Abstract
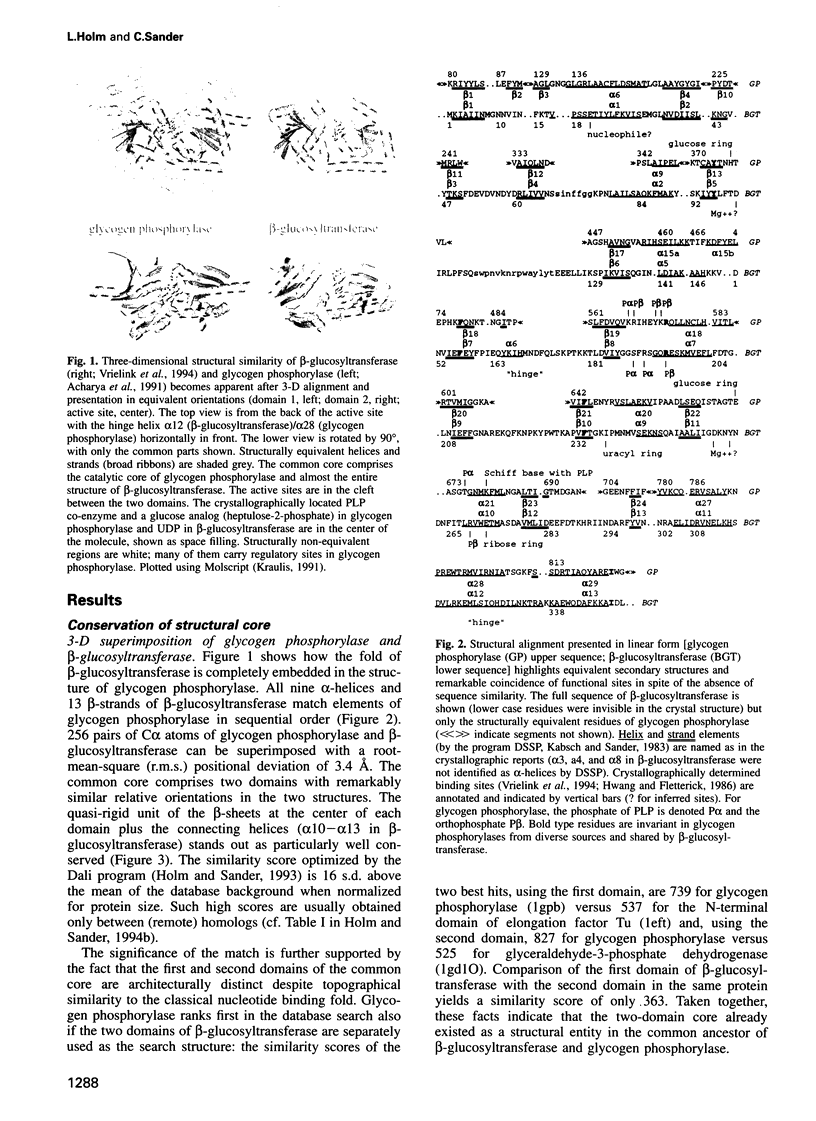
We report here an unexpected similarity in three-dimensional structure between glucosyltransferases involved in very different biochemical pathways, with interesting evolutionary and functional implications. One is the DNA modifying enzyme beta-glucosyltransferase from bacteriophage T4, alias UDP-glucose:5-hydroxymethyl-cytosine beta-glucosyltransferase. The other is the metabolic enzyme glycogen phosphorylase, alias 1.4-alpha-D-glucan:orthophosphate alpha-glucosyltransferase. Structural alignment revealed that the entire structure of beta-glucosyltransferase is topographically equivalent to the catalytic core of the much larger glycogen phosphorylase. The match includes two domains in similar relative orientation and connecting helices, with a positional root-mean-square deviation of only 3.4 A for 256 C alpha atoms. An interdomain rotation seen in the R- to T-state transition of glycogen phosphorylase is similar to that observed in beta-glucosyltransferase on substrate binding. Although not a single functional residue is identical, there are striking similarities in the spatial arrangement and in the chemical nature of the substrates. The functional analogies are (beta-glucosyltransferase-glycogen phosphorylase): ribose ring of UDP-pyridoxal ring of pyridoxal phosphate co-enzyme; phosphates of UDP-phosphate of co-enzyme and reactive orthophosphate; glucose unit transferred to DNA-terminal glucose unit extracted from glycogen. We anticipate the discovery of additional structurally conserved members of the emerging glucosyltransferase superfamily derived from a common ancient evolutionary ancestor of the two enzymes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2247–2249. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Duke E. M., Wakatsuki S., Hadfield A., Johnson L. N. Laue and monochromatic diffraction studies on catalysis in phosphorylase b crystals. Protein Sci. 1994 Aug;3(8):1178–1196. doi: 10.1002/pro.5560030804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui T., Shimomura S., Nakano K. Potato and rabbit muscle phosphorylases: comparative studies on the structure, function and regulation of regulatory and nonregulatory enzymes. Mol Cell Biochem. 1982 Feb 19;42(3):129–144. doi: 10.1007/BF00238507. [DOI] [PubMed] [Google Scholar]
- Gommers-Ampt J. H., Van Leeuwen F., de Beer A. L., Vliegenthart J. F., Dizdaroglu M., Kowalak J. A., Crain P. F., Borst P. beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993 Dec 17;75(6):1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- Hobohm U., Sander C. Enlarged representative set of protein structures. Protein Sci. 1994 Mar;3(3):522–524. doi: 10.1002/pro.5560030317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Sander C. Evaluation of protein models by atomic solvation preference. J Mol Biol. 1992 May 5;225(1):93–105. doi: 10.1016/0022-2836(92)91028-n. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Fast and simple Monte Carlo algorithm for side chain optimization in proteins: application to model building by homology. Proteins. 1992 Oct;14(2):213–223. doi: 10.1002/prot.340140208. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Parser for protein folding units. Proteins. 1994 Jul;19(3):256–268. doi: 10.1002/prot.340190309. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993 Sep 5;233(1):123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Searching protein structure databases has come of age. Proteins. 1994 Jul;19(3):165–173. doi: 10.1002/prot.340190302. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. The FSSP database of structurally aligned protein fold families. Nucleic Acids Res. 1994 Sep;22(17):3600–3609. [PMC free article] [PubMed] [Google Scholar]
- Hudson J. W., Golding G. B., Crerar M. M. Evolution of allosteric control in glycogen phosphorylase. J Mol Biol. 1993 Dec 5;234(3):700–721. doi: 10.1006/jmbi.1993.1621. [DOI] [PubMed] [Google Scholar]
- Hwang P. K., Fletterick R. J. Convergent and divergent evolution of regulatory sites in eukaryotic phosphorylases. Nature. 1986 Nov 6;324(6092):80–84. doi: 10.1038/324080a0. [DOI] [PubMed] [Google Scholar]
- Johnson L. N., Acharya K. R., Jordan M. D., McLaughlin P. J. Refined crystal structure of the phosphorylase-heptulose 2-phosphate-oligosaccharide-AMP complex. J Mol Biol. 1990 Feb 5;211(3):645–661. doi: 10.1016/0022-2836(90)90271-M. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kiel J. A., Boels J. M., Beldman G., Venema G. Glycogen in Bacillus subtilis: molecular characterization of an operon encoding enzymes involved in glycogen biosynthesis and degradation. Mol Microbiol. 1994 Jan;11(1):203–218. doi: 10.1111/j.1365-2958.1994.tb00301.x. [DOI] [PubMed] [Google Scholar]
- Kirsch J. F., Eichele G., Ford G. C., Vincent M. G., Jansonius J. N., Gehring H., Christen P. Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. J Mol Biol. 1984 Apr 15;174(3):497–525. doi: 10.1016/0022-2836(84)90333-4. [DOI] [PubMed] [Google Scholar]
- Murzin A. G. Can homologous proteins evolve different enzymatic activities? Trends Biochem Sci. 1993 Nov;18(11):403–405. doi: 10.1016/0968-0004(93)90132-7. [DOI] [PubMed] [Google Scholar]
- Neidhart D. J., Kenyon G. L., Gerlt J. A., Petsko G. A. Mandelate racemase and muconate lactonizing enzyme are mechanistically distinct and structurally homologous. Nature. 1990 Oct 18;347(6294):692–694. doi: 10.1038/347692a0. [DOI] [PubMed] [Google Scholar]
- Rath V. L., Fletterick R. J. Parallel evolution in two homologues of phosphorylase. Nat Struct Biol. 1994 Oct;1(10):681–690. doi: 10.1038/nsb1094-681. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994 May;19(1):55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Sprang S. R., Madsen N. B., Withers S. G. Multiple phosphate positions in the catalytic site of glycogen phosphorylase: structure of the pyridoxal-5'-pyrophosphate coenzyme-substrate analog. Protein Sci. 1992 Sep;1(9):1100–1111. doi: 10.1002/pro.5560010904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang S. R., Withers S. G., Goldsmith E. J., Fletterick R. J., Madsen N. B. Structural basis for the activation of glycogen phosphorylase b by adenosine monophosphate. Science. 1991 Nov 29;254(5036):1367–1371. doi: 10.1126/science.1962195. [DOI] [PubMed] [Google Scholar]
- Tomaschewski J., Gram H., Crabb J. W., Rüger W. T4-induced alpha- and beta-glucosyltransferase: cloning of the genes and a comparison of their products based on sequencing data. Nucleic Acids Res. 1985 Nov 11;13(21):7551–7568. doi: 10.1093/nar/13.21.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrielink A., Rüger W., Driessen H. P., Freemont P. S. Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 1994 Aug 1;13(15):3413–3422. doi: 10.1002/j.1460-2075.1994.tb06646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990 Mar;8(1):52-6, 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- Wilmanns M., Hyde C. C., Davies D. R., Kirschner K., Jansonius J. N. Structural conservation in parallel beta/alpha-barrel enzymes that catalyze three sequential reactions in the pathway of tryptophan biosynthesis. Biochemistry. 1991 Sep 24;30(38):9161–9169. doi: 10.1021/bi00102a006. [DOI] [PubMed] [Google Scholar]
- Withers S. G., Madsen N. B., Sprang S. R., Fletterick R. J. Catalytic site of glycogen phosphorylase: structural changes during activation and mechanistic implications. Biochemistry. 1982 Oct 12;21(21):5372–5382. doi: 10.1021/bi00264a039. [DOI] [PubMed] [Google Scholar]
- Withers S. G., Madsen N. B., Sykes B. D., Takagi M., Shimomura S., Fukui T. Evidence for direct phosphate-phosphate interaction between pyridoxal phosphate and substrate in the glycogen phosphorylase catalytic mechanism. J Biol Chem. 1981 Nov 10;256(21):10759–10762. [PubMed] [Google Scholar]






