Abstract
Cystic fibrosis (CF) patients have chronic airway infection and frequent exposure to antibiotics, which often leads to the emergence of resistant organisms. Achromobacter xylosoxidans is a new emergent pathogen in CF spectrum. From 2005 to 2010 we had an outbreak in A. xylosoxidans prevalence in our CF center, thus, the present study was aimed at deeply investigating virulence traits of A. xylosoxidans strains isolated from infected CF patients. To this purpose, we assessed A. xylosoxidans genome variability by randomly amplified polymorphic DNA (RAPD), biofilm production, antibiotic resistances, and motility. All A. xylosoxidans strains resulted to be biofilm producers, and were resistant to antibiotics usually employed in CF treatment. Hodge Test showed the ability to produce carbapenemase in some strains. Strains who were resistant to β-lactamics antibiotics, showed the specific band related to metal β-lactamase (blaIMP-1), and some of them showed to possess the integron1. Around 81% of A. xylosoxidans strains were motile. Multivariate analysis showed that RAPD profiles were able to predict Forced Expiratory Volume (FEV1%) and biofilm classes. A significant prevalence of strong biofilm producers strains was found in CF patients with severely impaired lung functions (FEV1% class 1). The outbreak we had in our center (prevalence from 8.9 to 16%) could be explained by an enhanced adaptation of A. xylosoxidans in the nosocomial environment, despite of aggressive antibiotic regimens that CF patients usually undergo.
Keywords: Achromobacter xylosoxidans, Cystic fibrosis, antimicrobial resistance, motility, RAPD, biofilm
Introduction
In sputa of patients with Cystic fibrosis (CF), the Gram-negative Pseudomonas aeruginosa and the Gram-positive Staphylococcus aureus are the most frequently found pathogens. Recently, new bacterial species have also emerged, such as Burkholderia cepacia complex, Stenotrophomonas maltophilia, and Achromobacter xylosoxidans (Dakin et al., 2002; Rajan and Saiman, 2002; Saiman and Siegel, 2004; Vonberg and Gastmeier, 2005; Spicuzza et al., 2009; Hansen et al., 2010). A. xylosoxidans is an environmental Gram-negative bacillus with harmful properties in immuno-compromised hosts. It is difficult to be correctly identified, and suffers from a confusing nomenclature (Liu et al., 2002; Hauser et al., 2011). A. xylosoxidans is increasingly being isolated from sputa of CF patients (Zemel et al., 2000; Liu et al., 2002): its current prevalence ranges from 2 to 11% (Tan et al., 2002; Raso et al., 2008; Magni et al., 2010; Lambiase et al., 2011). Recently, a Brazilian study reported a prevalence of 21.8% (Pereira et al., 2011). Infection is frequently transient, although approximately 2.3% of CF patients are chronically infected (Tan et al., 2002). Relatively little is known about the clinical significance of A. xylosoxidans in CF patients, but it was found that infection occurs in CF patients with advanced lung disease (Lambiase et al., 2011): thus, additional studies are necessary to determine the aetio-pathological role A. xylosoxidans in CF. To this end, we decided to study some characteristics of A. xylosoxidans in selected strains isolated from CF patients, such as: genomic variability, biofilm production, antibiotic resistance, and motility. The last three bacterial features are considered important prerequisites for in vivo colonization and infection, and recent evidence report on their interdependence (Molin and Tolker-Nielsen, 2003; Hoffman et al., 2005; Shrout et al., 2011; Boles and Horswill, 2012). During chronic lung infection of CF patients, bacteria can survive under the challenging selective pressure imposed by both immune system and antibiotic regimens, developing both antibiotic resistances and biofilm formation (Macfarlane et al., 2011; Sibley and Surette, 2011; Sibley et al., 2011; Bragonzi et al., 2012). Furthermore, many bacterial species become highly motile thanks to enhanced production of flagella and pili (Henrichsen, 1983; Jarrell and McBride, 2008), also within established biofilms (Houry et al., 2012), increasing the efficiency in nutrient acquisition, the ability in avoiding toxic compounds, and the colonization of new available patches (Amiel et al., 2010; Sibley and Surette, 2011). Due to the doubled prevalence of CF patients colonized by A. xylosoxidans in our CF center, the present study was aimed at characterizing 57 strains of A. xylosoxidans isolated from CF patients and non-CF patients by: (i) genomic variability assessed by RAPD; (ii) ability to produce biofilm on abiotic surface; (iii) antibiotic resistance patterns; (iv) motility assay. Multivariate analysis was employed to cross-correlate all collected data.
Materials and methods
Patients
From January 2008 to January 2010, our laboratory isolated 225 strains of A. xylosoxidans from respiratory samples (i.e., sputum and tracheal aspirated) of 80 CF patients attending the CF Centre of the Pediatric Department of Policlinico Umberto I of Rome and from 5 non-CF patients. For the present study, we selected 52 strains from 34 CF patients, taken within the aforementioned group of 80, for whom A. xylosoxidans strains were isolated more than once. In addition, five strains were isolated from 4 non-CF patients: three strains from respiratory samples of 2 patients with a genetic respiratory disorder (Kartagener syndrome), and other two strains from 2 blood samples. Thus, the total number of selected A. xylosoxidans strains for this study was 57 from overall 38 patients. We divided the 34 CF selected patients into the following three Forced Expiratory Volume (FEV1%) classes (following the European Respiratory Society's criteria): class 3, mild obstruction or normal (≥70%); class 2, moderate obstruction (>40 and <70%); and class 1, severe obstruction (≤40%). A further subdivision was based on the chronic or intermittent presence of A. xylosoxidans. Chronicity was considered when there was a sputum culture positive for A. xylosoxidans in at least three occasions over a 6-month period, as previously suggested (Tan et al., 2002). In Table 1 are summarized patients' demographics and clinical features. All patients were involved in the study after providing written consent. The study protocol was approved by the Committee on Ethical Practice of the Policlinico Umberto I, Rome, Italy.
Table 1.
Patients' demographics and strains characteristics.
| Patient | Age (year) | FEV1% class | Weight (kg) | Height (m) | BMI | Strainc | Biofilm class | Chronicity | ResistANcese | Swimming (mm) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 20 | 2 | 48.0 | 1.70 | 16.6 | 129 | M | 1 | ATM, GM, NN | 17.7 |
| 1 | 47.0 | 1.70 | 16.2 | 157 | W | 1 | ATM, GM, NN | 23.7 | ||
| 2 (K)a | 42 | nab | 76.5 | 1.82 | 23.1 | 232 | S | 1 | AN, ATM, FEP, CIP, GM, MER, PIP, TIM, NN | 13.0 |
| 3 | 56 | 2 | 80.0 | 1.65 | 29.4 | 66 | W | 0 | AN, ATM, FEP, CIP, GM | 26.3 |
| 4 | 6 | na | 19.2 | 1.10 | 15.9 | 180 | M | 1 | AN, ATM, FEP, CIP, GM, NN | 24.3 |
| 5 | 44 | 1 | 60.0 | 1.74 | 19.8 | 128 | S | 1 | AN, ATM, FEP, CIP, CS, GM, MN, NN | na |
| 2 | 60.0 | 1.74 | 19.8 | 147 | W | 1 | AN, FEP, CIP, GM, NN | 22.7 | ||
| 3 | 60.0 | 1.74 | 19.8 | 248 | S | 1 | AN, ATM, FEP, CIP, GM, NN | na | ||
| 6 | 36 | 1 | 41.0 | 1.55 | 17.1 | 35 | Md | 0 | AN, ATM, FEP, CIP, GM, NN | 35.7 |
| 2 | 50.0 | 1.55 | 20.8 | 241 | S | 0 | ATM, FEP, CIP, GM, MER, MN, NN | 28.3 | ||
| 7 | 34 | 1 | 44.0 | 1.65 | 16.2 | 145 | Sd | 0 | AN, ATM, FEP, CIP, CS, GM, MN, NN | na |
| 8 | 32 | 3 | 62.8 | 1.69 | 21.9 | 53 | W | 1 | AN, ATM, GM | 52.3 |
| 2 | 61.0 | 1.69 | 21.3 | 228 | S | 1 | AN, ATM, FEP, CIP, GM, PIP, NN | na | ||
| 2 | 61.0 | 1.69 | 21.3 | 229 | W | 1 | AN, ATM, FEP, CIP, GM, MN, NN | 23.7 | ||
| 9 | 23 | 1 | 45.0 | 1.78 | 14.2 | 209g | S | 1 | ATM, MER | 31.0 |
| 1 | 45.0 | 1.78 | 14.2 | 209p | S | 1 | FEP, CIP, MER | 28.0 | ||
| 10 | 15 | 3 | 40.8 | 1.51 | 17.9 | 144 | S | 1 | AN, ATM, FEP, GM, PIP, NN | 29.3 |
| 11 | 48 | 1 | 52.0 | 1.63 | 19.6 | 4 | S | 1 | AN, ATM, FEP, CAZ, CIP, GM, IPM, MER, PIP, TIM, NN | 23.7 |
| na | 55.0 | 1.64 | 20.5 | 276 | S | 1 | AN, ATM, FEP, CIP, GM, NN | 56.3 | ||
| na | 55.0 | 1.64 | 20.4 | 277 | S | 1 | AN, ATM, FEP, CIP, GM, NN | 52.3 | ||
| 12 | 35 | 1 | 68.4 | 1.72 | 23.1 | 77 | M | 0 | AN, ATM, FEP, CIP, GM, NN | 39.7 |
| 1 | 68.4 | 1.72 | 23.1 | 287 | M | 0 | AN, ATM, FEP, CIP, CS, GM, MN, NN | 50.3 | ||
| 13 | 5 | na | 21.7 | 1.10 | 17.8 | 162 | W | 0 | AN, ATM, CS, GM | 52.3 |
| 14 | 19 | 2 | 58.3 | 1.70 | 20.2 | 84 | S | 1 | ATM, GM | na |
| 15 | 7 | 3 | 19.6 | 1.20 | 13.6 | 101p | M | 0 | AN, ATM, CS, GM, NN | 9.7 |
| 16 | 23 | 1 | 40.0 | 1.56 | 16.4 | 266 | S | 1 | GM, NN | 14.7 |
| 1 | 40.0 | 1.56 | 16.4 | 278 | S | 1 | AN, GM, NN | 14.0 | ||
| 17 | 24 | 1 | 52.0 | 1.61 | 20.1 | 252 | S | 0 | AN, ATM, CIP, GM, MN, NN | na |
| 18 | 21 | 2 | 60.0 | 1.67 | 21.5 | 222 | W | 0 | ATM, CIP | 56.3 |
| 19 | 34 | 1 | na | na | na | 116 | S | 1 | AN, ATM, CIP, GM, NN | 23.0 |
| na | 48.5 | 1.70 | 16.8 | 267 | S | 1 | AN, ATM, FEP, CIP, CS, GM, NN | 21.3 | ||
| 20 | 8 | 2 | 21.3 | 1.21 | 14.5 | 207 | M | 0 | GM | 30.7 |
| 21 | 37 | 2 | 58.0 | 1.67 | 20.8 | 251 | S | 1 | AN, ATM, FEP, CAZ, CIP, GM, IPM, TIM, NN | 61.7 |
| 22 | 6 | 3 | 23.0 | 1.23 | 15.2 | 153 | S | 0 | – | na |
| 23 (B)a | 61 | na | na | na | na | 291 | S | 0 | ATM, GM | 31.0 |
| 24 (B)a | 51 | na | na | na | na | 230 | S | 0 | AN, ATM, CS, GM | 43.7 |
| 25 | 22 | 2 | 68.0 | 1.88 | 19.2 | 167p | S | 0 | AN, ATM, FEP, CIP, GM, MN, PIP, NN | na |
| 26 | 21 | 3 | 53.0 | 1.68 | 18.8 | 226 | M | 1 | AN, ATM, FEP, CIP, GM, MN, NN | 42.7 |
| 27 | 24 | 3 | 40.0 | 1.51 | 17.5 | 133 | S | 0 | AN, ATM, FEP, CIP, CS, GM, NN | na |
| 2 | 40.0 | 1.51 | 17.5 | 231 | M | 0 | AN, ATM, FEP, CS, GM | 9.3 | ||
| 28 | 26 | 1 | 48.0 | 1.56 | 19.7 | 68 | S | 0 | AN, ATM, GM, NN | 35.0 |
| 29 | 22 | 3 | 67.0 | 1.82 | 20.2 | 247 | S | 0 | ATM | 40.3 |
| 30 | 21 | 3 | 62.6 | 1.73 | 20.9 | 225 | S | 0 | AN, ATM, FEP, CIP, GM, NN | 55.7 |
| 31 | 44 | 1 | 49.0 | 1.56 | 20.1 | 24 | S | 0 | ATM, GM, MN, NN | 20.3 |
| 32 | 14 | 3 | 56.0 | 1.60 | 21.8 | 49 | M | 1 | AN, ATM, GM | 54.0 |
| 3 | 56.5 | 1.60 | 22.1 | 50 | M | 1 | ATM | 53.0 | ||
| 33 | 43 | 2 | 52.0 | 1.67 | 18.6 | 275 | M | 1 | AN, ATM, FEP, CIP, GM, NN | 8.3 |
| 34 | 1 | na | 8.6 | 0.72 | 16.3 | 215 | M | 0 | ATM, NN | 21.7 |
| 35 | 27 | 2 | 55.0 | 1.61 | 21.2 | 90 | M | 0 | AN, ATM, FEP, CIP, CS, GM, NN | 28.3 |
| 1 | 61.6 | 1.60 | 24.1 | 237 | S | 0 | AN, ATM, FEP, CIP, GM, NN | 50.3 | ||
| 36 (K) | 34 | na | 59.0 | 1.63 | 22.2 | 175 | S | 0 | AN,. ATM, FEP, CIP, GM, NN | 33.7 |
| 37 | 36 | 1 | 49.6 | 1.61 | 19.1 | 213 | S | 1 | AN, FEP, CIP, GM, MN | 11.7 |
| 38 (K) | 26 | na | na | na | na | 67 | S | 0 | AN, ATM, GM | 43.0 |
| 39 | 30 | 3 | 52.7 | 1.65 | 19.3 | 249 | M | 1 | AN, FEP, CS, IPM, MER, PIP | 22.7 |
| 3 | 52.7 | 1.65 | 19.3 | 250 | S | 1 | AN, ATM, FEP, CIP, GM, IPM, TIM, NN | na | ||
| 3 | 54.9 | 1.65 | 20.2 | 218g | M | 1 | AN, ATM, FEP, CIP, GM, NN | 55.0 | ||
| 3 | 54.9 | 1.65 | 20.2 | 218p | S | 1 | FEP, CIP, IPM, MN | na |
K, Kartagener syndrome; B, blood culture.
na, not available.
Letters g and p stand for different morphologies of the A. xylosoxidans colony grown on BCSA.
Biofilm class was assessed after PLS-DA model.
Resistances were determined by automated Vitek 2 system: ATM, aztreonam; GM, gentamicin; AN, amikacin; NN, tobramycin; FEP, cefepime; CIP, ciprofloxacin; CS, colistin; MN, minociclin; MER, meropenem; PIP, piperacillin; IPM, imipenem; TIM, ticarcillin-clavulanic acid; CAZ, ceftazidime; and TZP, piperacillin + tazobactam.
Microbial identification
API 20NE system (bioMérieux, Marcy l'Etoile, France) and automated Vitek2 system (bioMérieux, Marcy l'Etoile, France) were used for strains identification. Oxidase activity was checked with dimethyl-paraphenylenediamine disks (bioMérieux, Marcy l'Etoile, France). Results obtained from API 20NE tests and oxidase reactions were further interpreted with the Apilab Plus software package (bioMérieux, Marcy l'Etoile, France). In order to avoid misidentification of Gram negative bacilli, isolated from CF patients, we performed species-specific PCR for all A. xylosoxidans strains. Collected strains were cryopreserved at −80°C before use. Antibiotic resistances assays for all A. xylosoxidans strains were provided by automated Vitek2 system.
DNA extraction
A. xylosoxidans DNA extraction was performed by use of a Wizard genomic DNA purification kit (Promega Corporation, Madison, WI) following manufacturer's instructions. DNA was finally quantified by spectrophotometer at 260 nm, and its quality assayed by 260/280 nm ratio.
Species-specific PCR assay
Species-specific PCR was performed as described elsewhere (Hogardt et al., 2007). Negative and positive control PCRs were employed for every experiment. PCR products were visualized by electrophoresis in a 2% agarose gel (Invitrogen Corporation, CA), stained with ethidium bromide (EtBr; Invitrogen Corporation), and captured with a DigiDoc-It (UVP, Cambridge, United Kingdom) photographic system. Bands of 163 bp were considered positive for A. xylosoxidans identification.
RAPD typing
The RAPD amplification mixture and cycling conditions were described elsewhere (Lambiase et al., 2006). The primer used was the 270 (5′-TGCGCGCGGG-3′). RAPD products were separated by electrophoresis in 1.5% agarose gel (Invitrogen Corporation, CA). Molecular size markers (Invitrogen Corporation) and negative control were included in all gels. Gels were stained with EtBr (at 0.5 μM) (Invitrogen Corporation) and captured with a DigiDoc-It (UVP) photographic system.
Biofilm production assay
Overnight cultures of A. xylosoxidans in Trypticase Soy Broth (Becton Dickinson) at 37°C in dynamic conditions (90 rpm), were diluted into fresh TSB to reach OD550 = 1 (corresponding to 1 × 109 CFU/ml). After diluting 1:100 the bacterial culture, 200 μl were used to inoculate sterile flat-bottom polystyrene tissue culture 96-wells plates, followed by a 48 h incubation at 37°C. Non-adherent bacteria were removed by washing three times with sterile Phosphate Buffered Saline (PBS). Wells were stained at room temperature for 5 min with 200 μl of 1% Crystal Violet solution, then rinsed with distilled water and dried at 37°C for 30 min. Stained biofilm were dissolved adding 250 μl of 33% glacial acetic acid for 15 min. The optical density (OD) of each well was measured at 570 nm using a microtiter-plate reader (Multiskan EX, Thermo Scientific, Massachusetts, USA). A. xylosoxidans strains were divided following Stepanovic method (Stepanovic et al., 2007), into different biofilm producers classes, named: N, no biofilm producer; W, weak biofilm producer; M, moderate biofilm producer; and S, strong biofilm producer.
Motility assay
Swimming assay was performed in “Swim plates” [tryptone broth (10 g/l tryptone (Difco)-5 g/l NaCl) containing 0.3% (wt/vol) agarose (GIBCO/BRL)]. LB agar Overnight culture of isolated strains identified as A. xylosoxidans, were used to inoculate Swim plates, and incubated at 30°C for 12–14 h (Rashid et al., 2000). Diameter of resulting concentric ring, expressed in millimetres, was used to define the motility of a specific A. xylosoxidans strain.
PCR detections of resistance determinants
The presence of Class 1 Integron was tested by PCR, by means of specific primers: 5′-CS (GGC ATC CAA GCA GCA AGC) and 3′-CS (AAA GCA GAC TTG ACC TGA) (Levesque et al., 1995; Neuwirth et al., 2006). Carbapenemase determinants blaIMP, blaVIM, and β-lactamase determinants blaVEB plus blaOXA-1 were also assayed by PCR. Primer used were: IMP-1 F (5′CAT GGT TTG GTG GTT CTT GT 3′) and IMP-1 R (5′ATA ATT TGG CGG ACT TTG GC 3′) for blaIMP (Yum et al., 2002); VIM-F (5′-AGT GGT GAG TATCCG ACA G-3′) and VIM-R (5′-ATG AAA GTG CGTGGA GAC-3′) for blaVIM1 (Tsakris et al., 2000); VIM-2A (5′-ATGTTCAAACTTTTGAGTAGTAAG-3′) and VIM-2B (5′CTACTCAACGACTGAGCG-3′) for blaVIM-2-like genes (Poirel et al., 2000); VEB-1 F (5′ CCA GAT AGG AGT ACA GAC 3′) and VEB-1 R (5′ GAC TCT GCA ACA AAT ACG C 3′) for blaVEB1 (Neuwirth et al., 2006); OXA-1 F (5′CTT GAT TGA AGG GTT GGG CG-3′) and OXA-1 R (5′AGC CGT TAA AAT TAA GCC C-3′) for blaOXA-1 (Shin et al., 2005).
Data analysis
Agglomerative hierarchical classification (AHC)
AHC, an unsupervised method, was performed on RAPD profiles by means of a binary matrix generated by the presence/absence of RAPD bands, using Doc-It LS software (UVP), and the subsequent dendrogram was generated with XLStat 7.5 (Addinsoft), using a Euclidean distance dissimilarity matrix and the agglomeration method of Ward.
Factorial discriminant analysis (FDA)
FDA, a supervised method closely linked to multivariate analysis of variance, was employed by means of XLStat 7.5 software (Addinsoft). Explanatory variables were automatically verified to be linearly independent by calculating the multiple correlation of each variable with all the others. Fisher's test was used to compare patients' clinical features or characteristics of selected A. xylosoxidans strains with RAPD profiles: a P value less than or equal to 0.05 was considered statistically significant.
Results
Achromobacter xylosoxidans isolates
Overall 39 patients referring to the Regional CF Centre, “Policlinico Umberto I” hospital (Rome), were enrolled. Thirty-four above the 39 patients were affected by CF, while the remaining non-CF subjects had a different disease (three patients with Kartagener syndrome, that involves an impairment in epithelial cilia movement, and other two patients from an oncologic unit). Patients with CF and Kartagener syndrome gave sputa samples, while subjects from oncologic unit gave blood samples. All CF patients were not permanently convalescing at the Policlinico Umberto I hospital, but underwent a visit each 6 months, or when specifically required by the physician. Patients' antibiotic treatments were as follows (taking into consideration the closer to A. xylosoxidans strains isolation from each CF patient): colimycin 41.18% (14/34), sulfametoxazole + trimethoprim 17.65% (6/34), tobramycin 11.76% (4/34), amoxicillin + clavulanic acid 8.82% (3/34), levofloxacin 8.82% (3/34), amikacin 5.88% (2/52), ceftriaxone 2.94% (1/34), linezolid 2.94% (1/34). All CF patients went through aforementioned antibiotic treatments for a period of 6 months before A. xylosoxidans sampling under these conditions: colimycin aerosolized 2 flasks/day every 2 days, sulfametoxazole + trimethoprim orally provided every 2 days, tobramycin aerosolized 300 mg/day, amoxicillin + clavulanic acid orally provided every 2 days, levofloxacin provided intravenously 300 mg/day, amikacin aerosolized 1 flask/day, ceftriaxone provided intravenously 1 dose/day, linezolid orally provided every 2 days. Patients' demographics, along with number and characteristics of A. xylosoxidans isolates, were reported in Table 1. The isolation range was 1–4 bacterial strains per patient, with a total of 57 A. xylosoxidans strains. At the same time of isolation, data about the Forced Expiratory Value (FEV), expressed as FEV1%, and the Body Mass Index (BMI), were collected. In the present study, spanning from January 2008 to January 2010, we had 80 patients A. xylosoxidans-positive patients above a total of 500, with a prevalence of 16%: 34/80 were chronically infected, and we were able to isolate 225 strains. In respect to our previous study (Magni et al., 2010), we had a significantly higher prevalence of A. xylosoxidans-positive patients: from 8.9 to 16% (χ2 = 10.18, P = 0.0014).
RAPD-PCR
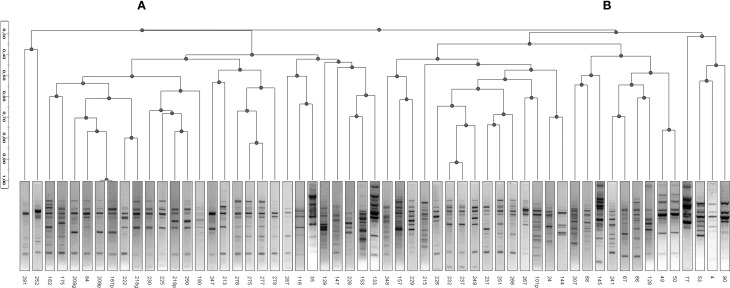
The 57 strains of A. xylosoxidans were characterized for their genomic asset by the Randomly Amplified Polymorphic DNA (RAPD) technique, useful to look for genetic relationships among the isolated strains and for discerning strains within a species. In Figure 1 was reported the dendrogram obtained by Ascendent Hierarchical Clusterization (AHC), a non-supervised statistical method to ascertain genomic relatedness among RAPD profiles obtained. The dendrogram showed two major clusters (A and B): the first cluster (A) grouped 29/57 A. xylosoxidans isolates, while the second one (B) grouped 28/57 bacterial strains (Figure 1). Intra-cluster mean similarity was calculated by Dice index, as described in Materials and Methods, and gave a result of 45.9 ± 0.8% for cluster A, while it was 41.7 ± 0.8% for cluster B, and this difference was significant (Mann-Whitney, P = 0.00078).
Figure 1.
Ascendent Hierarchical Clusterization (AHC). A dendrogram based on RAPD profiles of A. xylosoxidans strains was generated by means of inverse Euclidean distance dissimilarity matrix and agglomeration method of Ward. Two clusters are visible (A and B) based upon a threshold set at 28% of similarity. No known variable was responsible in defining such a cluster formation (for all variables, P > 0.05).
Biofilm production
In our assay conditions, all strains were able to produce biofilm on abiotic surface (96-well plate) (Table 1). On the basis of biofilm amount produced, assessed at OD570 accordingly to Stepanovic (Stepanovic et al., 2007), A. xylosoxidans strains were divided into three different groups: strong (S), moderate (M), and weak (W). As reported in Table 1, 33/57 (57.9%) strains were strong biofilm producers, 15/57 (26.3%) strains felt into moderate class, while 7/57 (12.3%) strains were weak producers. Two strains above 57 (3.5%), namely 35 and 145, showed OD590 values intermediate between strong and moderate produces. Those strains were consequently placed in the class moderate and strong, respectively, on the basis of their RAPD profiles (see results of statistical correlations).
Motility assay
Forty-six above 57 A. xylosoxidans strains (80.7%) showed a swimming phenotype (Table 1), with the formation of a concentric ring with different diameters, expressed in millimetres. Eleven A. xylosoxidans isolates resulted negative for swimming motility. No swarming as well as no twitching phenotype were found among the isolated strains of A. xylosoxidans (data not shown).
Antibiotic resistance phenotype
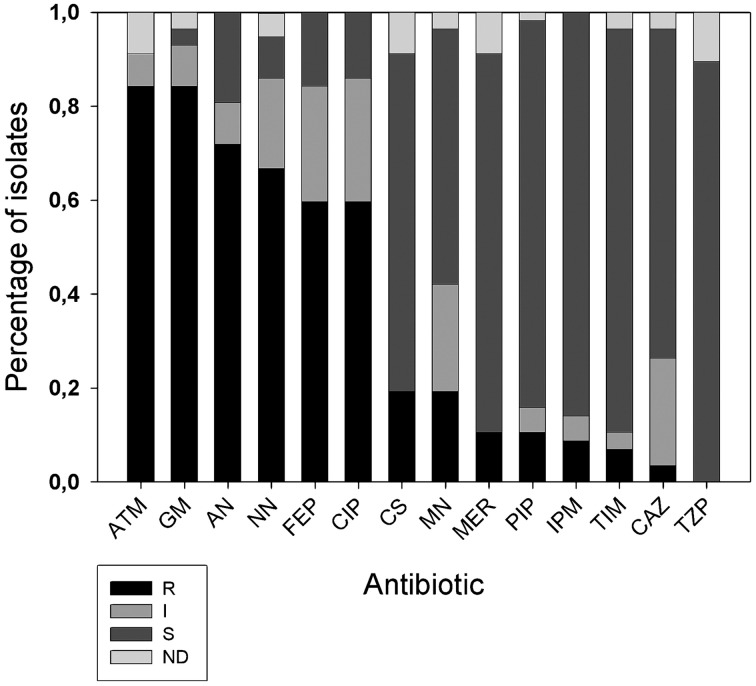
All 57 A. xylosoxidans isolates were tested for 14 antibiotic resistances by means of Vitek2 instrument. Results showed (Figure 2) high frequency of resistances to aztreonam (84%), gentamicin (84%), amikacin (75%), tobramycin (73%), cefepime (60%) e ciprofloxacin (60%). Hodge Test showed that 2/11 of the carbapenem resistant strains were able to produce carbapenemase. 3/57 strains resistant to β-lactam antibiotics showed to possess the specific band of the integron1, and 7 the specific band related to metal β-lactamase (blaIMP−1). Of these last 7 strains, three showed to possess the specific band of the integron1 meropenem (MER), piperacillin (PIP), imipenem (IPM), ticarcillin clavulanate (TIM), ceftazidime (CAZ). Taking into consideration the antibiotic exposure of the 52 selected A. xylosoxidans strains among CF patients, we found these percentages of prevalence: Amoxicillin + clavulanic acid 9.62% (5/52), levofloxacin 9.62% (5/52), ceftriaxone 3.85% (2/52), tobramycin 11.54% (6/52), colimycin 38.46% (20/52), linezolid 3.85% (2/52), sulfametoxazole + trimethoprim 19.23% (10/52), amikacin 3.85% (2/52).
Figure 2.
Distribution of antibiotic resistances among A. xylosoxidans isolates. Fourteen antibiotics (x axis) were tested with Vitek 2 instrument: ATM, aztreonam; GM, gentamicin; AN, amikacin; NN, tobramycin; FEP, cefepime; CIP, ciprofloxacin; CS, colistin; MN, minociclin; MER, meropenem; PIP, piperacillin; IPM, imipenem; TIM, ticarcillin clavulanate; CAZ, ceftazidime; and TZP, piperacillin+tazobactam. On y axis is the fraction of isolates normalized to one. R, resistant; I, intermediate; S, susceptible; ND, indeterminate.
Data correlations
RAPD—FEV1%
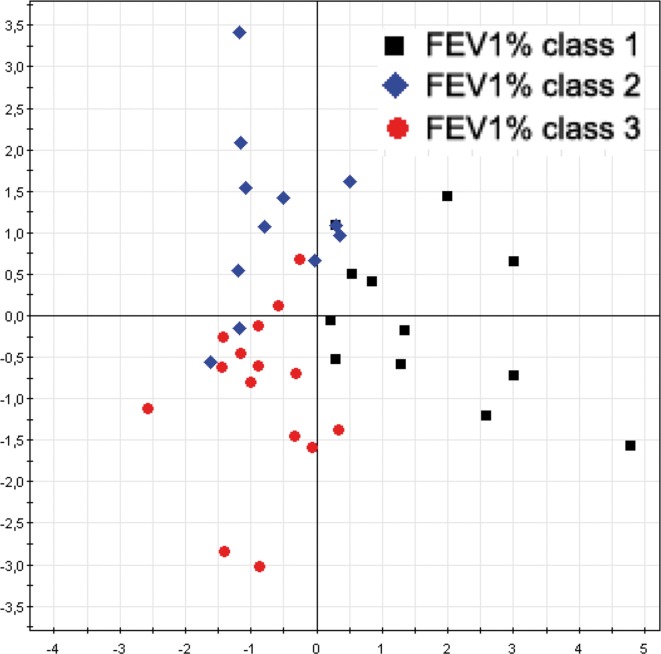
In order to confirm the results already obtained in a previous our study (Magni et al., 2010) we performed a correlation analysis between RAPD profiles of the new 57 A. xylosoxidans strains and patients' FEV1% classes. A multivariate statistical approach, performed with Factorial Discriminant Analysis (FDA) algorithm, showed a separation between the FEV1% classes (Figure 3), with a model predictability of 84.62% (Fisher's P = 5.2*10−10). The analysis correctly classified 9/12 (75.0%) of A. xylosoxidans strains isolated from CF patients with predicted FEV1% class 1, 14/15 (93.3%) of the strains isolated from CF patients with predicted FEV1% classes 3, and 10/12 (83.3%) of the strains isolated from CF patients with predicted FEV1% classes 2. Eighteen A. xylosoxidans strains with no predicted FEV1% class were classified among the three FEV1% classes on the basis of their RAPD profiles: 4 to class 1, 7 to class 2, and 7 to class 3. The FEV1% class assignment of these last 18 strains were in agreement with the clinical status of corresponding CF patients, as assessed by pediatric physicians.
Figure 3.
A. xylosoxidans RAPD profiles grouped by FEV1% class. The overall data variation was described by the factorial axes t[1] and t[2]. The predictability of the model, in dividing FEV1% classes, was 84.62% (Fisher's P = 5.2*10−10).
RAPD—biofilm
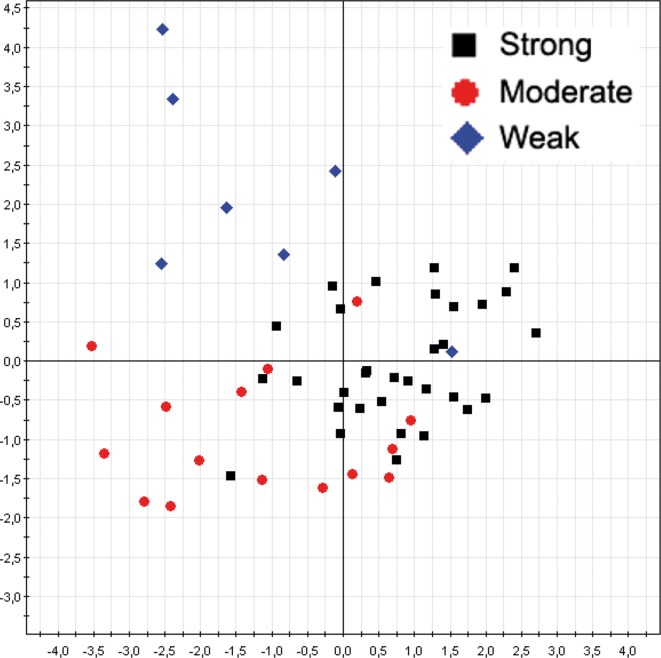
The same multivariate statistical approach of PLS-DA was employed to correlate A. xylosoxidans strains RAPD profiles and corresponding biofilm classes. The model showed a separation between the biofilm classes (Figure 4), with a model predictability of 85.45% (Fisher's P = 5.4*10−11). The analysis correctly classified 31/33 (93.9%) of strong biofilm producers, 10/15 (66.7%) of medium one, and 6/7 (85.7%) of weak biofilm producers. Two A. xylosoxidans strains without biofilm classification were classified upon their RAPD profiles in this way: one to strong class, and one to weak class. Furthermore, we studied the distribution of A. xylosoxidans strains, in relation to their biofilm production ability, between FEV1% classes. A significant prevalence of strong biofilm producers strains was found in FEV1% class 1 CF patients (FEV1% class 1 vs. FEV1% class 2, P = 0.0007, χ2 = 11.44; FEV1% class 1 vs. FEV1% class 3, P = 0.0046, χ2 = 8.02) (Table 2). No difference was found in the mean number of resistance traits among strains colonizing CF patients with different FEV1% value (data not shown). Similarly, no differences were found comparing mean number of resistance traits and biofilm producing ability among strains colonizing CF patients in a chronic or intermittent way (data not shown).
Figure 4.
A. xylosoxidans RAPD profiles grouped by biofilm classes. The overall data variation was described by the factorial axes t[1] and t[2]. The predictability of the model, in dividing biofilm classes, was 85.45% (Fisher's P = 5.4*10−11).
Table 2.
Distribution of collected strains among biofilm and FEV1% classes.
| Biofilm class | |||
|---|---|---|---|
| FEV1% class | S (n = 33) | M (m = 17) | W (n = 7) |
| 1 | 14 | 5 | 1 |
| 2 | 6 | 4 | 4 |
| 3 | 8 | 4 | 1 |
| 0 | 5 | 4 | 1 |
Discussion
In a previous paper (Magni et al., 2010) we showed an A. xylosoxidans prevalence of 8.9% in our Regional CF center, referred to a time window from January 2005 to January 2007. In that paper we reported that above overall 450 patients, 40 were affected by A. xylosoxidans, 16 were chronically infected, and we isolated 106 strains. In the present study, spanning from January 2008 to January 2010, we found an almost doubled prevalence of A. xylosoxidans-positive patients (16%), meaning a possible outbreak in our center. Due to the doubled prevalence found, we focused our attention on well-known virulence traits able to enhance bacterial colonization. It was proposed how biofilm formation, antibiotic resistance acquisition, and motility could be interdependent (Molin and Tolker-Nielsen, 2003; Hoffman et al., 2005), and recent evidence support such hypothesis (Shrout et al., 2011; Boles and Horswill, 2012). We studied the abovementioned virulence factors in 57 CF and non-CF clinical isolates of A. xylosoxidans, in order to investigate their role in this emerging pathogen: this is the first report on biofilm formation and motility of A. xylosoxidans. Genomic fingerprint assessed by RAPD analysis showed a significant division into two major clusters (Figure 1). Within such clusters all features considered (BMI, FEV1%, chronicity, biofilm production, resistance determinants, motility) were randomly distributed: probably, other features not taken into account in this study are accountable for cluster formation. We showed a significant association between RAPD profiles, FEV1% and biofilm classes, meaning that different CF lung habitats would be able to select particular A. xylosoxidans genomic variants and biofilm producers. As reported in Table 2, we found a significant presence of strong biofilm producers within FEV1% class 1, underlining the influence of biofilm on clinical exacerbations in CF. Biofilm development is a common trait in CF patients at the surface of airway mucosae (Sibley and Surette, 2011; Sibley et al., 2011; Bragonzi et al., 2012): bacteria inside biofilms are well protected, developing antibiotic resistances even by means of horizontal genetic transfer (HGT) (Molin and Tolker-Nielsen, 2003). Interestingly, all our A. xylosoxidans strains having specific resistance integrons were strong biofilm producers, supporting the involvement of HGT in biofilm development (Ghigo, 2001; Traglia et al., 2012). In this view, the unique environmental A. xylosoxidans genome sequenced thus far (Strnad et al., 2011) showed the presence of two large plasmids with resistance genes. As found in P. aeruginosa, mechanisms other than HGT should be employed in biofilm formation, such as sub-inhibitory concentrations of aminoglycosides involving alterations of c-di-GMP levels (Hoffman et al., 2005). Interestingly, more than 70% of A. xylosoxidans strains were resistant to the three aminoglycosides (gentamicin, amikacin, tobramycin) tested by Vitek 2 instrument (Figure 2), paving the way of a double strategy (HGT and c-di-GMP) enrolled by A. xylosoxidans in producing biofilms within the lung environment to circumvent antibiotic pressure. We had a high prevalence of patients that underwent colimycin/colistin usage (41%) before A. xylosoxidans strain isolation, and we observed only a 20% of resistance to colistin assessed by Vitek2 (Figure 3): this discrepancy should be due to the short time of exposure (3 months) of the pathogen to the antibiotic. One could argue that a prolonged exposure should enhance the prevalence of this resistance determinant within the A. xylosoxidans population, but colimycin/colistin should be a good candidate to hinder an outbreak. Once established, bacteria are able to swim within the biofilm itself (Boles and Horswill, 2012; Houry et al., 2012), enhancing colonization of new patches and nutrient acquisition. Even if around 81% of our A. xylosoxidans strains showed swimming ability in “Swim plates,” no positive or negative correlation was found with biofilm production assessed by 96-wells plates (R2 = 5.7*10−4): thus, further studies will investigate on A. xylosoxidans motility within established biofilms by means of fluorescent dyes. This study evidenced how biofilm formation, antibiotic resistances acquisition, and motility showed by A. xylosoxidans species could enhance its survival in CF patients' lung.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Amiel E., Lovewell R. R., O'toole G. A., Hogan D. A., Berwin B. (2010). Pseudomonas aeruginosa evasion of phagocytosis is mediated by loss of swimming motility and is independent of flagellum expression. Infect. Immun. 78, 2937–2945 10.1128/IAI.00144-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles B. R., Horswill A. R. (2012). Swimming cells promote a dynamic environment within biofilms. Proc. Natl. Acad. Sci. U.S.A. 109, 12848–12849 10.1073/pnas.1210297109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragonzi A., Farulla I., Paroni M., Twomey K. B., Pirone L., Lore N. I., et al. (2012). Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS ONE 7:e52330 10.1371/journal.pone.0052330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakin C. J., Numa A. H., Wang H., Morton J. R., Vertzyas C. C., Henry R. L. (2002). Inflammation, infection, and pulmonary function in infants and young children with cystic fibrosis. Am. J. Respir. Crit. Care Med. 165, 904–910 10.1164/ajrccm.165.7.2010139 [DOI] [PubMed] [Google Scholar]
- Ghigo J. M. (2001). Natural conjugative plasmids induce bacterial biofilm development. Nature 412, 442–445 10.1038/35086581 [DOI] [PubMed] [Google Scholar]
- Hansen C. R., Pressler T., Nielsen K. G., Jensen P. O., Bjarnsholt T., Hoiby N. (2010). Inflammation in Achromobacter xylosoxidans infected cystic fibrosis patients. J. Cyst. Fibros. 9, 51–58 10.1016/j.jcf.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Hauser A. R., Jain M., Bar-Meir M., McColley S. A. (2011). Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin. Microbiol. Rev. 24, 29–70 10.1128/CMR.00036-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen J. (1983). Twitching motility. Annu. Rev. Microbiol. 37, 81–93 10.1146/annurev.mi.37.100183.000501 [DOI] [PubMed] [Google Scholar]
- Hoffman L. R., D'argenio D. A., Maccoss M. J., Zhang Z., Jones R. A., Miller S. I. (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436, 1171–1175 10.1038/nature03912 [DOI] [PubMed] [Google Scholar]
- Hogardt M., Hoboth C., Schmoldt S., Henke C., Bader L., Heesemann J. (2007). Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 195, 70–80 10.1086/509821 [DOI] [PubMed] [Google Scholar]
- Houry A., Gohar M., Deschamps J., Tischenko E., Aymerich S., Gruss A., et al. (2012). Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 109, 13088–13093 10.1073/pnas.1200791109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K. F., McBride M. J. (2008). The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6, 466–476 10.1038/nrmicro1900 [DOI] [PubMed] [Google Scholar]
- Lambiase A., Catania M. R., Del Pezzo M., Rossano F., Terlizzi V., Sepe A., et al. (2011). Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 30, 973–980 10.1007/s10096-011-1182-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambiase A., Raia V., Del Pezzo M., Sepe A., Carnovale V., Rossano F. (2006). Microbiology of airway disease in a cohort of patients with cystic fibrosis. BMC Infect. Dis. 6:4 10.1186/1471-2334-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque C., Piche L., Larose C., Roy P. H. (1995). PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39, 185–191 10.1128/AAC.39.1.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Coenye T., Burns J. L., Whitby P. W., Stull T. L., Lipuma J. J. (2002). Ribosomal DNA-directed PCR for identification of Achromobacter (Alcaligenes) xylosoxidans recovered from sputum samples from cystic fibrosis patients. J. Clin. Microbiol. 40, 1210–1213 10.1128/JCM.40.4.1210-1213.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S., Bahrami B., Macfarlane G. T. (2011). Mucosal biofilm communities in the human intestinal tract. Adv. Appl. Microbiol. 75, 111–143 10.1016/B978-0-12-387046-9.00005-0 [DOI] [PubMed] [Google Scholar]
- Magni A., Trancassini M., Varesi P., Iebba V., Curci A., Pecoraro C., et al. (2010). Achromobacter xylosoxidans genomic characterization and correlation of randomly amplified polymorphic DNA profiles with relevant clinical features [corrected] of cystic fibrosis patients. J. Clin. Microbiol. 48, 1035–1039 10.1128/JCM.02060-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin S., Tolker-Nielsen T. (2003). Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14, 255–261 10.1016/S0958-1669(03)00036-3 [DOI] [PubMed] [Google Scholar]
- Neuwirth C., Freby C., Ogier-Desserrey A., Perez-Martin S., Houzel A., Pechinot A., et al. (2006). VEB-1 in Achromobacter xylosoxidans from cystic fibrosis patient, France. Emerg. Infect. Dis. 12, 1737–1739 10.3201/eid1211.060143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R. H., Carvalho-Assef A. P., Albano R. M., Folescu T. W., Jones M. C., Leao R. S., et al. (2011). Achromobacter xylosoxidans: characterization of strains in Brazilian cystic fibrosis patients. J. Clin. Microbiol. 49, 3649–3651 10.1128/JCM.05283-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Naas T., Nicolas D., Collet L., Bellais S., Cavallo J. D., et al. (2000). Characterization of VIM-2, a carbapenem-hydrolyzing metallo-beta-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44, 891–897 10.1128/AAC.44.4.891-897.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan S., Saiman L. (2002). Pulmonary infections in patients with cystic fibrosis. Semin. Respir. Infect. 17, 47–56 10.1053/srin.2002.31690 [DOI] [PubMed] [Google Scholar]
- Rashid M. H., Rumbaugh K., Passador L., Davies D. G., Hamood A. N., Iglewski B. H., et al. (2000). Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 97, 9636–9641 10.1073/pnas.170283397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raso T., Bianco O., Grosso B., Zucca M., Savoia D. (2008). Achromobacter xylosoxidans respiratory tract infections in cystic fibrosis patients. APMIS 116, 837–841 10.1111/j.1600-0463.2008.00995.x [DOI] [PubMed] [Google Scholar]
- Saiman L., Siegel J. (2004). Infection control in cystic fibrosis. Clin. Microbiol. Rev. 17, 57–71 10.1128/CMR.17.1.57-71.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K. S., Han K., Lee J., Hong S. B., Son B. R., Youn S. J., et al. (2005). Imipenem-resistant Achromobacter xylosoxidans carrying blaVIM-2-containing class 1 integron. Diagn. Microbiol. Infect. Dis. 53, 215–220 10.1016/j.diagmicrobio.2005.06.018 [DOI] [PubMed] [Google Scholar]
- Shrout J. D., Tolker-Nielsen T., Givskov M., Parsek M. R. (2011). The contribution of cell-cell signaling and motility to bacterial biofilm formation. MRS Bull. 36, 367–373 10.1557/mrs.2011.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley C. D., Grinwis M. E., Field T. R., Eshaghurshan C. S., Faria M. M., Dowd S. E., et al. (2011). Culture enriched molecular profiling of the cystic fibrosis airway microbiome. PLoS ONE 6:e22702 10.1371/journal.pone.0022702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley C. D., Surette M. G. (2011). The polymicrobial nature of airway infections in cystic fibrosis: Cangene Gold Medal Lecture. Can. J. Microbiol. 57, 69–77 10.1139/W10-105 [DOI] [PubMed] [Google Scholar]
- Spicuzza L., Sciuto C., Vitaliti G., Di Dio G., Leonardi S., La Rosa M. (2009). Emerging pathogens in cystic fibrosis: ten years of follow-up in a cohort of patients. Eur. J. Clin. Microbiol. Infect. Dis. 28, 191–195 10.1007/s10096-008-0605-4 [DOI] [PubMed] [Google Scholar]
- Stepanovic S., Vukovic D., Hola V., Di Bonaventura G., Djukic S., Cirkovic I., et al. (2007). Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115, 891–899 10.1111/j.1600-0463.2007.apm_630.x [DOI] [PubMed] [Google Scholar]
- Strnad H., Ridl J., Paces J., Kolar M., Vlcek C., Paces V. (2011). Complete genome sequence of the haloaromatic acid-degrading bacterium Achromobacter xylosoxidans A8. J. Bacteriol. 193, 791–792 10.1128/JB.01299-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K., Conway S. P., Brownlee K. G., Etherington C., Peckham D. G. (2002). Alcaligenes infection in cystic fibrosis. Pediatr. Pulmonol. 34, 101–104 10.1002/ppul.10143 [DOI] [PubMed] [Google Scholar]
- Traglia G. M., Almuzara M., Merkier A. K., Adams C., Galanternik L., Vay C., et al. (2012). Achromobacter xylosoxidans: an emerging pathogen carrying different elements involved in horizontal genetic transfer. Curr. Microbiol. 65, 673–678 10.1007/s00284-012-0213-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakris A., Pournaras S., Woodford N., Palepou M. F., Babini G. S., Douboyas J., et al. (2000). Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38, 1290–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonberg R. P., Gastmeier P. (2005). Isolation of infectious cystic fibrosis patients: results of a systematic review. Infect. Control Hosp. Epidemiol. 26, 401–409 10.1086/502558 [DOI] [PubMed] [Google Scholar]
- Yum J. H., Yi K., Lee H., Yong D., Lee K., Kim J. M., et al. (2002). Molecular characterization of metallo-beta-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: identification of two new integrons carrying the bla(VIM-2) gene cassettes. J. Antimicrob. Chemother. 49, 837–840 10.1093/jac/dkf043 [DOI] [PubMed] [Google Scholar]
- Zemel B. S., Jawad A. F., Fitzsimmons S., Stallings V. A. (2000). Longitudinal relationship among growth, nutritional status, and pulmonary function in children with cystic fibrosis: analysis of the Cystic Fibrosis Foundation National CF Patient Registry. J. Pediatr. 137, 374–380 10.1067/mpd.2000.107891 [DOI] [PubMed] [Google Scholar]