colorectal cancer for clinical decision-making regarding both treatment and follow-up. Current pathological staging fails to predict recurrence accurately in many patients undergoing curative surgery for localized colorectal cancer. In fact, 10%-20% of patients with Stage II colorectal cancer (theoretically cured by a proper oncological operation), and 30%-40% of those with Stage III colorectal cancer, develop recurrence.16 Where the “blame” lies in this is controversial; however, factors such as inadequate surgical margins or lymph node extraction, insufficient pathological examination (i.e., under-staging), and tumor biology have all been reported to play a role in recurrent disease.17 In the realm of colorectal cancer, microarray technology has been used to investigate gene expression profiles, yet no established signature has been found that is reliably useful for clinical practice. Even less data exists for predicting prognosis and recurrence. Studies on CRC have been poorly reproducible, possibly because CRC is composed of distinct molecular entities that may develop through multiple pathways on the basis of different molecular features.18 Currently, most scientific information is in the preclinical stages, however, there is promise that one or more of the following laboratory findings may play a larger role in determining treatment guidance as well as cancer recurrence.
Carcinoembryonic Antigen (CEA)
Serum carcinoembryonic antigen (CEA) is still the most widely used tumor marker in patients with colorectal cancer. Pretreatment CEA levels are useful for prediction of prognosis, and postoperative serial assays of CEA level provide an opportunity for early detection of recurrent disease by following trends and basing additional diagnostic testing on early elevations.19 As such, serum CEA level is still considered the laboratory value of choice to monitor the risk of local or distant recurrence after surgical resection. Consistent elevation in CEA is a concerning sign of disease recurrence and it remains the only widely used serum tumor marker that has been shown to correlate sufficiently with colorectal tumor activity to be used reliably during follow-up.20 However, there is not uniform acceptance of this notion. Amri and colleagues questioned whether preoperative continuous CEA values could be used as a predictor of disease-free survival, recurrence, and overall survival, in surgical colon cancer cases. Specifically, CEA levels were measured at different intervals up to the point of colorectal cancer resection. They showed that using CEA levels continuously had no relationship with metastatic recurrence and a weak association with the overall risk of metastatic disease and death. The association of preoperative CEA with recurrence was not statistically significant using logistic regression (P=0.5). Cox regression also showed no change in hazard ratio of shorter survival duration (HR=1, P<0.001) and shorter disease-free interval (HR=1, P<0.001).21 At this time, preoperative CEA may be a useful risk estimator, but has limited significance for predictions of long-term outcomes in individual cases or the risk of recurrence. Furthermore, it highlights the need for better markers of early recurrence, as drawbacks to CEA's use include a lack of consensus on the frequency or duration of its testing, the potential for unnecessary additional diagnostic testing (i.e., lack of cost-effectiveness), as well as its utility in guiding invasive intervention, especially in light of advances in imaging techniques such as PET/CT.22,23
Microsatellite Instability (MSI)
Among the molecular markers that have been extensively investigated for colon cancer characterization and prognosis, microsatellite instability is caused by a defective function of the DNA mismatch repair system. MSI also shows some of the most promising data related to colon cancer recurrence. Patients with MSI-high colon cancers, defined as those with multiple insertion or deletion mutations in at least 2 of 5 loci of DNA, rarely develop distant metastasis and have a longer overall survival (OS) than stage-matched microsatellite stable (i.e., those cancers with no instability in at least 5 loci of DNA) or microsatellite-low (1 of 5 loci with insertion or deletion mutations) colon cancer patients.24, 25,26 Tikidzhieva et al. found that MSI status modulates the risk of disease relapse in colon cancer patients in a time-varying manner, with a significantly reduced risk of disease relapse in MSI-high cancer patients compared with MSI-stable cancer patients after 12 months of follow-up; however, they did show an increased in relapse during the first 12 months in patients with a MSI-high status, which subsequently decreased after years 2 and 3 from their procedure (5-year TTR estimates of 0.82 for MSI-high vs. 0.66 for MSS patients (P = 0.03).27 This is a similar finding by Lin et al. in that MSI-high is an independent favorable prognostic factor for survival in colon cancer.28 Garcia and associates found that MSI-low was an independent risk factor for recurrent distant disease in Stage II and III disease (HR 1.83; 95% CI 1.06-3.15).29 On the other hand, in a retrospective review of 322 patients, MSI status (combined with CpG island methylator phenotype (CIMP) was not associated with 3-year disease-free survival (P=0.33) after adjuvant FOLFOX in Stage III disease.25 Therefore, while microsatellite stability should likely be tested in all tumors, it is yet to be determined what its ultimate role will be in predicting recurrence, and how clinically this will be implemented. One option may involve stratification of intensive (MSI-low) versus standard (MSI-high) surveillance strategy based on MSI status. Another future use could involve the use of specific testing based on MSI status. For example, as MSI-low has been associated with recurrent distant disease, whole body MRI or PET/CT may be a component of surveillance for that individual patient.
Metastasis-Associated in Colon Cancer 1 (MACC1)
The metastasis-associated in colon cancer 1 (MACC1) gene has promising features: it is frequently overexpressed in metastatic colon cancer, and elevated levels of expression in the primary lesion are associated with poor prognosis.30,31 MACC1 mRNA is expressed in both primary colon cancer and colon cancer metastases. MACC1 promotes tumor cell motility and invasion, which would then theoretically lead to local-regional and systemic dissemination of the disease. These observations have been extended also to other cancer types, including lung adenocarcinomas, hepatocellular carcinomas, gastric carcinomas, and ovarian tumors.32-34 Isella et al. demonstrated that low-MACC1 (good prognosis) patients had a significantly lower recurrence rate (2 of 13; 15.4%) than high-MACC1 (poor prognosis) patients (35 of 51; 68.6%; P=0.01).35 Moreover, the prognostic power of MACC1 expression seems to be unrelated to adjuvant therapy (HR=6.5 in patients treated with adjuvant therapy, HR=4.3 for patients not treated). This reinforces the notion that MACC1 is a pure prognostic indicator that could be exploited to inform rational therapeutic decisions after surgical intervention. In regards to rectal cancer, Kawamura et al. also demonstrated the poor prognostic characteristics associated with high expression of MACC1. Specifically, MACC1 was associated with reduced relapse-free survival when cutoff values above 0.261 were identified amongst individuals followed for an average of 5 years.36 Ultimately, this biomarker could pave the way for the inclusion of MACC1 expression analysis in a scoring system (including molecular, clinical, and pathological features) that could help the clinician in assigning aggressive, mild, or even non-adjuvant regimens to resected patients.
Plastin3
Tumor cells can be shed and found to be circulating within the peripheral circulatory system. These are termed circulating tumor cells (CTCs). There are multiple ways to identify these cells; however, the current systems available may miss the cells in a transition phase-- known as the epithelial - mesenchymal transition--a process through which these cells lose some of the identifying characteristics that CTCs possess.37 The Plastin3 gene is not repressed during this transformation and therefore can be used to identify those CTCs that are in transition. Plastin3 (PLS3) is located on chromosome Xq23 and functions to polymerize actin fibers38. Yokobori et al showed that patients with PLS3-positive CTCs in peripheral blood had significantly shorter disease-free survival than PLS3-negative patients. Multivariate analysis showed that PLS3-positive CTCs in peripheral blood are independently associated with poor prognosis (HR = 2.17; 95% CI = 1.38-3.40) and recurrence (HR = 2.32; 95% CI = 1.42-3.74).39 This finding is of utmost importance, as it shows that early tumor cell dissemination can be detected by the established PLS3 assay and is relevant for the subsequent course of the disease in patients with CRC. The current belief is that if patients with low recurrence rates can be detected by PLS3 expression in peripheral blood, adjuvant chemotherapy for some patients may be unnecessary. Conversely, this could serve as a marker for identifying high-risk groups with node-negative disease that may benefit from (but not normally receive) adjuvant therapy.
Retinoic Acid-induced protein 3 (RAI3)
One of the many plasma membrane proteins overexpressed in colon cancer, RAI3 represents a novel biomarker that may have prognostic benefits. Zougman and colleagues were able to confirm through the use of immunohistochemistry that cytoplasmic RAI3 expression was significantly associated with disease recurrence in Stage I-III patients. High RAI3 expression was associated with a three-fold risk of recurrence compared to patients with low expression [HR = 3.076; (95% CI = 1.738-5.445); P < 0.001].40 With this, they concluded that simple and reproducible immunohistochemistry of cytoplasmic RAI3 expression could potentially separate colon cancer patients into high and low-risk groups.40 Similar to the other genetic markers, this risk stratification can be incorporated into algorithms for individualizing adjuvant therapy or surveillance protocols.
MicroRNA-29c
A mature microRNA (miRNA) is a small, noncoding RNA that contains approximately 20 nucleotides and can post-transcriptionally regulate the expression of several target genes at same time.41 The tumorogenesis of CRC involves multi-step genomic changes, including the activation of oncogenes and inactivation of tumor suppressor genes.41 Multiple miRNAs have been suggested to play a role in the development of cancers, including carcinogenesis, progression, and recurrence.42 Only a few studies have investigated circulating miRNAs in patients with CRC.41, 43 Yang et al. looked at 107 patients with Stage II and III CRC who did not undergo neoadjuvant therapy, 56 of which were non-early relapse patients and 51 were early relapsed patients. They defined early versus non-early relapse with recurrence <1 year or greater than 1 year, respectively. Serum samples were then collected and measured before they received surgery. They found miRNA-29c levels were lower in the samples from early relapsed patients by 2-fold compared to that in samples of non-early relapsed patients. They concluded that the expression of miRNA-29c is an independent predictor for detecting the early relapse of CRC. The combination of results from Yang et al. in the realms of cellular, animal, and human studies consistently indicate that miRNA-29c exerts an anti-tumorogenesis effect, and it may be a useful biomarker for the prediction of early relapse in CRC patients after an operation.44,45 As with the previous biomarkers, miRNA-29c has the potential to identify high-risk CRC patients after radical resection. This can ultimately result in an enhanced surveillance regimen or even an intensive therapeutic program for these patients.
Individual biomarkers may be the wave of the future in regards to identifying the risk of recurrence; however, this will ultimately become very inefficient and likely expensive. Multigene assays can potentially mitigate these obstacles and give a more reliable prediction of recurrence to the patient and physician. One such panel is the Oncotype DX Colon Cancer Recurrence Score (Genomic Health, Redwood City, CA). This modality was developed by using tumor gene expression data from 1,851 patients with resected colon cancer in four independent trials.46 It has been confirmed by multiple prospective trials and validated as a predictor of recurrence in patients with stage II colon CA.47
The Recurrence Score (RS) obtained using Oncotype DX is calculated using a quantitative reverse transcriptase polymerase chain reaction assay that measures the expression of 12 genes (seven recurrence genes and five reference genes) in formalin-fixed paraffin-embedded (FFPE) primary colon tumor tissue.46 The RS is determined by identifying specific changes in tumor biology when unregulated. Six genes are in specific biologic pathways: cell cycle control (MKI67, MYC, MYBL2) and stromal response (FAP, BGN, INHBA).46 The seventh gene is considered a marker of genotoxicity (GADD45B) that may regulate the activity of the stromal response genes. Venook and colleagues used the 12-gene RS to show how it was significantly associated with risk of tumor recurrence in patients with Stage II colon cancer [HR=1.52 for a 25-unit increase in RS (95% CI, 1.09 to 2.12; P=0.013)].48 They also demonstrated how the 12-gene RS was prognostic for outcomes even when the usual markers of clinical and pathologic factors are removed, (i.e. T-Stage, number of nodes examined, and grade).48These biomarkers and multigene assays are clearly the future of identifying recurrence in patients with colon cancer. While they are novel today, it is important to recognize that with the extensive ongoing research in this field, the individual markers may soon be obsolete as more specific markers are identified. In reality, the assays available now use a relatively small number of genes and markers. As technology progresses, so likely will the numbers of available markers to further push the envelope when it comes to both tailoring therapy and identifying those patients who are at a much greater risk to recur.
Endoscopy for Recurrence
Imaging modalities such as chest/abdominal/pelvis CT scans and MRI are the standard recommendations for the follow-up evaluation of colon cancer treatment. Colonoscopy is also a modality to detect intra-luminal recurrence, and follow-up colonoscopy is currently the standard of care for patients with a history of colon cancer. Novel techniques have surfaced which are suggesting even better detection rates than the standard white light colonoscopic evaluation.
Chromoendoscopy
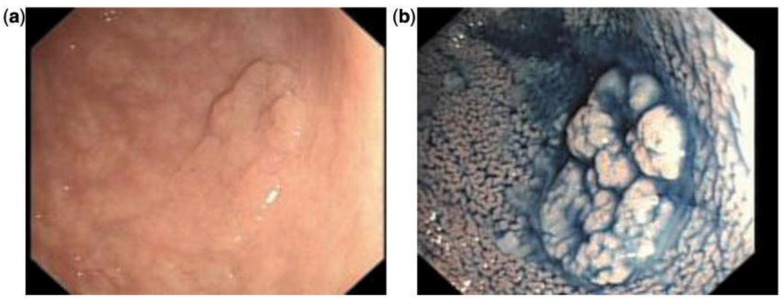
Chromoendoscopy is a diagnostic method in which a chemical substance is sprayed onto the mucosal surface of the gastrointestinal tract to highlight specific areas or distinguish among different types of epithelia (Figure 2).49 One of the first, large prospective trials used vital staining with indigo carmine on all visible lesions in 100 consecutive patients without visible inflammatory changes. If findings on macroscopic examinations were unremarkable, the sigmoid colon and rectum were stained with indigo carmine over a defined segment (0-30 cm) and inspected for lesions visible only after staining. Using this technique, 178 additional lesions in the sigmoid colon were identified as being detectable only after dye spray.50
Figure 2.
Indigo carmine chromoendoscopy showing mucosal alteration in the sigmoid: (a) before staining and (b) after staining.49 (With permission Oxford University Press).
Fuji intelligent chromoendoscopy (Fujinon) or i-Scan (Pentax) utilizes the light reflected from the intestinal mucosa, which is then modified by 'post-processor computer algorithms' that allow different enhancements of the vasculature, surface architecture or tissue patterns. Although these techniques provide accurate classification and differentiation of colorectal polyps and neoplastic lesions, a clear advantage in adenoma detection rate over chromocolonoscopy or high-resolution white light endoscopy has not yet been shown.51, 52 If these technologies were added to the surveillance algorithm in patients with resected colon cancer, identifying a local recurrence may be detected earlier and possibly even easier.
Autofluorescence imaging (AFI)
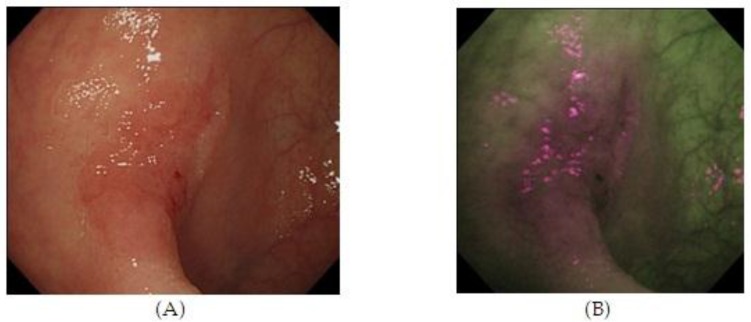
Autofluorescence imaging (AFI) endoscopy uses short wavelengths of light to stimulate endogenous substances, so called 'fluorophores' in the tissue to emit fluorescent light of a longer wavelength. Due to different content of such fluorophores, normal and neoplastic tissues differ in their autofluorescence spectra. Whereas normal mucosa appears green, neoplastic areas are 'flagged up' in violet (Figure 3).53,54 Interestingly, AFI does not detect hyperplastic polyps as a clear magenta area.55 Based on this property, AFI is expected to be used for discriminating colon neoplasms from non-neoplasms, which would be a clear benefit for assessing the scar or staple line from a recently resected colonic neoplastic process. However, studies have not consistently demonstrated the superiority of this method compared with high resolution endoscopy alone, especially for detection of colonic dysplasia.54 Thus, AFI guided biopsies are not currently able to replace random biopsies during surveillance endoscopies, but may be in the future as the technology evolves.
Figure 3.
High Resolution Endoscopy (A) and Auto Fluorescence Imaging (B) of a flat and depressed type of colon cancer with submucosal invasion not seen on HRE. 55 (With Permission, InTech Publishing).
Confocal Laser endomicroscopy (CLE)
Confocal Laser endomicroscopy (CLE) is a technique that offers in-vivo imaging of the mucosal layer at cellular and subcellular resolutions without the fixation artifacts one experiences with histological specimens. In this system, fluorescent dyes are applied either locally or systemically, and subsequently excited by a low-power laser. The intensity of the fluorescent energy is then captured as an image. Studies have shown the high accuracy (96.7%) with which CLE allows differentiation from low-grade to high-grade intra-epithelial colorectal neoplasia. Moreover, chromoendoscopy together with targeted CLE is able to increase the diagnostic yield of intra-epithelial neoplasia by 4.75-fold (P = 0.005).56,57 Unfortunately, several factors are currently working against CLE, one of which includes the cost of a CLE unit and questionable reimbursement rates. As CLE is relatively new, the concept that a histologic specimen is not needed remains theoretical. Most endoscopists will still remove tissue to confirm diagnosis rather than potentially leave cancer behind. Once better results are confirmed through trials adoption of this technique may occur.
In summary, endoscopy is an essential component to post-resection colorectal cancer to diagnosing and excluding intra-luminal recurrence. It makes sense that if we are able to identify a recurrence earlier due to advances of technology, potentially this will allow earlier re-intervention, and overall survival will be improved. The techniques described are currently in the niche-use or testing stages, and yet to be scrutinized with randomized controlled trials. The benefit of these new technologies is not necessarily their individual use, but their combination which increases the ability to detect early recurrence during surveillance. Imagine one scope in which all the aforementioned mucosal and video enhancements are available to the endoscopist. Finally, as with all technologies, cost remains a significant barrier in the early stages until competition or another breakthrough becomes a factor to drive down prices. As long as identifying recurrence is the goal, novel technologies will help to push surgeons and endoscopists to move closer to identifying recurrent cancers as early as possible.
Conclusion
Assessment of tumor recurrence is an integral part of this long-term survival and plays a pivotal role in the selection of those that will undergo re-intervention or adjuvant therapy and those that should not. The role of chemotherapy regimens, genomic databases/biomarkers, and imaging modalities, are the most widely discussed topics in the literature right now. There is great hope that these innovations will soon be at the disposal of those physicians who treat these patients on a daily basis.
Disclaimers
The views expressed are those of the author(s) and do not reflect the official policy of the Department of the Army, Department of the Navy, the Department of Defense or the U.S. Government.
Table 1.
Emerging Tools to Detect Tumor Recurrence.
| Clinical | Labs | Endoscopy |
|---|---|---|
| Adjuvant! Online | CEA | Chromoendoscopy |
| MSKCC | RAI3 | AFI |
| BBN | MACC1 | CLE |
| MSI | ||
| Plastin3 | ||
| miRNA | ||
| OncotypeDx |
MSKCC - Memorial Sloan Kettering Colon Cancer, BBN - Bayesian Belief Network, AFI - Autofluorescence imaging, CLE - Confocal Laser Endoscopy, CEA - Carcinoembryonic antigen, RAI3 - Retinoic Acid-induced protein 3, MACC1 - metastasis associated in colon cancer 1, MSI - Microsatellite Instability, miRNA - Micro RNA
References
- 1.Chang GJ, Kaiser AM, Mills S, Rafferty JF, Buie WD. Practice parameters for the management of colon cancer. Dis Colon Rectum. 2012;55(8):831–843. doi: 10.1097/DCR.0b013e3182567e13. [DOI] [PubMed] [Google Scholar]
- 2.Tjandra JJ, Chan MK. Follow-up after curative resection of colorectal cancer: a meta-analysis. Dis Colon Rectum. 2007;50(11):1783–1799. doi: 10.1007/s10350-007-9030-5. [DOI] [PubMed] [Google Scholar]
- 3.Seo SI, Lim SB, Yoon YS. et al. Comparison of recurrence patterns between </=5 years and >5 years after curative operations in colorectal cancer patients. J Surg Oncol. 2013;108(1):9–13. doi: 10.1002/jso.23349. [DOI] [PubMed] [Google Scholar]
- 4.Cortet M, Grimault A, Cheynel N, Lepage C, Bouvier A, Faivre J. Patterns of recurrence of obstructing colon cancers after surgery for cure: a population-based study. Colorectal Dis. 2013. [DOI] [PubMed]
- 5.Paty PB, Nash GM, Baron P. et al. Long-term results of local excision for rectal cancer. Ann Surg. 2002;236(4):522–529. doi: 10.1097/00000658-200210000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petersen SH, Harling H, Kirkeby LT, Wille-Jorgensen P, Mocellin S. Postoperative adjuvant chemotherapy in rectal cancer operated for cure. Cochrane Database Syst Rev. 2012;3:CD004078. doi: 10.1002/14651858.CD004078.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Des Guetz G, Uzzan B, Morere JF, Perret G, Nicolas P. Duration of adjuvant chemotherapy for patients with non-metastatic colorectal cancer. Cochrane Database Syst Rev. 2010:CD007046. doi: 10.1002/14651858.CD007046.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stojadinovic A, Bilchik A, Smith D. et al. Clinical decision support and individualized prediction of survival in colon cancer: bayesian belief network model. Ann Surg Oncol. 2013;20(1):161–174. doi: 10.1245/s10434-012-2555-4. [DOI] [PubMed] [Google Scholar]
- 9.Ravdin PM, Siminoff LA, Davis GJ. et al. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol. 2001;19(4):980–991. doi: 10.1200/JCO.2001.19.4.980. [DOI] [PubMed] [Google Scholar]
- 10.Adjuvant! Online. Decision making tools for health care professionals. Accessed September 1. 2013.
- 11.Colorectal Cancer Nomogram: Post-Surgery. MSKCC; http://www.mskcc.org/mskcc/html/83364.cfm. [Google Scholar]
- 12.Weiser MR, Landmann RG, Kattan MW. et al. Individualized prediction of colon cancer recurrence using a nomogram. J Clin Oncol. 2008;26(3):380–385. doi: 10.1200/JCO.2007.14.1291. [DOI] [PubMed] [Google Scholar]
- 13.Memorial Sloan-Kettering Cancer Center, Colorectoral Cancer Nomogram. http://nomograms.mskcc.org/Colorectal/index.aspx.
- 14.Stojadinovic A, Nissan A, Eberhardt J, Chua TC, Pelz JO, Esquivel J. Development of a Bayesian Belief Network Model for personalized prognostic risk assessment in colon carcinomatosis. Am Surg. 2011;77(2):221–230. [PubMed] [Google Scholar]
- 15.Stojadinovic A, Peoples GE, Libutti SK. et al. Development of a clinical decision model for thyroid nodules. BMC Surg. 2009;9:12. doi: 10.1186/1471-2482-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aghili M, Izadi S, Madani H, Mortazavi H. Clinical and pathological evaluation of patients with early and late recurrence of colorectal cancer. Asia Pac J Clin Oncol. 2010;6(1):35–41. doi: 10.1111/j.1743-7563.2010.01275.x. [DOI] [PubMed] [Google Scholar]
- 17.Avital I, Langan RC, Summers TA. et al. Evidence-based Guidelines for Precision Risk Stratification-Based Screening (PRSBS) for Colorectal Cancer: Lessons learned from the US Armed Forces: Consensus and Future Directions. J Cancer. 2013;4(3):172–192. doi: 10.7150/jca.5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bravo HC, Pihur V, McCall M, Irizarry RA, Leek JT. Gene expression anti-profiles as a basis for accurate universal cancer signatures. BMC Bioinformatics. 2012;13:272. doi: 10.1186/1471-2105-13-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandes LC, Kim SB, Saad SS, Matos D. Value of carcinoembryonic antigen and cytokeratins for the detection of recurrent disease following curative resection of colorectal cancer. World J Gastroenterol. 2006;12(24):3891–3894. doi: 10.3748/wjg.v12.i24.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anthony T, Simmang C, Hyman N. et al. Practice parameters for the surveillance and follow-up of patients with colon and rectal cancer. Dis Colon Rectum. 2004;47(6):807–817. doi: 10.1007/s10350-004-0519-x. [DOI] [PubMed] [Google Scholar]
- 21.Amri R, Bordeianou LG, Sylla P, Berger DL. Preoperative carcinoembryonic antigen as an outcome predictor in colon cancer. J Surg Oncol. 2013;108(1):14–18. doi: 10.1002/jso.23352. [DOI] [PubMed] [Google Scholar]
- 22.Goldstein MJ, Mitchell EP. Carcinoembryonic antigen in the staging and follow-up of patients with colorectal cancer. Cancer Invest. 2005;23(4):338–351. doi: 10.1081/cnv-58878. [DOI] [PubMed] [Google Scholar]
- 23.Makis W, Kurzencwyg D, Hickeson M. 18F-FDG PET/CT superior to serum CEA in detection of colorectal cancer and its recurrence. Clin Imaging. 2013. [DOI] [PubMed]
- 24.Boland CR, Thibodeau SN, Hamilton SR. et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58(22):5248–5257. [PubMed] [Google Scholar]
- 25.Han SW, Lee HJ, Bae JM. et al. Methylation and microsatellite status and recurrence following adjuvant FOLFOX in colorectal cancer. Int J Cancer. 2013;132(9):2209–2216. doi: 10.1002/ijc.27888. [DOI] [PubMed] [Google Scholar]
- 26.Gryfe R, Kim H, Hsieh ET. et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342(2):69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 27.Tikidzhieva A, Benner A, Michel S. et al. Microsatellite instability and Beta2-Microglobulin mutations as prognostic markers in colon cancer: results of the FOGT-4 trial. Br J Cancer. 2012;106(6):1239–1245. doi: 10.1038/bjc.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin SP, Lee YT, Yang SH. et al. Colon cancer stem cells resist antiangiogenesis therapy-induced apoptosis. Cancer Lett. 2013;328(2):226–234. doi: 10.1016/j.canlet.2012.08.036. [DOI] [PubMed] [Google Scholar]
- 29.Garcia M, Choi C, Kim HR. et al. Association between recurrent metastasis from stage II and III primary colorectal tumors and moderate microsatellite instability. Gastroenterology. 2012;143(1):48–50. doi: 10.1053/j.gastro.2012.03.034. e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boardman LA. Overexpression of MACC1 leads to downstream activation of HGF/MET and potentiates metastasis and recurrence of colorectal cancer. Genome Med. 2009;1(4):36. doi: 10.1186/gm36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirahata A, Sakata M, Kitamura Y. et al. MACC 1 as a marker for peritoneal-disseminated gastric carcinoma. Anticancer Res. 2010;30(9):3441–3444. [PubMed] [Google Scholar]
- 32.Stein U, Dahlmann M, Walther W. MACC1 - more than metastasis? Facts and predictions about a novel gene. J Mol Med (Berl) 2010;88(1):11–18. doi: 10.1007/s00109-009-0537-1. [DOI] [PubMed] [Google Scholar]
- 33.Stein U, Walther W, Arlt F. et al. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat Med. 2009;15(1):59–67. doi: 10.1038/nm.1889. [DOI] [PubMed] [Google Scholar]
- 34.Shimokawa H, Uramoto H, Onitsuka T. et al. Overexpression of MACC1 mRNA in lung adenocarcinoma is associated with postoperative recurrence. J Thorac Cardiovasc Surg. 2011;141(4):895–898. doi: 10.1016/j.jtcvs.2010.09.044. [DOI] [PubMed] [Google Scholar]
- 35.Isella C, Mellano A, Galimi F. et al. MACC1 mRNA levels predict cancer recurrence after resection of colorectal cancer liver metastases. Ann Surg. 2013;257(6):1089–1095. doi: 10.1097/SLA.0b013e31828f96bc. [DOI] [PubMed] [Google Scholar]
- 36.Kawamura M, Saigusa S, Toiyama Y. et al. Correlation of MACC1 and MET expression in rectal cancer after neoadjuvant chemoradiotherapy. Anticancer Res. 2012;32(4):1527–1531. [PubMed] [Google Scholar]
- 37.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 38.Delanote V, Vandekerckhove J, Gettemans J. Plastins: versatile modulators of actin organization in (patho)physiological cellular processes. Acta Pharmacol Sin. 2005;26(7):769–779. doi: 10.1111/j.1745-7254.2005.00145.x. [DOI] [PubMed] [Google Scholar]
- 39.Yokobori T, Iinuma H, Shimamura T. et al. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res. 2013;73(7):2059–2069. doi: 10.1158/0008-5472.CAN-12-0326. [DOI] [PubMed] [Google Scholar]
- 40.Zougman A, Hutchins GG, Cairns DA. et al. Retinoic acid-induced protein 3: identification and characterisation of a novel prognostic colon cancer biomarker. Eur J Cancer. 2013;49(2):531–539. doi: 10.1016/j.ejca.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 41.Schepeler T, Reinert JT, Ostenfeld MS. et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68(15):6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 42.Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42(8):1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 43.Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012;36(1):e61–67. doi: 10.1016/j.canep.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Kuo TY, Hsi E, Yang IP, Tsai PC, Wang JY, Juo SH. Computational analysis of mRNA expression profiles identifies microRNA-29a/c as predictor of colorectal cancer early recurrence. PLoS One. 2012;7(2):e31587. doi: 10.1371/journal.pone.0031587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang IP, Tsai HL, Hou MF. et al. MicroRNA-93 inhibits tumor growth and early relapse of human colorectal cancer by affecting genes involved in the cell cycle. Carcinogenesis. 2012;33(8):1522–1530. doi: 10.1093/carcin/bgs166. [DOI] [PubMed] [Google Scholar]
- 46.O'Connell MJ, Lavery I, Yothers G. et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28(25):3937–3944. doi: 10.1200/JCO.2010.28.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gray RG, Quirke P, Handley K. et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29(35):4611–4619. doi: 10.1200/JCO.2010.32.8732. [DOI] [PubMed] [Google Scholar]
- 48.Venook AP, Niedzwiecki D, Lopatin M. et al. Biologic determinants of tumor recurrence in stage II colon cancer: validation study of the 12-gene recurrence score in cancer and leukemia group B (CALGB) 9581. J Clin Oncol. 2013;31(14):1775–1781. doi: 10.1200/JCO.2012.45.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trivedi PJ, Braden B. Indications, stains and techniques in chromoendoscopy. QJM. 2013;106(2):117–131. doi: 10.1093/qjmed/hcs186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Rijn JC, Reitsma JB, Stoker J, Bossuyt PM, van Deventer SJ, Dekker E. Polyp miss rate determined by tandem colonoscopy: a systematic review. Am J Gastroenterol. 2006;101(2):343–350. doi: 10.1111/j.1572-0241.2006.00390.x. [DOI] [PubMed] [Google Scholar]
- 51.dos Santos CE, Lima JC, Lopes CV. et al. Computerized virtual chromoendoscopy versus indigo carmine chromoendoscopy combined with magnification for diagnosis of small colorectal lesions: a randomized and prospective study. Eur J Gastroenterol Hepatol. 2010;22(11):1364–1371. doi: 10.1097/MEG.0b013e32833a5d63. [DOI] [PubMed] [Google Scholar]
- 52.Aminalai A, Rosch T, Aschenbeck J. et al. Live image processing does not increase adenoma detection rate during colonoscopy: a randomized comparison between FICE and conventional imaging (Berlin Colonoscopy Project 5, BECOP-5) Am J Gastroenterol. 2010;105(11):2383–2388. doi: 10.1038/ajg.2010.273. [DOI] [PubMed] [Google Scholar]
- 53.Curvers WL, van Vilsteren FG, Baak LC. et al. Endoscopic trimodal imaging versus standard video endoscopy for detection of early Barrett's neoplasia: a multicenter, randomized, crossover study in general practice. Gastrointest Endosc. 2011;73(2):195–203. doi: 10.1016/j.gie.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 54.Kuiper T, van den Broek FJ, Naber AH. et al. Endoscopic trimodal imaging detects colonic neoplasia as well as standard video endoscopy. Gastroenterology. 2011;140(7):1887–1894. doi: 10.1053/j.gastro.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 55.Autofluorescence Imaging for Diagnosing Intestinal Disorders, Colonoscopy; Prof Paul Miskovitz (Ed), ISBN: 978-953-307-568-6, InTech. Fujiya M, Moriichi K, Ueno N, Saitoh Y, Kohgo Y; http://www.intechopen.com/books/colonoscopy/autofluorescence-imaging-for-diagnosing-intestinal-disorders. [Google Scholar]
- 56.Sanduleanu S, Driessen A, Gomez-Garcia E, Hameeteman W, de Bruine A, Masclee A. In vivo diagnosis and classification of colorectal neoplasia by chromoendoscopy-guided confocal laser endomicroscopy. Clin Gastroenterol Hepatol. 2010;8(4):371–378. doi: 10.1016/j.cgh.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 57.Kiesslich R, Goetz M, Lammersdorf K. et al. Chromoscopy-guided endomicroscopy increases the diagnostic yield of intraepithelial neoplasia in ulcerative colitis. Gastroenterology. 2007;132(3):874–882. doi: 10.1053/j.gastro.2007.01.048. [DOI] [PubMed] [Google Scholar]