Abstract
The genus Mycobacterium is comprised of more than 150 species that reside in a wide variety of habitats. Most mycobacteria are environmental organisms that are either not associated with disease or are opportunistic pathogens that cause non-transmissible disease in immunocompromised individuals. In contrast, a small number of species, such as the tubercle bacillus, Mycobacterium tuberculosis, are host-adapted pathogens for which there is no known environmental reservoir. In recent years, gene disruption studies using the host-adapted pathogen have uncovered a number of “virulence factors,” yet genomic data indicate that many of these elements are present in non-pathogenic mycobacteria. This suggests that much of the genetic make-up that enables virulence in the host-adapted pathogen is already present in environmental members of the genus. In addition to these generic factors, we hypothesize that molecules elaborated exclusively by professional pathogens may be particularly implicated in the ability of M. tuberculosis to infect, persist, and cause transmissible pathology in its host species, Homo sapiens. One approach to identify these molecules is to employ comparative analysis of mycobacterial genomes, to define evolutionary events such as horizontal gene transfer (HGT) that contributed M. tuberculosis-specific genetic elements. Independent studies have now revealed the presence of HGT genes in the M. tuberculosis genome and their role in the pathogenesis of disease is the subject of ongoing investigations. Here we review these studies, focusing on the hypothesized role played by HGT loci in the emergence of M. tuberculosis from a related environmental species into a highly specialized human-adapted pathogen.
Keywords: Mycobacterium tuberculosis, Mycobacterium kansasii, M. tuberculosis-specific genes, horizontal gene transfer, comparative genomics
INTRODUCTION
Through modification of their genome content, bacteria can evolve to exploit different ecological niches. While vertical events such as gene duplication, chromosomal rearrangement and gene decay can affect the shape and structure of a genome (Ventura et al., 2007), horizontal gene transfer (HGT) is an important mechanism for bacteria to acquire novel genetic material into their genomes (Lercher and Pal, 2008; Price et al., 2008), subsequently facilitating adaptation and diversification (Treangen and Rocha, 2011). HGT can be mediated by transformation (acquisition of naked DNA), transduction (DNA transfer via a bacteriophage), and conjugation (fusion of two bacterial cells enabling a unidirectional transfer of a plasmid or mobile element; Frost et al., 2005). HGT has been shown to profoundly impact the prokaryotic genome plasticity, allowing the acquisition of antibiotic resistance elements (Mohd-Zain et al., 2004; Palmer et al., 2010; Gray et al., 2013), virulence genes (Rosas-Magallanes et al., 2006; Li et al., 2012) and new metabolic pathways (Wilson et al., 2003; Baldwin et al., 2004; Chouikha et al., 2006; Noda-Garcia et al., 2013).
Mycobacterium, a genus of Actinobacteria, comprises mostly non-pathogenic species. Exceptionally, this genus contains a number of host-adapted pathogens, including the leprosy bacillus, Mycobacterium leprae, and the Johne’s bacillus, M. avium subspecies paratuberculosis, the latter defined by the presence of at least six genomic islands that were likely acquired by HGT (Alexander et al., 2009). In this review, we focus on the Mycobacterium tuberculosis complex (MTBC), agents of tuberculosis (TB) in their respective mammalian hosts. Among the various subspecies of the MTBC, M. tuberculosis sensu stricto is the cause of human TB, which infects over 2 billion people and causes an estimated 1.3 million deaths annually (World Health Organization, 2013).
When contrasting the genome of MTBC organisms with the most closely related environmental mycobacteria, M. marinum and M. kansasii, independent studies have identified M. tuberculosis-specific genetic factors putatively acquired by HGT (Becq et al., 2007; Veyrier et al., 2009; Supply et al., 2013), evidenced by the presence of clustering, vehicles of HGT (phage, transposons, toxin-antitoxin genes) and an aberrant GC content in their DNA (Zaneveld et al., 2008). For example, the M. tuberculosis genome codes for 55 proteins absent from M. kansasii, M. marinum and all other sequenced mycobacterial genomes (Veyrier et al., 2009). As 87% of these M. tuberculosis-specific genes are found in clusters, it has been postulated that these clusters may be pathogenicity islands that contribute to the unique virulence of M. tuberculosis (Hacker et al., 1997; Veyrier et al., 2009). As several of these M. tuberculosis-specific genes have been linked to host adaptation (Sassetti and Rubin, 2003; Pethe et al., 2004), this provides further support for the notion that HGT may have played a crucial role in the emergence of this pathogen. At the ecological level, M. tuberculosis uses humans as its sole known reservoir while environmental mycobacteria such as M. kansasii can be found in various aquatic habitats (McSwiggan and Collins, 1974; Steadham, 1980; Sartori et al., 2013; Thomson et al., 2013), further highlighting the impact of genome remodeling on bacterial biology.
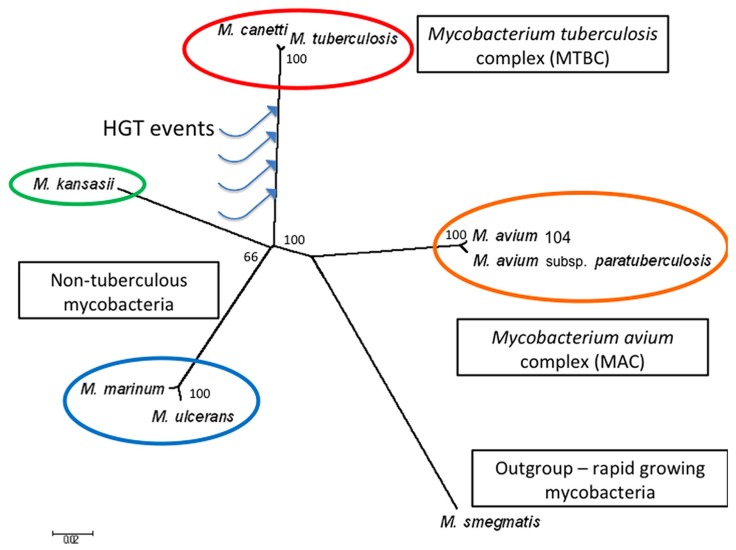
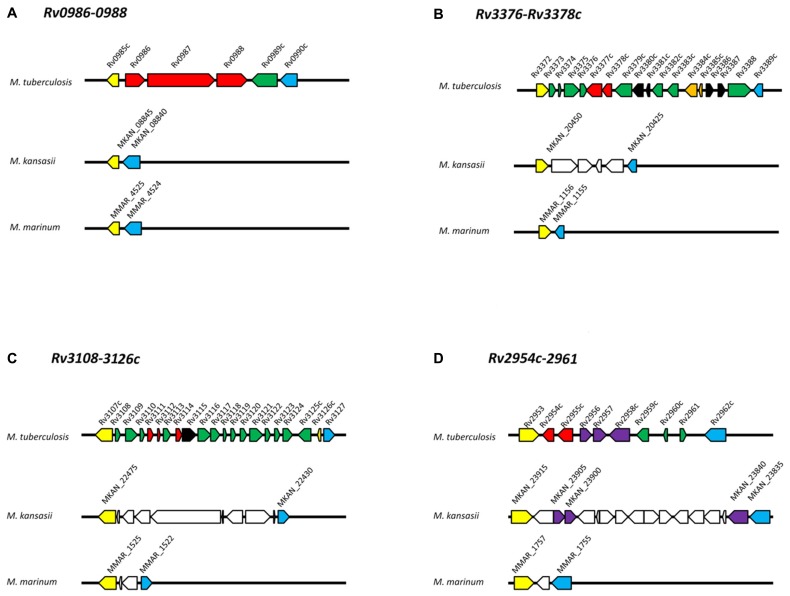
In this review, we briefly describe the early interplay between M. tuberculosis and the host during an infection, followed by bioinformatic data supporting the evidence for HGT and its potential contribution to the host-adapted lifestyle of this pathogen. To illustrate the relationships between various mycobacteria including the species discussed in this manuscript, in Figure 1 we present an un-rooted phylogenic tree based on 20 randomly selected genes conserved across eight mycobacteria, including seven slow-growing species (M. tuberculosis, M. canetti, M. kansasii, M. marinum, M. ulcerans, M. avium subsp. hominssuis, M. avium subsp. paratuberculosis) and a rapid-growing species (M. smegmatis) as the out-group. It is worth noting that the topology of this independently generated tree is congruent with the tree built from housekeeping genes (Veyrier et al., 2009), providing additional support for the evolutionary relationships between these species. The genes used for tree generation are provided in the figure legend. The organization of each M. tuberculosis-specific locus discussed is illustrated in Figure 2, in comparison with the flanking genomic regions in the related organisms M. kansasii and M. marinum.
FIGURE 1.
Phylogeny of M. tuberculosis and closely related Mycobacterium species. The un-rooted phylogenetic tree was generated by MEGA6.0 using 20 randomly selected genes conserved across eight Mycobacterium species (Schwab et al., 2009). The blue arrows schematically represent where putative HGT events may have occurred, resulting in M. tuberculosis-specific genomic islands. The scale bar indicates 0.02 substitutions per nucleotide position, and the bootstrap values calculated using the neighbor-joining method (expressed as a percentage of 1000 replicates) are shown at the branch points. The fast growing species M. smegmatis is used as the out-group. Genes used are listed below (represented as M. tuberculosis genes): Rv0001-dnaA, Rv0041-leuS, Rv0236A–Rv0236A, Rv0248c–Rv0248c, Rv0285-PE5, Rv0287-esxG, Rv0288-esxH, Rv1085c–Rv1085c, Rv0197–Rv0197, Rv1304-atpB, Rv1305-atpE, Rv1894c–Rv1894c, Rv2172c–Rv2172c, Rv2392-cysH, Rv2440c-obg, Rv2477c–Rv2477c, Rv3019c-esxR, Rv3045-adhC, Rv3392c-cmaA1, Rv3502c- hsd4A.
FIGURE 2.
Genomic organization of operons in M. tuberculosis, M. kansasii, and M. marinum. (A) Rv0986-0988; (B) Rv3376-3378c; (C) Rv3108-3126c; (D) Rv2954c-2961. Protein-coding genes are represented by arrows and orthologous genes are indicated by arrows of the same color. Yellow and blue arrows mark the “boundary” of each M. tuberculosis-specific locus. Red arrows indicate genes discussed in this review. Dark green arrows indicate M. tuberculosis genes with no orthologs within the corresponding M. kansasii and M. marinum genomic regions. White arrows in M. kansasii and M. marinum genomes indicate genes present but not orthologs to M. tuberculosis genes. Black arrows indicate transposases; orange arrows indicate toxin-antitoxin genes. Genome organizations for M. tuberculosis, M. kansasii, M. marinum and gene clusters were obtained from the Kyoto Encyclopedia of Genes and Genomes (http://www.kegg.jp/) based on databases available at the Sanger Institute, Tuberculist, and McGill University, respectively. Orthologs were verified by comparing each predicted protein against the H37Rv genome using the BLAST program. Only proteins with >50% coverage, 60% identity and E-value <e-20 were used.
M. tuberculosis AND THE HOST ENVIRONMENT
When M. tuberculosis enters the pulmonary alveoli via the aerosol route, it is thought to first encounter alveolar macrophages. Following phagocytosis by these macrophages, the bacterium finds itself in the phagosomal compartment which is, among other attributes, iron limiting, carbon poor, hypoxic, nitrosative, and oxidative (Schnappinger et al., 2003). M. tuberculosis is able to resist these bactericidal effects by synthesizing antioxidants, repairing DNA and proteins, maintaining intracellular pH and cell wall integrity (Buchmeier et al., 2000; Master et al., 2002; Boshoff et al., 2003; Darwin and Nathan, 2005; Vandal et al., 2008, 2009; Colangeli et al., 2009). Moreover, M. tuberculosis can cope with hypoxic and other growth-limiting environments using a number of tactics such as activating the dormancy regulon, promoting alternate metabolic pathways and iron metabolism (Leistikow et al., 2010; Marrero et al., 2010; Ryndak et al., 2010; Griffin et al., 2011). In addition, M. tuberculosis is able to prevent the fusion of phago-lysosome (Sun et al., 2010; Wong et al., 2011), permeabilize the phagosomal membrane (Manzanillo et al., 2012), and escape into the cytosol. The cytosol likely offers a less hostile, thus more permissive environment, where the bacteria can replicate and induce the infected macrophages to undergo necrosis instead of apoptosis, a strategy that allows the bacteria to infect neighboring cells, thereby enabling the perpetuation of the infection process (van der Wel et al., 2007; Divangahi et al., 2009; Behar et al., 2010).
Rv0986–0988
In the seminal work that first described evidence of HGT in the M. tuberculsosis genome, Becq et al. (2007) detected a 5.6 kb M. tuberculosis-specific Island with a reduced GC content (53%) compared to the average for the M. tuberculosis genome (65.6%; Cole et al., 1998). Further molecular and in silico analyses demonstrated that this operon is present in other members of the MTBC, including M. bovis, M. africanum, and M. microti (Rosas-Magallanes et al., 2006). Based on phylogenetic analyses, it was proposed that three genes within this locus, Rv0986–8, had been acquired from phylogenetically distant γ-proteobacteria via plasmid transfer. The fact that the orthologs of these three genes are consistently together suggest that one single HGT event occurred during the acquisition of this operon by the ancestor of M. tuberculosis (Rosas-Magallanes et al., 2006).
Rv0986 is predicted to encode an adenosine triphosphate (ATP)-binding protein that is orthologous to the Agrobacterium tumefaciens attE polypeptide, and form an ABC transporter with Rv0987 (Braibant et al., 2000; Rosas-Magallanes et al., 2006). The A. tumefaciens attE gene is located on a plasmid which harbors the attE-H operon. This operon has been proposed to encode an ABC transporter that secretes a host cell adhesion factor (Matthysse et al., 1996, 2000). Intriguingly, the N- and C- terminal sequences of Rv0987 share 40% similarity with attF and attG, and the neighboring Rv0988 shows 50% similarity with attH (Rosas-Magallanes et al., 2006).
The Rv0986–8 operon has been implicated in M. tuberculosis virulence as mutants with disruption in Rv0986 and 0987 exhibit reduced ability to inhibit phagosome acidification in macrophages (Pethe et al., 2004). Furthermore, these mutants had impaired binding to host cells, and this phenotype could be rescued by complementing with a cosmid carrying M. tuberculosis DNA encompassing the Rv0986–8 operon (Rosas-Magallanes et al., 2007). Although Rv0986 and Rv0987 mutants were not shown to be attenuated in mouse lungs and spleens (Rosas-Magallanes et al., 2007), the Rv0986 mutant was subsequently shown to be less virulent in the context of central nervous system infection (Be et al., 2008). Recently Rv0986–8 have been found to be regulated by EspR, a transcription factor that also regulates the ESX-1 secretion system (Blasco et al., 2012), a major virulence mediator of M. tuberculosis (Brodin et al., 2006).
Rv3376–Rv3378c
Another genomic island potentially acquired by HGT is the 3.1 kb region encompassing Rv3376–8c (Becq et al., 2007; Veyrier et al., 2009). This island exhibits a reduced GC content (54.7%) and is associated with the presence of transposases, known to mediate HGT events (Becq et al., 2007; Veyrier et al., 2009). The closest genera harboring such genes are Agrobacterium and Rhizobium (Becq et al., 2007). More recently, Mann and Peters, (2012) speculated that, while Rv3377c amino acid sequence shares homology with proteins from another actinomycete, Micromonospora, Rv3378c has no ortholog in any other organism with the exception of a hypothetical protein in amoeba. These observations suggest that these MTBC-specific genes originated in different sources (Mann and Peters, 2012).
Biochemical characterization has revelated that Rv3377c and Rv3378c encode a halimadienyl diphosphate (HPP) synthase and a diterpene synthase, respectively. In a step-wise fashion, these enzymes catalyze the cyclization of the precursor, geranylgeranyl diphosphate (GGPP), and the hydrolysis of the HPP intermediate to produce isotuberculosinol (isoTB), a diterpene species (Nakano et al., 2005; Mann et al., 2009a, b). Terpenes are one of the most widespread and chemically diverse compounds found in nature. They are hydrocarbons made up of five, or multiples of five, carbon units (Simion, 2005). In plants and fungi, they are commonly found as essential as well as secondary metabolites involved in signaling and defense (Buckingham, 1994). While members of the Actinobcteria group synthesize a plethora of natural products (Baltz, 2008), very few are known to encode diterpene (C20) synthases (Dairi et al., 2001; Hamano et al., 2002; Dürr et al., 2006; Smanski et al., 2011).
During macrophage infection, M. tuberculosis mutants with disruption in either Rv3377c or Rv3378c showed marked defect in arresting phagosome acidification as well as intracellular survival, suggesting that these genes are involved in the modulation of early infection process (Pethe et al., 2004). Intriguingly, these genes are not transcriptionally altered during macrophage infection (Stewart et al., 2005; Rohde et al., 2007; Waddell and Butcher, 2007), implying that the synthesis of isoTB is regulated at the protein level, potentially triggered by ambient magnesium levels (Mann et al., 2009a, 2011). In addition, isoTB has shown to inhibit phagosome acidification by 0.5 pH units as well as proteolytic activity (Mann et al., 2009b). Recently Rv3378c has been characterized as a tuberculosinyl transferase that converts isoTB into the proposed end product, tuberculosinyladenosine (TbAd; Layre et al., 2014). The cellular mechanism by which Rv3377–8c modifies phagosome function thus remains to be further investigated.
Rv3108–3126c
Rv3108–3126c is a 15.1 kb, MTBC-specific genomic island that has a reduced GC content (56.7%) and contains genes encoding insertion sequences and a transposase, features typical of HGT (Becq et al., 2007). The proposed donor species include Burkholderia, Corynebacterium, and Pseudomonas. This island contains two potential virulence genes; Rv3111 and Rv3114.
A transposon mutant of Rv3111 (moaC1) was first shown to be attenuated in replicating in macrophages (Rosas-Magallanes et al., 2007). In a more recent high-throughput genetic screen, mutants disrupted for the genes moaC1/moaD1(Rv3112), implicated in molybdenum cofactor biosynthesis, were found to be trafficked to acidified intracellular compartments rapidly (Brodin et al., 2010), potentially providing an explanation for the impaired intracellular growth. Molybdopterin is the main building block of the molybdenum cofactor (MoCo) and can be found in enzymes that catalyze redox reactions in carbon, nitrogen, and sulfur utilization (Williams et al., 2011). moeB1, a homologous gene potentially involved in MoCo biosynthesis, has also been shown to be required for arresting phagosome acidification (MacGurn and Cox, 2007). moaC1 mutant itself has exhibited reduced virulence in macrophages as well as primate lungs (Rosas-Magallanes et al., 2007; Dutta et al., 2010). Further investigation of how MoCo-mediated redox reactions alter the intraphagosomal environment should provide more insight on the cellular processes employed by M. tuberculosis to adapt to the mammalian host environment.
Rv3114 has been shown to be required for M. tuberculosis persistence in the mouse spleen (Sassetti and Rubin, 2003) and is temporally regulated during infection (Talaat et al., 2004). It encodes a putative nucleoside deaminase involved in nucleotide metabolism (Akhter et al., 2008).
Rv2954c–2961
Rv2954c–2961 make-up a 7 kb genomic island with a low GC content (53.6%) and a transposase gene (Becq et al., 2007). A phylogenetic analysis of multiple mycobacterial genome sequences proposed a step-wise acquisition of the genes within this locus. Specifically, some genes are present in slow-growing, but not rapid-growing mycobacteria, suggesting that they were acquired by the common ancestor of the slow-growing species. Conversely, other genes in this island are specific to M. tuberculosis and are therefore inferred to have been acquired after the common ancestor with M. kansasii and M. marinum (Veyrier et al., 2009).
Genes within this island are involved in the synthesis and modification of phenolic glycolipids (PGLs), complex lipids located in the outermost layer of the mycobacterial cell envelope. PGLs are composed of long-chain fatty acid backbones with a phenol ring and methylated sugars, including two rhamnosyl and a terminal fucosyl residue (Onwueme et al., 2005). PGLs are only produced by the members of the MTBC and related slow-growing mycobacteria, yet even among the mycobacteria that make PGL; there are species-specific modifications in the carbohydrate moiety [Daffe and Laneelle, 1988; Onwueme et al., 2005; schematically illustrated in (Veyrier et al., 2011)]. The PGLs have been implicated in mycobacterial pathogenicity such as oxidative stress resistance (Chan et al., 1989), cell tropism (Ng et al., 2000; Rambukkana, 2001), and immunomodulation (Reed et al., 2004; Guenin-Mace et al., 2009; Cambier et al., 2014).
The major type of PGLs produced by M. tuberculosis is denoted PGL-tb (Simeone et al., 2007). While most genes involved in the synthesis of the lipid core and carbohydrates are characterized, the enzymes responsible for O-methylating the fucosyl residue remained elusive until recently. It is now known that Rv2954c–Rv2956 code for the methyltransferases that are responsible for O-methylation of the terminal fucosyl residue (Veyrier et al., 2009; Simeone et al., 2013). These three proteins catalyze the O-methylation of the hydroxyl groups of the terminal fucosyl residue of PGL-tb in a sequential process (Simeone et al., 2013).
In other pathogenic mycobacteria that lack the Rv2954c, Rv2955c, and Rv2956 orthologs such as M. marinum and M. leprae, their PGLs do not contain the terminal O-methylated fucosyl residue (Onwueme et al., 2005). Although M. kansasii does not possess Rv2954c or 2955c, it does encode an enzyme that is highly similar to Rv2956 (84%), and its PGL contains four sugar residues (Riviere et al., 1987; Onwueme et al., 2005). Interestingly, Rv2954c and Rv2955c have been found to be virulence genes during macrophage infection (Rosas-Magallanes et al., 2007), and Rv2954c was induced upon exposure to lung surfactant (Schwab et al., 2009).
As the enzymes encoded within this island have been observed to catalyze the transfer of functional groups from one molecule to another, they may play an important role at “decorating” existing mycobacterial products and fine-tuning host responses toward the organism to optimize its intracellular survival (Veyrier et al., 2011).
CONCLUSION
Phylogenetic analyses have been used as robust and reliable tools for identifying potential HGT loci in M. tuberculosis and other pathogenic mycobacteria. However, the biological relevance of most of these genomic regions remains to be delineated. In this review we examine four examples of how such putative HGT genes can affect the physiology of the pathogen and its interaction with the host. The functional characterization of these and other putative HGT-associated genes will allow us to understand whether and how HGT events have contributed to the pathogenesis of M. tuberculosis, ultimately guiding the development of new diagnostic tests and vaccines against this particularly successful pathogen.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Joyce Wang is supported by a studentship award from the Fonds de la Recherche en Sante du Quebec (FRSQ). Marcel A. Behr is a Chercheur-National of the FRSQ and a William Dawson Scholar of McGill University. Mycobacterial genomic work in the Behr lab is funded by an operating grant from the Canadian Institutes for Health Research, MOP-97813.
REFERENCES
- Akhter Y., Yellaboina S., Farhana A., Ranjan A., Ahmed N., Hasnain S. E. (2008). Genome scale portrait of cAMP-receptor protein (CRP) regulons in mycobacteria points to their role in pathogenesis. Gene 407 148–158 10.1016/j.gene.2007.10.017 [DOI] [PubMed] [Google Scholar]
- Alexander D. C., Turenne C. Y., Behr M. A. (2009). Insertion and deletion events that define the pathogen Mycobacterium avium subsp paratuberculosis. J. Bacteriol. 191 1018–1025 10.1128/JB.01340-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A., Sokol P. A., Parkhill J., Mahenthiralingam E. (2004). The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72 1537–1547 10.1128/IAI.72.3.1537-1547.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R. H. (2008). Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 8 557–563 10.1016/j.coph.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Be N. A., Lamichhane G., Grosset J., Tyagi S., Cheng Q. J., Kim K. S., et al. (2008). Murine model to study the invasion and survival of Mycobacterium tuberculosis in the central nervous system. J. Infect. Dis. 198 1520–1528 10.1086/592447 [DOI] [PubMed] [Google Scholar]
- Becq J., Gutierrez M. C., Rosas-Magallanes V., Rauzier J., Gicquel B., Neyrolles O., et al. (2007). Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol. Biol. Evol. 24 1861–1871 10.1093/molbev/msm111 [DOI] [PubMed] [Google Scholar]
- Behar S. M., Divangahi M., Remold H. G. (2010). Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat. Rev. Microbiol. 8 668–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco B., Chen J. M., Hartkoorn R., Sala C., Uplekar S., Rougemont J., et al. (2012). Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog. 8:e1002621 10.1371/journal.ppat.1002621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff H. I., Reed M. B., Barry C. E., III, Mizrahi V. (2003). DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113 183–193 10.1016/S0092-8674(03)00270-8 [DOI] [PubMed] [Google Scholar]
- Braibant M., Gilot P., Content J. (2000). The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 24 449–467 10.1111/j.1574-6976.2000.tb00550.x [DOI] [PubMed] [Google Scholar]
- Brodin P., Majlessi L., Marsollier L., de Jonge M. I., Bottai D., Demangel C., et al. (2006). Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect. Immun. 74 88–98 10.1128/IAI.74.1.88-98.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P., Poquet Y., Levillain F., Peguillet I., Larrouy-Maumus G., Gilleron M., et al. (2010). High content phenotypic cell-based visual screen identifies Mycobacterium tuberculosis acyltrehalose-containing glycolipids involved in phagosome remodeling. PLoS Pathog. 6:e1001100 10.1371/journal.ppat.1001100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier N., Blanc-Potard A., Ehrt S., Piddington D., Riley L., Groisman E. A. (2000). A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol. Microbiol. 35 1375–1382 10.1046/j.1365-2958.2000.01797.x [DOI] [PubMed] [Google Scholar]
- Buckingham J. (1994). Dictionary of Natural Products Vol. 1–7 London: Chapman & Hall [Google Scholar]
- Cambier C. J., Takaki K. K., Larson R. P., Hernandez R. E., Tobin D. M., Urdahl K. B., et al. (2014). Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505 218–222 10.1038/nature12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Fujiwara T., Brennan P., McNeil M., Turco S. J., Sibille J. C., et al. (1989). Microbial glycolipids: possible virulence factors that scavenge oxygen radicals. Proc. Natl. Acad. Sci. U.S.A. 86 2453–2457 10.1073/pnas.86.7.2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouikha I., Germon P., Brée A., Gilot P., Moulin-Schouleur M., Schouler C. (2006). A selC-associated genomic island of the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is involved in carbohydrate uptake and virulence. J. Bacteriol. 188 977–987 10.1128/JB.188.3.977-987.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangeli R., Haq A., Arcus V. L., Summers E., Magliozzo R. S., McBride A., et al. (2009). The multifunctional histone-like protein Lsr2 protects mycobacteria against reactive oxygen intermediates. Proc. Natl. Acad. Sci. U.S.A. 106 4414–4418 10.1073/pnas.0810126106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393 537–544 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- Daffe M., Laneelle M. A. (1988). Distribution of phthiocerol diester, phenolic mycosides and related compounds in mycobacteria. J. Gen. Microbiol. 134 2049–2055 [DOI] [PubMed] [Google Scholar]
- Dairi T., Hamano Y., Kuzuyama T., Itoh N., Furihata K., Seto H. (2001). Eubacterial diterpene cyclase genes essential for production of the isoprenoid antibiotic terpentecin. J. Bacteriol. 183 6085–6094 10.1128/JB.183.20.6085-6094.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin K. H., Nathan C. F. (2005). Role for nucleotide excision repair in virulence of Mycobacterium tuberculosis. Infect. Immun. 73 4581–4587 10.1128/IAI.73.8.4581-4587.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divangahi M., Chen M., Gan H., Desjardins D., Hickman T. T., Lee D. M., et al. (2009). Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat. Immunol. 10 899–906 10.1038/ni.1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr C., Schnell H. J., Luzhetskyy A., Murillo R., Weber M., Welzel K., et al. (2006). Biosynthesis of the terpene phenalinolactone in Streptomyces sp. Tu6071: analysis of the gene cluster and generation of derivatives. Chem. Biol. 13 365–377 10.1016/j.chembiol.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Dutta N. K., Mehra S., Didier P. J., Roy C. J., Doyle L. A., Alvarez X., et al. (2010). Genetic requirements for the survival of tubercle bacilli in primates. J. Infect. Dis. 201 1743–1752 10.1086/652497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Leplae R., Summers A. O., Toussaint A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3 722–732 10.1038/nrmicro1235 [DOI] [PubMed] [Google Scholar]
- Gray T. A., Krywy J. A., Harold J., Palumbo M. J., Derbyshire K. M. (2013). Distributive conjugal transfer in mycobacteria generates progeny with meiotic-like genome-wide mosaicism, allowing mapping of a mating identity locus. PLoS Biol. 11:e1001602 10.1371/journal.pbio.1001602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J. E., Gawronski J. D., Dejesus M. A., Ioerger T. R., Akerley B. J., Sassetti C. M. (2011). High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7:e1002251 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenin-Mace L., Simeone R., Demangel C. (2009). Lipids of pathogenic mycobacteria: contributions to virulence and host immune suppression. Transbound. Emerg. Dis. 56 255–268 10.1111/j.1865-1682.2009.01072.x [DOI] [PubMed] [Google Scholar]
- Hacker J., Blum-Oehler G., Mühldorfer I, Tschäpe H. (1997). Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23 1089–1097 10.1046/j.1365-2958.1997.3101672.x [DOI] [PubMed] [Google Scholar]
- Hamano Y., Kuzuyama T., Itoh N., Furihata K., Seto H., Dairi T. (2002). Functional analysis of eubacterial diterpene cyclases responsible for biosynthesis of a diterpene antibiotic, terpentecin. J. Biol. Chem. 277 37098–37104 10.1074/jbc.M206382200 [DOI] [PubMed] [Google Scholar]
- Layre E., Lee H. J., Young D. C., Jezek Martinot A., Buter J., Minnaard A. J., et al. (2014). Molecular profiling of Mycobacterium tuberculosis identifies tuberculosinyl nucleoside products of the virulence-associated enzyme Rv3378c. Proc. Natl. Acad. Sci. U.S.A. 111 2978–2983 10.1073/pnas.1315883111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leistikow R. L., Morton R. A., Bartek I. L., Frimpong I., Wagner K., Voskuil M. I. (2010). The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J. Bacteriol. 192 1662–1670 10.1128/JB.00926-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher M. J., Pal C. (2008). Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol. Biol. Evol. 25 559–567 10.1093/molbev/msm283 [DOI] [PubMed] [Google Scholar]
- Li M., Du X., Villaruz A. E., Diep B. A., Wang D., Song Y., et al. (2012). MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat. Med. 18 816–819 10.1038/nm.2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn J. A., Cox J. S. (2007). A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect. Immun. 75 2668–2678 10.1128/IAI.01872-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M., Peters R. J. (2012). Isotuberculosinol: the unusual case of an immunomodulatory diterpenoid from Mycobacterium tuberculosis. Medchemcomm 3 899–904 10.1039/c2md20030a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M., Prisic S., Hu H., Xu M., Coates R. M., Peters R. J. (2009a). Characterization and inhibition of a class II diterpene cyclase from Mycobacterium tuberculosis: implications for tuberculosis. J. Biol. Chem. 284 23574–23579 10.1074/jbc.M109.023788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M., Xu M., Chen X., Fulton D. B., Russell D. G., Peters R. J. (2009b). Edaxadiene: a new bioactive diterpene from Mycobacterium tuberculosis. J. Am. Chem. Soc. 131 17526–17527 10.1021/ja9019287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M., VanderVen B. C., Peters R. J. (2011). Magnesium depletion triggers production of an immune modulating diterpenoid in Mycobacterium tuberculosis. Mol. Microbiol. 79 1594–1601 10.1111/j.1365-2958.2011.07545.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo P. S., Shiloh M. U., Portnoy D. A., Cox J. S. (2012). Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11 469–480 10.1016/j.chom.2012.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero J., Rhee K. Y., Schnappinger D., Pethe K., Ehrt S. (2010). Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U.S.A. 107 9819–9824 10.1073/pnas.1000715107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Master S. S., Springer B., Sander P., Boettger E. C., Deretic V., Timmins G. S. (2002). Oxidative stress response genes in Mycobacterium tuberculosis: role of ahpC in resistance to peroxynitrite and stage-specific survival in macrophages. Microbiology 148(Pt 10) 3139–3144 [DOI] [PubMed] [Google Scholar]
- Matthysse A. G., Yarnall H. A., Young N. (1996). Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J. Bacteriol. 178 5302–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse A. G., Yarnall H., Boles S. B., McMahan S. (2000). A region of the Agrobacterium tumefaciens chromosome containing genes required for virulence and attachment to host cells. Biochim. Biophys. Acta 1490 208–212 10.1016/S0167-4781(99)00250-X [DOI] [PubMed] [Google Scholar]
- McSwiggan D. A., Collins C. H. (1974). The isolation of M. kansasii and M. xenopi from water systems. Tubercle 55 291–297 10.1016/0041-3879(74)90038-5 [DOI] [PubMed] [Google Scholar]
- Mohd-Zain Z., Turner S. L., Cerdeño-Tárraga A. M., Lilley A. K., Inzana T. J., Duncan A. J., et al. (2004). Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 186 8114–8122 10.1128/JB.186.23.8114-8122.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano C., Okamura T., Sato T., Dairi T., Hoshino T. (2005). Mycobacterium tuberculosis H37Rv3377c encodes the diterpene cyclase for producing the halimane skeleton. Chem. Commun. (Camb.) 1016–1018 10.1039/B415346D [DOI] [PubMed] [Google Scholar]
- Ng V., Zanazzi G., Timpl R., Talts J. F., Salzer J. L., Brennan P. J., et al. (2000). Role of the cell wall phenolic glycolipid-1 in the peripheral nerve predilection of Mycobacterium leprae. Cell 103 511–524 10.1016/S0092-8674(00)00142-2 [DOI] [PubMed] [Google Scholar]
- Noda-Garcia L., Camacho-Zarco A. R., Medina-Ruïz S., Gaytán P., Carrillo-Tripp M., Fülöp V., et al. (2013). Evolution of substrate specificity in a recipient’s enzyme following horizontal gene transfer. Mol. Biol. Evol. 30 2024–2034 10.1093/molbev/mst115 [DOI] [PubMed] [Google Scholar]
- Onwueme K. C., Vos C. J., Zurita J., Ferreras J. A., Quadri L. E. (2005). The dimycocerosate ester polyketide virulence factors of mycobacteria. Prog. Lipid Res. 44 259–302 10.1016/j.plipres.2005.07.001 [DOI] [PubMed] [Google Scholar]
- Palmer K. L., Kos V. N., Gilmore M. S. (2010). Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 13 632–639 10.1016/j.mib.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethe K., Swenson D. L., Alonso S., Anderson J., Wang C., Russell D. G. (2004). Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc. Natl. Acad. Sci. U.S.A. 101 13642–13647 10.1073/pnas.0401657101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2008). Horizontal gene transfer and the evolution of transcriptional regulation in Escherichia coli. Genome Biol. 9:R4. 10.1186/gb-2008-9-1-r4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambukkana A. (2001). Molecular basis for the peripheral nerve predilection of Mycobacterium leprae. Curr. Opin. Microbiol. 4 21–27 10.1016/S1369-5274(00)00159-4 [DOI] [PubMed] [Google Scholar]
- Reed M. B., Domenech P., Manca C., Su H., Barczak A. K., Kreiswirth B. N., et al. (2004). A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431 84–87 10.1038/nature02837 [DOI] [PubMed] [Google Scholar]
- Riviere M., Fournie J. J., Puzo G. (1987). A novel mannose containing phenolic glycolipid from Mycobacterium kansasii. J. Biol. Chem. 262 14879–14884 [PubMed] [Google Scholar]
- Rohde K. H., Abramovitch R. B., Russell D. G. (2007). Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2 352–364 10.1016/j.chom.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Rosas-Magallanes V., Deschavanne P., Quintana-Murci L., Brosch R., Gicquel B., Neyrolles O. (2006). Horizontal transfer of a virulence operon to the ancestor of Mycobacterium tuberculosis. Mol. Biol. Evol. 23 1129–1135 10.1093/molbev/msj120 [DOI] [PubMed] [Google Scholar]
- Rosas-Magallanes V., Stadthagen-Gomez G., Rauzier J., Barreiro L. B., Tailleux L., Boudou F., et al. (2007). Signature-tagged transposon mutagenesis identifies novel Mycobacterium tuberculosis genes involved in the parasitism of human macrophages. Infect. Immun. 75 504–507 10.1128/IAI.00058-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryndak M. B., Wang S., Smith I., Rodriguez G. M. (2010). The Mycobacterium tuberculosis high-affinity iron importer, IrtA, contains an FAD-binding domain. J. Bacteriol. 192 861–869 10.1128/JB.00223-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori F. G., Leandro L. F., Montanari L. B., de Souza M. G., Pires R. H., Sato D. N., et al. (2013). Isolation and identification of environmental mycobacteria in the waters of a hemodialysis center. Curr. Microbiol. 67 107–111 10.1007/s00284-013-0341-6 [DOI] [PubMed] [Google Scholar]
- Sassetti C. M., Rubin E. J. (2003). Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U.S.A. 100 12989–12994 10.1073/pnas.2134250100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D., Ehrt S., Voskuil M. I., Liu Y., Mangan J. A., Monahan I. M., et al. (2003). Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198 693–704 10.1084/jem.20030846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab U., Rohde K. H., Wang Z., Chess P. R., Notter R. H., Russell D. G. (2009). Transcriptional responses of Mycobacterium tuberculosis to lung surfactant. Microb. Pathog. 46 185–193 10.1016/j.micpath.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone R., Constant P., Guilhot C., Daffé M., Chalut C. (2007). Identification of the missing trans-acting enoyl reductase required for phthiocerol dimycocerosate and phenolglycolipid biosynthesis in Mycobacterium tuberculosis. J. Bacteriol. 189 4597–4602 10.1128/JB.00169-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone R., Huet G., Constant P., Malaga W., Lemassu A., Laval F., et al. (2013). Functional characterisation of three o-methyltransferases involved in the biosynthesis of phenolglycolipids in Mycobacterium tuberculosis. PLoS ONE 8:e58954 10.1371/journal.pone.0058954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simion G. (2005). “Terpenoids,” in Encyclopedia of Chromatography 2nd Edn ed. Cazes J. CRC Press; 1676–1681 10.1201/NOE0824727857.ch368 [DOI] [Google Scholar]
- Smanski M. J., Yu Z., Casper J., Lin S., Peterson R. M., Chen Y., et al. (2011). Dedicated ent-kaurene and ent-atiserene synthases for platensimycin and platencin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 108 13498–13503 10.1073/pnas.1106919108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steadham J. E. (1980). High-catalase strains of Mycobacterium kansasii isolated from water in Texas. J. Clin. Microbiol. 11 496–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. R., Patel J., Robertson B. D., Rae A., Young D. B. (2005). Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 1:269–278 10.1371/journal.ppat.0010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Wang X., Lau A., Liao T. Y., Bucci C., Hmama Z. (2010). Mycobacterial nucleoside diphosphate kinase blocks phagosome maturation in murine RAW 264.7 macrophages. PLoS ONE 5:e8769 10.1371/journal.pone.0008769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P., Marceau M., Mangenot S., Roche D., Rouanet C., Khanna V., et al. (2013). Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat. Genet. 45 172–179 10.1038/ng.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaat A. M., Lyons R., Howard S. T., Johnston S. A. (2004). The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. U.S.A. 101 4602–4607 10.1073/pnas.0306023101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson R. M., Carter R., Tolson C., Coulter C., Huygens F., Hargreaves M. (2013). Factors associated with the isolation of Nontuberculous mycobacteria (NTM) from a large municipal water system in Brisbane, Australia. BMC Microbiol. 13:89 10.1186/1471-2180-13-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T. J, Rocha E. P. C. (2011). Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 7:e1001284 10.1371/journal.pgen.1001284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandal O. H., Pierini L. M., Schnappinger D., Nathan C. F., Ehrt S. (2008). A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat. Med. 14 849–854 10.1038/nm.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandal O. H., Roberts J. A., Odaira T., Schnappinger D., Nathan C. F., Ehrt S. (2009). Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J. Bacteriol. 191 625–631 10.1128/JB.00932-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel N., Hava D., Houben D., Fluitsma D., van Zon M., Pierson J., et al. (2007). M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129 1287–1298 10.1016/j.cell.2007.05.059 [DOI] [PubMed] [Google Scholar]
- Ventura M., Canchaya C., Tauch A., Chandra G., Fitzgerald G. F., Chater K. F., et al. (2007). Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylura. Microbiol. Mol. Biol. Rev. 71 495–548 10.1128/MMBR.00005-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrier F. J., Dufort A., Behr M. A. (2011). The rise and fall of the Mycobacterium tuberculosis genome. Trends Microbiol. 19 156–161 10.1016/j.tim.2010.12.008 [DOI] [PubMed] [Google Scholar]
- Veyrier F., Pletzer D., Turenne C., Behr M. A. (2009). Phylogenetic detection of horizontal gene transfer during the step-wise genesis of Mycobacterium tuberculosis. BMC Evol. Biol. 9:196 10.1186/1471-2148-9-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell S. J., Butcher P. D. (2007). Microarray analysis of whole genome expression of intracellular Mycobacterium tuberculosis. Curr. Mol. Med. 7 287–296 10.2174/156652407780598548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. J., Kana B. D., Mizrahi V. (2011). Functional analysis of molybdopterin biosynthesis in mycobacteria identifies a fused molybdopterin synthase in Mycobacterium tuberculosis. J. Bacteriol. 193 98–106 10.1128/JB.00774-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. S., Herrick J. B., Jeon C. O., Hinman D. E., Madsen E. L. (2003). Horizontal transfer of phnAc dioxygenase genes within one of two phenotypically and genotypically distinctive naphthalene-degrading guilds from adjacent soil environments. Appl. Environ. Microbiol. 69 2172–2181 10.1128/AEM.69.4.2172-2181.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D., Bach H., Sun J., Hmama Z., Av-Gay Y. (2011). Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc. Natl. Acad. Sci. U.S.A. 108 19371–19376 10.1073/pnas.1109201108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2013). Global Tuberculosis Report 2013. Geneva, Switzerland: WHO/HTM/TB/2013.11 [Google Scholar]
- Zaneveld J. R., Nemergut D. R., Knight R. (2008). Are all horizontal gene transfers created equal? Prospects for mechanism-based studies of HGT patterns. Microbiology 154 1–15 10.1099/mic.0.2007/011833-0 [DOI] [PubMed] [Google Scholar]