Abstract
Plant vacuoles play critical roles in development, growth and stress responses. In mature cells, vacuolar membranes (VMs) display several types of structures, which are formed by invagination and folding of VMs into the lumenal side and can gradually move and change shape. Although such VM structures are observed in a broad range of tissue types and plant species, the molecular mechanism underlying their formation and maintenance remains unclear. Here, we report that a novel HEAT-repeat protein, SHOOT GRAVITROPISM6 (SGR6), of Arabidopsis is involved in the control of morphological changes and dynamics of VM structures in endodermal cells, which are the gravity-sensing cells in shoots. SGR6 is a membrane-associated protein that is mainly localized to the VM in stem endodermal cells. The sgr6 mutant stem exhibits a reduced gravitropic response. Higher plants utilize amyloplast sedimentation as a means to sense gravity direction. Amyloplasts are surrounded by VMs in Arabidopsis endodermal cells, and the flexible and dynamic structure of VMs is important for amyloplast sedimentation. We demonstrated that such dynamic features of VMs are gradually lost in sgr6 endodermal cells during a 30 min observation period. Histological analysis revealed that amyloplast sedimentation was impaired in sgr6. Detailed live-cell imaging analyses revealed that the VM structures in sgr6 had severe defects in morphological changes and dynamics. Our results suggest that SGR6 is a novel protein involved in the formation and/or maintenance of invaginated VM structures in gravity-sensing cells.
Keywords: Arabidopsis, Gravitropism, HEAT-repeat protein, Vacuolar membrane
Introduction
The plant vacuole is a single membrane-bound organelle required for many physiological functions, including development, growth, senescence and defense response. In mature plant cells, the vacuole occupies 80–90% of the cell volume and harbors intricate structures that are formed by invagination of the outer boundary of the vacuolar membrane (VM) into the lumenal side (Oda et al. 2009). These structures continuously change their shape and dynamically move within the lumen. Transvacuolar strands (TVSs), one of the well-characterized dynamic VM structures, surround a relatively thick cytoplasm and actin cables, and penetrate the vacuole. TVSs are thought to function as a route for moving the organelles and cellular components. Bulbs and sheets are double-membrane structures that sandwich a thin cytoplasm, and several models have been proposed to explain how these bulbs are formed (Saito et al. 2002, Uemura et al. 2002). The bulbs and bulb-like structures are observed in cells of a broad range of plant species (Escobar et al. 2003, Vera-Estrella et al. 2004, Reisen et al. 2005, Mohanty et al. 2009). In epidermal cells of the Arabidopsis cotyledon, the number of bulbs decreases as the cell grows, suggesting that the bulbs function as a membrane and protein reservoir for rapidly expanding cells (Saito et al. 2002). However, the precise roles and conformational mechanisms of dynamic VM structures such as bulbs and sheets remain uncertain.
Invaginated VM structures and dynamic features of the VM play critical roles in the gravity-sensing process in gravitropism of the Arabidopsis inflorescence stem. Here, we use the term ‘invaginated VM structures’ to represent complex and flexible structures of the VM containing bulbs, internal sheets and TVSs. The endodermis in the stem is the primary gravity-sensing tissue, and cells contain a number of amyloplasts, which are starch-accumulating plastids. Molecular genetic analyses of Arabidopsis shoot gravitropism (sgr) mutants have indicated that sedimentation of amyloplasts in the direction of gravity is an important process during gravity sensing (Morita et al. 2002, Yano et al. 2003, Silady et al. 2004, Nakamura et al. 2011). Most of the volume of an endodermal cell is occupied by a large central vacuole. Amyloplasts are surrounded by the VM one by one or in clusters. However, VM structures and dynamics in endodermal cells of sgr2, sgr3, zigzag (zig)/sgr4 and gravitropism defective (grv) 2/sgr8/katamari (kam) 2 are quite different from those in the wild type (WT). Amyloplasts are not surrounded by VM and do not sediment in the direction of gravity in these mutants (Morita et al. 2002, Yano et al. 2003, Silady et al. 2004). SGR2 is a putative phosphatidic acid-preferring phospholipase A1 that localizes to the VM and unidentified compartments (Kato et al. 2002). SGR3 and ZIG/SGR4 are Qa-SNARE SYP22/VAM3 and Qb-SNARE VTI11, respectively (Kato et al. 2002, Yano et al. 2003). SYP22/VAM3 is localized to the vacuole, whereas VTI11 is localized to the trans-Golgi network (TGN), pre-vacuolar compartment (PVC) and vacuole (Sato et al. 1997, Saito and Ueda 2009). These two proteins form a SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex and mediate membrane fusion between the vesicle and the targeted organelle during membrane trafficking from the TGN to the PVC/vacuole. GRV2/SGR8/KAM2 is a homolog of receptor-mediated endocytosis 8 involved in endocytosis and retrograde transport from the endosomes to the Golgi in cells of Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens (Zhang et al. 2001, Chang et al. 2004, Fujibayashi et al. 2008, Shi et al. 2009). These studies have demonstrated that normal membrane trafficking to the vacuole and vacuolar formation are important for amyloplast sedimentation in gravity-sensing cells. Live-cell imaging of WT endodermal cells using vertical stage confocal microscopy has revealed invaginated VM structures and VM surrounding amyloplasts, which change their morphology concurrently with movement of the amyloplasts (Saito et al. 2005, Hashiguchi et al. 2013). Moreover, some amyloplasts actively move not only downward but also upward; however, the majority of amyloplasts are located on the lower side of endodermal cells (Saito et al. 2005, Nakamura et al. 2011). In contrast, no invaginated VM structures are observed in endodermal cells of zig mutants, and amyloplasts rarely move (Saito et al. 2005, Hashiguchi et al. 2013). These results suggest that the formation/maintenance of invaginated VM structures is important for amyloplast movement, including sedimentation.
The sgr6-1 mutant has a phenotype for gravitropism of the inflorescence stem but not for that of the root and hypocotyl (Yamauchi et al. 1997). Here, we show that the gene responsible for the sgr6 mutant is At2g36810 that encodes the HEAT (Huntingtin, Elongation factor 3, A-subunit of protein phosphatase 2A and TOR1) protein with an unknown function. Tissue-specific expression analysis of SGR6 indicated that SGR6 functions in the endodermis for shoot gravitropism. Interestingly, live-cell imaging of sgr6 endodermal cells demonstrated that most central vacuoles scarcely have invaginated VM structures and that amyloplasts have simultaneously lost their active movement. Moreover, amyloplast sedimentation in the direction of gravity was disrupted in the sgr6-1 living stem. Biochemical and subcellular localization analyses of SGR6 indicated that the protein is mainly localized to the VM in endodermal cells. These results suggest that SGR6 is a novel protein that is involved in the formation and/or maintenance of invaginated VM structures in gravity-sensing cells of Arabidopsis.
Results
sgr6 mutant phenotype
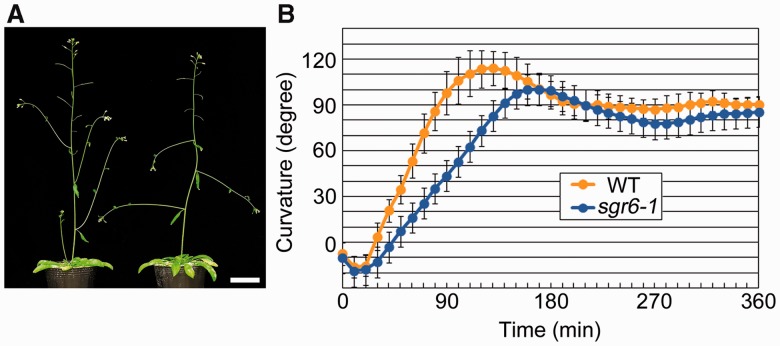
The sgr6-1 mutant has no obvious morphological abnormalities, except for lateral shoots that elongate horizontally (Fig. 1A). When the inflorescence stem of the WT is gravity stimulated by placing it horizontally, the stem bends 90° upward within 90 min (Fig. 1B). In contrast, the sgr6-1 stem requires 140 min to bend 90° (Fig. 1B), indicating that the sgr6-1 mutant stem exhibits a slower gravitropic response than the WT stem. Because the gravitropic response requires organ growth, we next examined stem growth. The sgr6-1 stem grew as well as the WT stem (Supplementary Fig. S1), indicating that the sgr6-1 gravitropic phenotype is not caused by a growth defect. It has been reported that the sgr6-1 mutant has no significant phenotype for root and hypocotyl gravitropism (Yamauchi et al. 1997). In addition, the sgr6-1 stem and hypocotyl show the same bending pattern as the WT stem and hypocotyl in response to unilateral light, suggesting that sgr6-1 has the ability to grow asymmetrically (Yamauchi et al. 1997).
Fig. 1.
Phenotypes of the shoot gravitropism 6 (sgr6)-1 mutant. (A) Six-week-old plants of the wild type (left) and sgr6-1 (right). Scale bar = 3 cm. (B) Gravitropic phenotype of the wild type (yellow circle) and sgr6-1 (blue circle). Twenty individuals of each genotype were examined. Bars represent the SD.
SGR6 is At2g36810
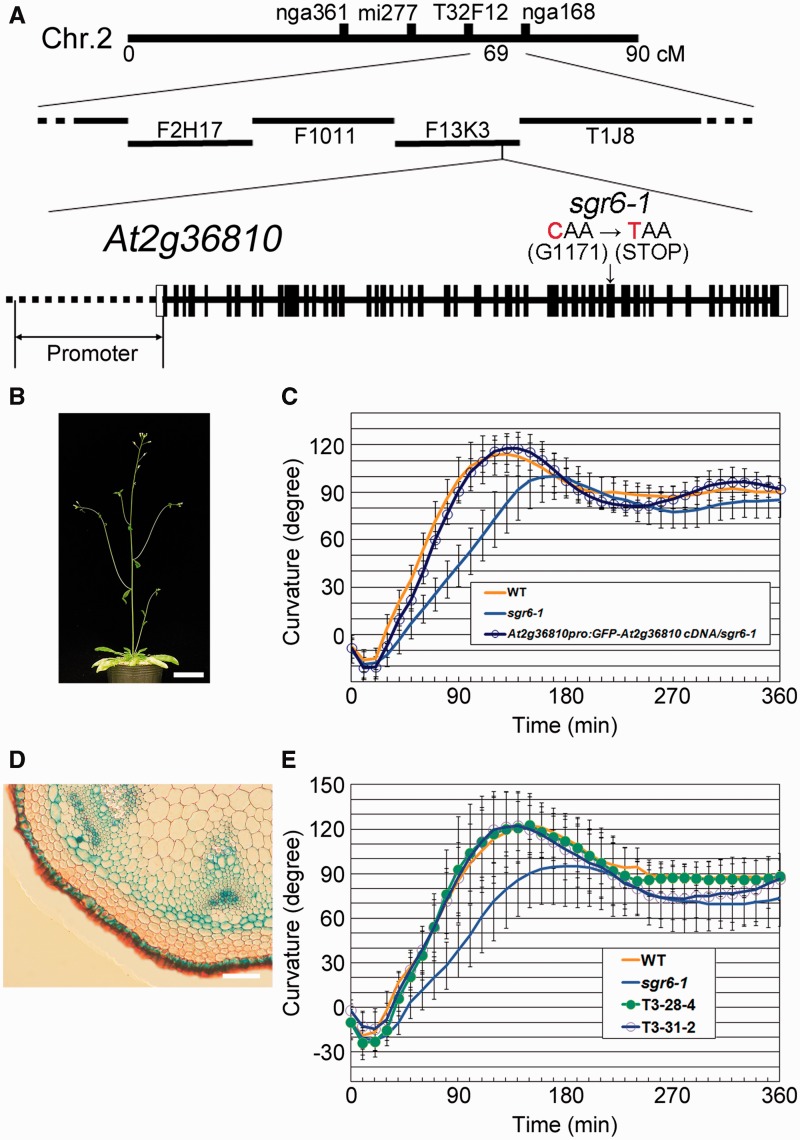
F2 progeny derived from a cross between sgr6-1 [Columbia (Col-0)] and Landsberg erecta (Ler) were used to map SGR6 roughly to chromosome II. After analyzing 446 independent F2 progeny, the locus was narrowed to a region between a polymorphic marker on bacterial artificial chromosome (BAC) F13K3 and that on BAC T1J8 (Fig. 2A). We found a single C to T substitution in the open reading frame of At2g36810, resulting in a nonsense mutation at glutamine (Q1171). A 5.1 kb WT At2g36810 cDNA fragment was cloned and fused with green fluorescent protein (GFP) cDNA at its N-terminal end. The fused gene was inserted downstream of the putative promoter fragment (2.9 kb) of At2g36810 and introduced into sgr6-1 plants for a complementation test. The stem of the resulting transgenic plant (SGR6pro:GFP-SGR6/sgr6-1) exhibited a normal lateral shoot growth direction and the primary shoot gravitropic response had kinetics similar to those of the WT (Fig. 2B, C).
Fig. 2.
SHOOT GRAVITROPISM 6 (SGR6) is At2g36810. (A) The SGR6 locus was mapped between the T32F12 and nga168 cleaved amplified polymorphic sequence markers. A nonsense mutation was found at G1171 in the open reading frame (ORF) of At2g36810 in the sgr6-1 mutant. The At2g36810 gene structure is schematically shown; exons are indicated by boxes (white boxes, untranslated regions; black boxes, coding regions) and introns are indicated by lines between boxes. The length of At2g36810, including the promoter region, was approximately 17 kb. (B) Six-week-old sgr6-1 T3 plants harboring the DNA fragments of the At2g36810 putative promoter region and the green fluorescent protein (GFP)-fused cDNA derived from the ORF of At2g36810. Scale bar = 3 cm. (C) Gravitropic phenotype of the plant shown in (B) (blue open circle). Twenty individuals of each genotype were examined. Bars represent the SD. (D) SGR6 expression pattern in inflorescence stems. Scale bar = 100 µm. (E) Gravitropic phenotype of two independent sgr6-1 T3 lines harboring the SCRpro:GFP-SGR6 DNA fragment. At least 14 individuals of each genotype were examined. Bars represent the SD.
We also isolated another three sgr6 alleles through several independent mutant screening procedures, such as sgr6-2 (Col-0 background), sgr6-3 and sgr6-4 (Ler background) (Supplementary Fig. S2). sgr6-2 had a G to TT substitution at the junction of the second intron and the third exon, causing a frameshift and a stop codon at asparagine (A45) (Supplementary Fig. S3). sgr6-3 had a single C to T substitution, resulting in a nonsense mutation at glutamine (Q1547) (Supplementary Fig. S3). sgr6-4 had an ATTTCT to CC substitution that caused a frameshift at serine (S220), resulting in substitutions of eight amino acids and a stop codon (Supplementary Fig. S3). Immunoblot analysis revealed that the SGR6 protein was not detected in sgr6-1, sgr6-2 or sgr6-4 (Supplementary Fig. S4), suggesting that sgr6-1, sgr6-2 and sgr6-4 are null alleles. In contrast, sgr6-3 produced a truncated form of SGR6, as predicted by its molecular lesion (Supplementary Fig. S4). The greatly reduced amount of protein in sgr6-3 suggested that the mutation may have affected the protein stability or protein synthesis efficiency. The sgr6-3 gravitropic response was comparable with that of sgr6-4 (Supplementary Fig. S2, 2F), suggesting that sgr6-3 is also a loss-of-function mutation. Taken together, these results demonstrate that SGR6 is At2g36810.
At2g36810 is a single gene in the Arabidopsis genome that encodes a protein of 1,703 amino acids with no functional or structural domain, except for 10 HEAT repeats (http://pfam.sanger.ac.uk/) (Supplementary Fig. S5A). As a result of a BLAST search (query: full-length amino acidic sequence of SGR6), SGR6 homologous genes were found in genomes of several animals and plants (Supplementary Fig. S5B, C); however, no homolog was found in yeast.
SGR6 functions in the endodermis for shoot gravitropism
Northern blot analysis suggested that SGR6 expression in roots, hypocotyls and stems was higher than that in other organs (Supplementary Fig. S6B). Moreover, reverse transcription–PCR (RT–PCR) analysis using total RNA derived from various organs of WT plants suggested that SGR6 was expressed in all organs examined, although the expression levels varied (Supplementary Fig. S6A). Transgenic plants that expressed β-glucuronidase (GUS) under the control of the SGR6 promoter were generated to investigate the SGR6 expression pattern at the tissue level. The SGR6 promoter showed strong activity in the root cap of primary roots, the vascular tissues of roots, hypocotyls and cotyledons, and the guard cells of cotyledons (Supplementary Fig. S6C–G). In the stem of the transgenic plants, SGR6 promoter activity was observed in the epidermis, endodermis, phloem, developing metaxylem and interfascicular cells (Fig. 2D). Because the endodermis is the major site for gravity sensing in the stem, we next investigated the relationship between expression in the endodermis and function of SGR6 during shoot gravitropism. Two independent T3 transgenic plants that expressed GFP–SGR6 under the control of the endodermis-specific SCARECROW (SCR) promoter exhibited a WT-like gravitropic response (Fig. 2E), indicating that the function of SGR6 in the endodermis is sufficient for a normal gravitropic response and that SGR6 functions in the endodermis for shoot gravitropism.
SGR6 is involved in amyloplast sedimentation in endodermal cells
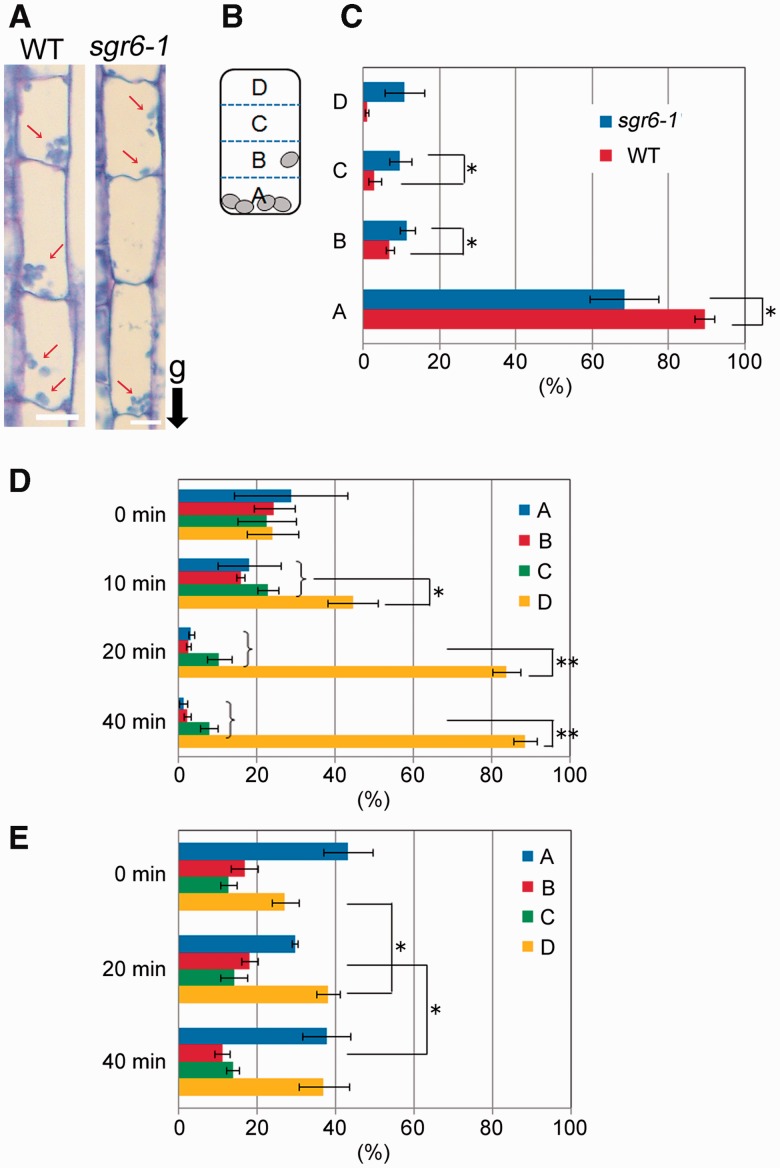
Endodermal cells in stems contain a number of amyloplasts, and sedimentation of amyloplasts in the direction of gravity is important for gravity sensing. First, we used a histological method to evaluate amyloplast sedimentation. The stem segments were fixed with the direction of gravity maintained constant, embedded and sectioned (Fig. 3A). An endodermal cell was partitioned into four regions for quantitative analysis (Fig. 3B), and the number of amyloplasts in each region was counted for 50 cells. Almost all amyloplasts in WT endodermal cells were localized at block A, the bottom side of the cell (Fig. 3C). In contrast, the ratio of amyloplasts in block A of sgr6-1 endodermal cells was significantly lower and the ratios in blocks B and C were higher than those of the WT.
Fig. 3.
Amyloplast sedimentation in endodermal cells. (A) Inflorescence stems (2–3 cm below the shoot apex) of wild type and shoot gravitropism 6 (sgr6)-1 mutant plants were cut and fixed with the direction of gravity maintained. Images are endodermal cells in longitudinal sections of cut stems. White arrows indicate amyloplasts. ‘g’ and a black arrow indicate the direction of gravity. (B) An endodermal cell was partitioned into four blocks for quantitative analysis (A, B, C and D from the lowest). (C) The number of amyloplasts in each block of 50 cells was counted, and the ratio of the number was calculated. Three individual stems of each genotype were analyzed. Bars represent the SDs. Asterisks indicate P-values of Student’s t-test (*P < 0.05). (D, E) Wild-type (D) and sgr6-1 (E) plants were inverted and incubated for 0, 10, 20 or 40 min. After the incubation, the stems were cut and fixed with the direction of gravity maintained. The ratio of amyloplasts was calculated as in (C). Three individual stems of each genotype were analyzed. Bars represent the SDs. Asterisks indicate P-values of Student’s t-test (*P < 0.05; **P < 0.01).
We inverted the plants at an angle of 180° to investigate amyloplast sedimentation using this method. The uppermost region in the endodermal cells of inverted plants was A, while the lowermost region was D. After inversion, the stems were immediately cut and fixed with the new gravity direction maintained constant. Amyloplasts were almost equally localized over four blocks in the endodermal cells of inverted WT stems (Fig. 3D, 0 min). Following this, the stems were incubated for 10, 20 or 40 min in the dark after inversion, cut and fixed. The ratios of block A, B and C gradually decreased in the WT as the incubation time increased, whereas the ratio of block D increased (Fig. 3D). In contrast, the ratio of each block in sgr6-1 did not change dramatically, as seen in the WT (Fig. 3E). This result suggests that SGR6 is involved in amyloplast sedimentation in the direction of gravity.
SGR6 is mainly localized to the VM in endodermal cells
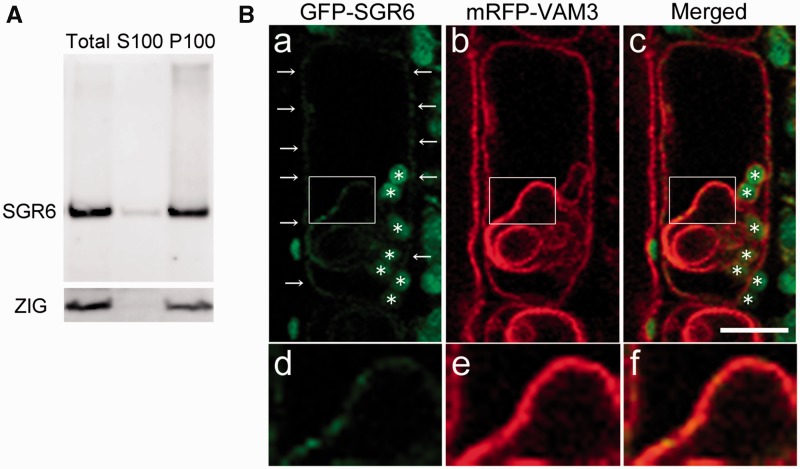
According to SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/), SGR6 is predicted to have no transmembrane domain. Moreover, SGR6 has no signal sequence (SignalP 4.1 Server, http://www.cbs.dtu.dk/services/SignalP-4.1/) and would not enter the lumenal side of organelles. A fractionation experiment revealed that most SGR6 existed in the membrane fraction (Fig. 4A). We next examined the SGR6 solubilization conditions. We found that SGR6 was not solubilized in high salt, urea or alkaline buffer at room temperature (Supplementary Fig. S7). However, part of SGR6 was solubilized by 1% Triton X-100 or 2% SDS at room temperature. These results suggest the possibility that SGR6 is rigidly associated with the membrane.
Fig. 4.
SHOOT GRAVITROPISM 6 (SGR6) is mainly localized to the vacuolar membrane (VM). (A) Subfractionation by ultracentrifugation. ZIG was detected as the control of the integral membrane protein. (B) Intracellular localization of SGR6. The endodermal cells in stems of SGR6pro:GFP-SGR6 and VAM3pro:mRFP-VAM3/sgr6-1 vam3-1 were observed. Green, green fluorescent protein (GFP) fluorescence; red, red fluorescent protein (RFP) fluorescence. Asterisks indicate autofluorescence of Chl from plastids. d, e and f are insets in a, b and c, respectively. Scale bar = 10 µm.
We first observed the endodermal cells of SGR6pro:GFP-SGR6/sgr6-1, the transgenic plant generated for the complementation test, to investigate which membrane existed in SGR6. The GFP signal was spotty along the periphery of the cells (Supplementary Fig. S8A, white arrows) and circular within the cells (Supplementary Fig. S8A, blue arrow). Moreover, relatively large dot-like signals occasionally emerged during the observations (Supplementary Fig. S8A, yellow arrow). Because Chl autofluorescence from amyloplasts was detected outside the peripheral GFP signal (Supplementary Fig. S8B, white arrow), GFP–SGR6 did not seem to exist on the plasma membrane. Moreover, it was difficult to determine whether GFP–SGR6 existed near the amyloplasts owing to Chl autofluorescence from amyloplasts.
We crossed SGR6pro:GFP-SGR6/sgr6-1 with several transgenic plants as follows: SCRpro:mRFP-er [the endoplasmic reticulum (ER) localization signal sequence]/WT (a marker line for the ER), SCRpro:ARA6-mRFP/WT (a marker line for the ARA6-positive endosome), SCRpro: mRFP-ARA7/WT (a marker line for the ARA7-positive endosome) and VAM3pro:mRFP-VAM3/vam3-1 (a marker line for the vacuole) (Uemura et al. 2010) and generated plants that harbored homozygous forms of the transgenes and the sgr6-1 mutation.
We first observed the endodermal cells of a plant derived from a cross between SGR6pro:GFP-SGR6/sgr6-1 and VAM3pro:mRFP-VAM3/vam3-1. The GFP–SGR6 signals along the cell periphery overlapped with those of monomeric red fluorescent protein (mRFP)–VAM3 (Fig. 4B, a–c). Moreover, the GFP signal within the cells merged with the invaginated structure of VM (Fig. 4B, d–f). Therefore, the circular form of the GFP–SGR6 signal shown in Supplementary Fig. S8A was thought to exist along the invaginated structure of the VM. The endodermal cell shown in Supplementary Fig. S9 is 30 s after the cell in Fig. 4B. A large dot-like SGR6 GFP signal was observed in the cell and merged with the VAM3 mRFP signal (Supplementary Fig. S9). THe ER exists adjacent to the VM in mature plant cells (Hanton and Brandizzi 2006). We next observed the endodermal cells of plants derived from a cross between SGR6pro:GFP-SGR6/sgr6-1 and SCRpro:mRFP-er/WT. Peripheral and circular GFP signals were observed in the endodermal cells; however, the signals did not merge with the mRFP signal from the ER (Supplementary Fig. S10). The VAM3/SYP22 signal that merged with that of SGR6 exists not only with the VM but also with the PVC (Sanderfoot et al. 1999). To examine whether the GFP–SGR6 dot-like signal was localized to the PVC, we observed endodermal cells of plants derived from a cross between SGR6pro:GFP-SGR6/sgr6-1 and SCRpro:ARA6-mRFP/WT or SCRpro: mRFP-ARA7/WT. However, the dot-like signal did not merge with such an organelle marker (Supplementary Figs. S11, S12). These results suggest that GFP–SGR6 is mainly localized to the VM.
SGR6 maintains invaginated VM structures in endodermal cells of the cut stem
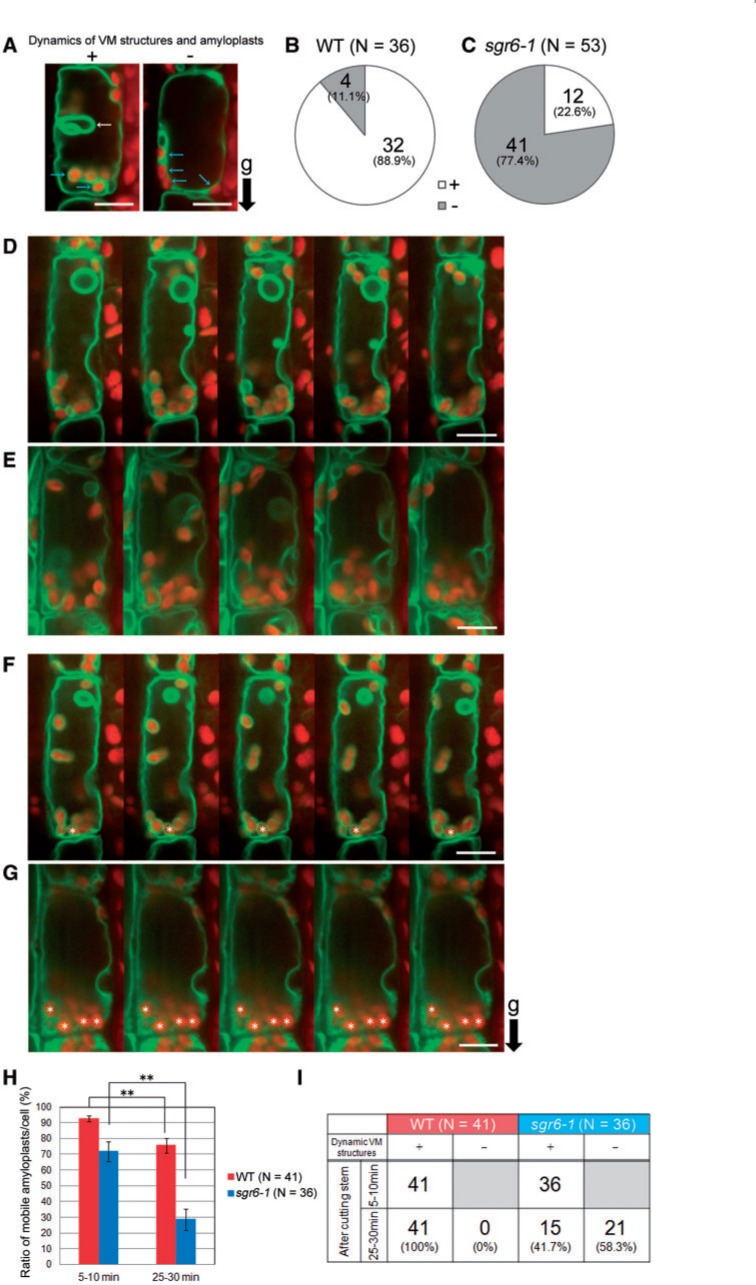
Based on the localization to the VM, we assumed that SGR6 is involved in amyloplast sedimentation through modulating the VM function(s). Consistently, genetic analyses of several sgr mutants and zig suppressor mutants have revealed that flexible VM structures are important for amyloplast sedimentation (Niihama et al. 2005, Niihama et al. 2009, Hashiguchi et al. 2010). We next observed the endodermis of a transgenic plant that expresses GFP–tonoplast intrinsic protein-γ (γ-TIP), a marker for lytic vacuoles, under control of the SCR promoter (Saito et al. 2005). A longitudinally cut stem segment was allowed to stand for 15 min on a vertical stage, and observations of each sample were finished within the next 15 min. Two types of endodermal cells were observed in both genotypes at a time point within 5–10 min after stem cutting, i.e. cells with invaginated VM structures and moving amyloplasts and those with simple VM structures and static amyloplasts (Fig. 5A; Supplementary Movies S1, S2). VMs in approximately 90% of WT endodermal cells showed invaginated structures, whereas they were observed in only 20% of sgr6-1 endodermal cells at the same time point (Fig. 5B, C). Amyloplasts rarely moved in the endodermal cells of both genotypes with no invaginated VM structures. Approximately 80% of sgr6-1 endodermal cells harbored mobility-less vacuoles and amyloplasts, and such a status of the vacuoles and amyloplasts lasted until 30 min after cutting the stem (data not shown). Following this, we focused on the endodermal cells containing invaginated VM structures and amyloplasts during a 2 min time window within 5–10 min after cutting the stem (Fig. 5D, E; Supplementary Movies S3, S4). Subsequently, the samples were held on the vertical stage and observed during a 2 min time window within 25–30 min after cutting the stems (Fig. 5F, G; Supplementary Movies S5, S6). The ratio of mobile amyloplasts per cell at 25–30 min after cutting the WT stem decreased significantly compared with that at 5–10 min after cutting (Fig. 5H). However, all endodermal cells maintained invaginated VM structures in the WT stem (Fig. 5I). In contrast, the ratio of mobile amyloplasts per cell in sgr6-1 decreased drastically (Fig. 5H), and approximately 60% of endodermal cells lost invaginated VM structures 25–30 min after stem cutting (Fig. 5I), indicating that VM structure in sgr6-1 is sensitive to the sample preparation process including stem cutting.
Fig. 5.
Dynamic vacuolar membrane (VM) structures and amyloplast dynamics in endodermal cells of cut stems. The endodermal cells of the transgenic plant expressing green fluorescent protein–tonoplast intrinsic protein-γ (GFP–γ-TIP), a lytic vacuole marker, under control of the SCARECROW (SCR) promoter, were observed using a vertical microscope. Green, GFP fluorescence; red, autofluorescence of Chl from plastids. (A) Endodermal cells that show or do not show invaginated VM structures and amyloplast dynamics at a time point within 5–10 min after cutting the stem. (B, C) Number of endodermal cells that show or do not show invaginated VM structures at a time point within 5–10 min after cutting the stem of the wild type (B) and sgr6-1 (C). At least eight stem segments of each genotype were observed. (D, E) Confocal images of wild-type (D) and sgr6-1 (E) endodermal cells observed for 2 min within 5–10 min after cutting the stem were aligned at approximately 30 s intervals. (F, G) Confocal images of wild-type (F) and sgr6-1 (G) endodermal cells observed for 2 min within 25–30 min after cutting the stem were aligned at approximately 30 s intervals. (H) The ratio of amyloplasts that moved along the x- or y-axis or that rotated during the 2 min observation. Endodermal cells that harbor invaginated VM structures at the starting point of observation within 5–10 min after cutting the stem were analyzed. At least 16 stem segments of each genotype were observed. Statistical differences between 5–10 min and 25–30 min were detected using Student’s t-test (**P < 0.01). Bars represent the SEs. (I) Number of endodermal cells that maintained invaginated VM structures during the 30 min observation. ‘g’ and a black arrow indicate the direction of gravity. Scale bars = 10 µm.
To observe the change in VM structures over time in more detail, the endodermal cells containing invaginated VM structures were placed horizontally and observed by spinning disk confocal microscopy. The 4D imaging of endodermal cells 10–20 min after cutting the stem showed that several VM structures were newly formed within the lumen (Fig. 6A, blue arrow) and changed shape (Fig. 6A, yellow arrows). Invaginated VM structures were emerged and changed shape one after another during the observations (Supplementary Movie S7). In contrast, sgr6-1 VM structures were quite different from those of the WT, i.e multiple layers of VMs were juxtaposed to each other (Fig. 6B, white arrows). VM structures and amyloplasts seemed gradually to lose their flexibility and mobility during the observation in sgr6-1 (Supplementary Movie S8). Moreover, VM structures with less motility gradually changed into numerous vesicular-like VM structures 20–30 min after cutting the stem (Fig. 6C, yellow arrows, Supplementary Movies S9, S10), whereas wild-type VM structures 30 min after cutting the stem differ little from those 10–20 min after cutting (data not shown). These results indicate that sgr6-1 affects the proper formation and morphological changes in VM structures and causes aberrant vesicular-like VM structures. Taken together, we conclude that SGR6 may act on formation and/or maintenance of invaginated VM structures and indirectly modulates amyloplast dynamics in endodermal cells.
Fig. 6.
Four-dimensional imaging of vacuolar membrane (VM) structures in endodermal cells. The endodermal cells of the transgenic plant expressing green fluorescent protein–tonoplast intrinsic protein-γ (GFP–γ-TIP), a lytic vacuole marker, under control of the SCR promoter, were observed using a spinning disk confocal microscopy. (A) Wild-type cell. Confocal images taken at 20 s intervals within 10–20 min after cutting the stem are aligned. Blue and yellow arrows indicate newly formed VM structures and dramatically changing VM structures, respectively. (B) sgr6-1 cells. Confocal images taken at 25 s intervals within 10–20 min after cutting the stem are aligned. White arrows indicate abnormally folded VM structures. (C) sgr6-1 cell. Confocal image taken 30 min after cutting the stem. Numerous vesicle-like VM structures have emerged. Yellow arrows indicate aberrant vesicle-like VM structures. Green, GFP; red, autofluorescence of Chl from plastids. Scale bars = 10 µm.
Discussion
How is SGR6 involved in shoot gravitropism?
Tissue-specific expression analysis indicated that SGR6 functions in the endodermis for shoot gravitropism (Fig. 2E). Then, how is SGR6 involved in the gravitropic response of the inflorescence stem in endodermal cells? Amyloplasts are hypothesized to be ‘statoliths’ or gravity susceptors that elicit a gravitropic response during the earliest part of gravitropism (Sack 1997). In agreement with this hypothesis, most sgr mutants reported to date have common defects in endodermal cell amyloplast sedimentation (Morita et al. 2002, Yano et al. 2003, Silady et al. 2004, Nakamura et al. 2011). In an attempt to clarify whether SGR6 is also related to amyloplast sedimentation, we first demonstrated that sgr6-1 endodermal cells were deficient in amyloplast sedimentation by histological analysis (Fig. 3E). In addition, vertical stage confocal microscopy showed that amyloplasts were not dynamic in approximately 80% of sgr6-1 endodermal cells 5–10 min after cutting the stem (Fig. 5C). This result suggests that SGR6 is involved in shoot gravitropism through amyloplast sedimentation in endodermal cells. Expression analysis of SGR6 indicated that the SGR6 promoter is active in various tissues including the guard cells of cotyledons (Supplementary Fig. S6G), though the GFP signal was not found in cells other than the endodermal cells of SGR6pro:GFP-SGR6/sgr6-1 under our observation conditions (data not shown). Although no obvious phenotype other than gravitropism was found in sgr6, it is possible that SGR6 may have some physiological roles under particular conditions such as environmental stresses.
What influences amyloplast sedimentation in sgr6-1?
Invaginated VM structures and actin microfilaments (AFs) are two major intracellular components that affect amyloplast dynamics in endodermal cells. The central vacuoles in endodermal cells continuously change morphologically (Saito et al. 2005, Hashiguchi et al. 2013). Part of the VM surrounds amyloplasts, and the flexible morphological changes in VM appear to be coupled with amyloplast movement. Together with analyses of several sgr mutants, it is obvious that such dynamic VM structural flexibility is important for amyloplasts to move freely and sediment in the direction of gravity. The interaction between amyloplasts and AFs also influences amyloplast movement in endodermal cells. Appropriate dissociation of amyloplasts from AFs due to SGR9 activity is important for normal amyloplast sedimentation (Nakamura et al. 2011).
Then, what influences amyloplast sedimentation in sgr6-1? Invaginated VM structures were maintained during 30 min of observation in cut stem samples from almost all WT endodermal cells (Fig. 5I). In contrast, most sgr6-1 endodermal cells did not maintain invaginated VM structures, and only 20% of cells showed flexibility of VM 5–10 min after cutting the stem (Fig. 5C). However, such cells lost their VM dynamics in the next 20–25 min (Fig. 5I). Together with the fact that SGR6 is a membrane-associated protein mainly localized to VMs (Figs. 3, 4), sgr6-1 may influence the formation/maintenance of invaginated VM structures in endodermal cells of living stems. Taken together, a decrease in the number of changes in the VM structure caused by SGR6 loss of function may affect amyloplast dynamics in endodermal cells, resulting in reduced shoot gravitropism.
Is it possible that AFs in endodermal cells are affected by sgr6-1? A mutation in Arabidopsis ACTIN-RELATED PROTEINS (ARP) 2, an important regulator of actin organization, causes vacuole fragmentation in trichome cells (Mathur et al. 2003). Moreover, treatment with latrunculin B (LatB), an inhibitor of actin polymerization, phenocopies the arp2 mutant (Mathur et al. 2003). LatB treatment also induces severe defects in vacuolar morphology and VM dynamics in lily pollen tubes (Ovečka et al. 2005). Thus, normal morphology and membrane motility of the vacuole require AFs. Thus, it is possible that sgr6-1 influences AFs localized to the VM periphery, causing the abnormality in invaginated VM structures in endodermal cells. Further investigation will be required to clarify the relationship between AFs and SGR6 function.
SGR6 molecular function
We noticed the unique behavior of SGR6 through protein analyses. Immunoblot analyses demonstrated that a portion of the SGR6 protein accumulated between stacking and running SDS–polyacrylamide gels (data not shown). When protein extracts mixed with Laemmli sample buffer were incubated overnight at room temperature instead of boiling, most SGR6 protein was detected at the predicted molecular weight position (Supplementary Fig. S4). Moreover, once the samples were ultracentrifuged, SGR6 tended to accumulate on the top side of the gel (Supplementary Fig. S7). Thus, SGR6 was likely to aggregate as a result of heat, high concentration or physical force.
SGR6 exists in the membrane fraction but has no transmembrane domain. Moreover, SGR6 is not likely to have the amino acids for post-translational lipid modifications such as myristoylation, palmitoylation and prenylation (MYR Predictor, http://mendel.imp.ac.at/myristate/SUPLpredictor.htm; Myristoylator, http://au.expasy.org/tools/myristoylator/; CSS-Palm, http://csspalm.biocuckoo.org; PrePS, http://mendel.imp.ac.at/PrePS/), suggesting that SGR6 is peripherally associated with membranes. Sodium carbonate treatment under alkaline conditions removes peripheral membrane proteins from microsomal membranes. However, SGR6 was not solubilized by alkaline buffer (Supplementary Fig. S7). Thus, SGR6 may have a specific membrane association characteristic.
SGR6 was predicted to have 10 HEAT repeats; however, no other conserved domain that could be used to speculate on the molecular function of SGR6 was found (Supplementary Fig. S5A). A single HEAT repeat unit is approximately 50 amino acid residues and repeatedly occurs in the protein. HEAT repeats are highly variable in length, amino acid sequence and three-dimensional structure, which makes it difficult to identify them by sequence comparison (Andrade et al. 2001). Therefore, a wider range of SGR6 than predicted could be actually occupied by a number of HEAT repeats. The HEAT motif is a tandem repeat sequence of HEAT repeats that forms elongated superhelices or solenoids and mediates protein–protein interactions (Andrade et al. 2001). HEAT proteins exist in all eukaryotic cells and are involved in a wide variety of cellular processes; however, it is quite difficult to predict the specific molecular function only on the basis of the HEAT motif. SGR6 had six predicted HEAT repeats in its C-terminal region of approximately 350 amino acids (Supplementary Fig. S5A). A truncated form of SGR6, which probably lacks the two HEAT repeats at its C-terminal end, was expressed in sgr6-3, an sgr6 allele (Supplementary Fig. S4). Moreover, SGR6 fused with GFP at its C-terminal end did not complement the sgr6-1 gravitropic phenotype (data not shown). If SGR6 requires protein–protein interaction(s) to function during gravitropism, the two HEAT repeats at its C-terminal end may be important for the interaction, although we cannot exclude the possibility that low SGR6 expression induces the sgr6-3 phenotype.
Detailed analyses of invaginated VM structures (Figs. 5, 6) suggest that SGR6 controls the proper formation of invaginated VM structures in the VM; loss of function of SGR6 leads to the deformation of VM structures and such deformed structures are unable to move as they do in the WT, and/or sgr6-1 causes abnormal VM dynamics and the less flexible VM cannot form properly. Together with the structural characteristics of SGR6, SGR6 may hold the interactor(s) on the VM to control the formation of the invaginated VM structure in endodermal cells. We found that SGR6 is a novel gene that may function in vacuolar dynamics in plant cells. Further analysis of SGR6 and its interactors may reveal new roles for vacuoles in plants.
Materials and Methods
Plant materials and growth conditions
The Col-0 or Ler accessions of A. thaliana (L.) Heynh. were used as the WT. sgr6-1 and sgr6-2 were isolated from an M2 population of Col-0 seeds mutagenized with ethyl methanesulfonate (EMS) and γ-rays, respectively. sgr6-3 and sgr6-4 were isolated from an M2 population of Ler seeds mutagenized with EMS and γ-rays, respectively.
Seeds were sterilized with 5% sodium hypochlorite solution and then plated on Murashige–Skoog plates at 4°C for 2 d. Seedlings were transplanted and grown on soil (mixture of vermiculite from Nittai and Metromix G550 and Sun Gro Horticulture covered with Ube Perlite Type I from Ube Kosan) under constant white light at 23°C.
Gravitropism assay
Intact 5-week-old plants, whose primary inflorescence stems were 5–8 cm in length, were horizontally placed under non-directional dim light at 23°C to examine the gravitropic response of the stems. Photographs were taken at the indicated times. The stem curvature was measured on digital images using ImageJ (http://rsb.info.nih.gov/ij/) as the angle formed between the growing direction of the apex and the horizontal baseline.
sgr6 mapping
We first isolated F2 progeny from a cross between Ler and sgr6-1, which contained a homologous sgr6-1 mutation and exhibited an obvious sgr6-1 phenotype. DNA was prepared from approximately 2,116 F2 progeny, and SGR6 was localized to chromosome II. We prepared cleaved amplified polymorphic sequence markers that recognized polymorphisms between Col and Ler for fine-scale mapping based on the information provided by The Arabidopsis Information Resource.
Molecular cloning and plant transformation
A 2.9 kb genomic DNA fragment of the At2g36810 putative promoter region was amplified by PCR (primer set: SGR6pro_F, SGR6pro_R) and cloned into pCR-BluntII-TOPO (Invitrogen) for complementation analysis. Total RNA was isolated from inflorescence stems using the RNeasy Plant Mini Kit (Qiagen). cDNAs were synthesized using SuperScript II reverse transcriptase (Invitrogen). SGR6 cDNA was amplified by PCR (primer set: cSGR6_F, cSGR6_R) using the synthesized cDNAs as templates and then cloned into the XbaI–SmaI site in the pHSG399 multicloning site. GFP was fused at the N-terminus of SGR6 cDNA on pHSG399. The resulting GFP-SGR6 fragment was cut and inserted into the SalI–SmaI site downstream of the putative At2g36810 promoter or the SCR promoter that was already subcloned into HindIII in the multicloning site of the pBI binary vector (Morita et al. 2002). The putative At2g36810 promoter fragment was amplified by PCR (primer set: promoter SGR6 F, promoter SGR6 R) and inserted into the SalI–BamHI site upstream of uidA in pBI101 for expression pattern analysis (Morita et al. 2002). The DNA sequence of each construct was confirmed by sequencing. The resulting constructs were transformed into Agrobacterium tumefaciens strain GV3101 (pMP90) and then introduced into Col-0 or sgr6-1 plants using the floral dip method (Clough and Bent 1998). T1 plants were selected for resistance against kanamycin (30 µg ml−1). The primer sequences used are indicated in Supplementary Table S1.
GUS staining
Tissues were fixed in 90% (v/v) ice-cold acetone for 30 min and incubated in GUS staining solution [100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.1% (v/v) TritonX-100 and 0.52 mg ml−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid] for 16 h at 37°C to detect GUS activity in the transgenic plants (SGR6pro:GUS) described above. After rinsing in 70% ethanol for 15 min, the tissues were cleared with a chloral hydrate–glycerol–water solution (8 g of chloral hydrate, 1 ml of glycerol and 2 ml of water) and observed for GUS staining under a light microscope. GUS-stained inflorescence stems were dehydrated in a graded ethanol series (30, 50, 70, 90 and 100%), embedded in Technovit 7100 (Heraeus Kulzery), and sectioned with a microtome for histological analyses.
Histological analyses
Stem segments (2–3 cm below the apex) were fixed, embedded in Technovit7100, and sectioned as described previously to analyze amyloplast localization (Morita et al. 2002). The plants were inverted at an angle of 180° and incubated for 0, 10, 20 or 40 min under non-directional dim light at 23°C to evaluate amyloplast sedimentation. Following this, the stems were cut and fixed with the direction of gravity maintained constant. The subsequent procedure was the same as that described above.
CLSM
Inflorescence stem segments (2–3 cm below the shoot apex) were excised from 5-week-old plants and sectioned longitudinally and manually. Fluorescence imaging of VM structures in endodermal cells with the direction of gravity maintained constant was performed as described previously (Nakamura et al. 2011) using a vertical stage confocal microscope (model BX50; Olympus) equipped with an oil-immersion objective (PlanApo N 60×/1.42NA, Olympus). Confocal images were taken at approximately 4 s intervals for 2 min and converted into a 32× movie. CLSM images were obtained (FV1000; Olympus) for subcellular localization analysis of SGR6. GFP and mRFP fluorescence was detected with 497–517 and 592–622 nm spectral settings, respectively, following 488 or 543 nm excitation. We used an all-in-one type confocal laser scanning microscope system, which includes a spinning disk-type scanner and Electron Multiplying CCD (CellVoyager CV1000; Yokogawa) for 4D imaging of VM structures in endodermal cells. GFP fluorescence and Chl autofluorescence were detected with 520–535 and 617–673 nm spectral settings, respectively, following 488 nm excitation. Twenty images of WT cells along the z-axis were obtained for 20 s, and imaging was repeated 30 times (10 min in total). Twenty-five images of sgr6-1 cells along the z-axis were obtained for 25 s, and imaging was repeated 25 times (10.4 min in total). The 20 and 25 images at each time point were combined, and the series of 30 and 25 images were converted into 120× and 125× movies, respectively, using ImageJ (http://rsb.info.nih.gov/ij/).
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Ministry of Education, Science, Sports, and Culture of Japan [a Grant-in-Aid for Scientific Research on Priority Areas (16085205 to M.T.M.)]; Japan Society for the Promotion of Science (JSPS) [Grant-in-Aid for JSPS Fellows (to Y.H.)]; Japan Science and Technology Agency [PRESTO project grant to M.T.M.].
Supplementary Material
Acknowledgments
We thank Shigeru Sakurai for useful advice on predicting the protein domains in SGR6, Masatoshi Yamaguchi and Yoshimi Nakano for interpretation of the SGR6 expression pattern in inflorescent stems, Yoichiro Fukao and Masayuki Fujiwara for valuable discussions about SGR6 biochemical analysis, and Ms. Seiko Ishihara, Ms. Nauko Inui and Ms. Kaori Kaminoyama for technical assistance. We dedicate this article to the memory of Daisuke Yano who passed away in Nara in November 2004.
Glossary
Abbreviations
- AF
actin microfilament
- CLSM
confocal laser scanning microscopy
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- HEAT
Huntingtin, Elongation factor 3, A-subunit of protein phosphatase 2A and TOR1
- mRFP
monomeric red fluorescent protein
- PM
plasma membrane
- PVC
pre-vacuolar compartment
- SCR
SCARECROW
- SGR
SHOOT GRAVITROPISM
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- TGN
trans-Golgi network
- γ-TIP
tonoplast intrinsic protein-γ
- TVS; transvacuolar strand; VM
vacuolar membrane
- WT
wild-type
Disclosures
The authors have no conflicts of interest to declare.
References
- Andrade MA, Petosa C, O’Donoghue SI, Müller CW, Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- Chang HC, Hull M, Mellman I. The J-domain protein Rme-8 interacts with Hsc70 to control clathrin-dependent endocytosis in Drosophila. J. Cell Biol. 2004;164:1055–1064. doi: 10.1083/jcb.200311084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Escobar NM, Haupt S, Thow G, Boevink P, Chapman S, Oparka K. High-throughput viral expression of cDNA–green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell. 2003;15:1507–1523. doi: 10.1105/tpc.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujibayashi A, Taguchi T, Misaki R, Ohtani M, Dohmae N, Takio K, et al. Human RME-8 is involved in membrane trafficking through early endosomes. Cell Struct. Funct. 2008;33:35–50. doi: 10.1247/csf.07045. [DOI] [PubMed] [Google Scholar]
- Hanton SL, Brandizzi F. Fluorescent proteins as markers in the plant secretory pathway. Microsc. Res. Tech. 2006;69:152–159. doi: 10.1002/jemt.20276. [DOI] [PubMed] [Google Scholar]
- Hashiguchi Y, Niihama M, Takahashi T, Saito C, Nakano A, Tasaka M, et al. Loss-of-function mutations of retromer large subunit genes suppress the phenotype of Arabidopsis zig mutant that lacks Qb-SNARE VTI11. Plant Cell. 2010;22:159–172. doi: 10.1105/tpc.109.069294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y, Tasaka M, Morita MT. Mechanism of higher plant gravity sensing. Amer. J. Bot. 2013;100:91–100. doi: 10.3732/ajb.1200315. [DOI] [PubMed] [Google Scholar]
- Kato T, Morita M, Fukaki H, Yamauchi Y, Uehara M, Niihama M, et al. SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell. 2002;14:33–46. doi: 10.1105/tpc.010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hülskamp M. Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell. 2003;15:1632–1645. doi: 10.1105/tpc.011676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty A, Luo A, DeBlasio S, Ling X, Yang Y, Tuthill DE, et al. Advancing cell biology and functional genomics in maize using fluorescent protein-tagged lines. Plant Physiol. 2009;149:601–605. doi: 10.1104/pp.108.130146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita MT, Kato T, Nagafusa K, Saito C, Ueda T, Nakano A, et al. Involvement of the vacuoles of the endodermis in the early process of shoot gravitropism in Arabidopsis. Plant Cell. 2002;14:47–56. doi: 10.1105/tpc.010216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Toyota M, Tasaka M, Morita MT. An Arabidopsis E3 ligase, SHOOT GRAVITROPISM9, modulates the interaction between statoliths and F-actin in gravity sensing. Plant Cell. 2011;23:1830–1848. doi: 10.1105/tpc.110.079442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niihama M, Takemoto N, Hashiguchi Y, Tasaka M, Morita MT. ZIP genes encode proteins involved in membrane trafficking of the TGN-PVC/vacuoles. Plant Cell Physiol. 2009;50:2057–2068. doi: 10.1093/pcp/pcp137. [DOI] [PubMed] [Google Scholar]
- Niihama M, Uemura T, Saito C, Nakano A, Sato MH, Tasaka M, et al. Conversion of functional specificity in Qb-SNARE VTI1 homologues of Arabidopsis. Curr. Biol. 2005;15:555–560. doi: 10.1016/j.cub.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Oda Y, Higaki T, Hasezawa S, Kutsuna N. New insights into plant vacuolar structure and dynamics. Int. Rev. Cell Mol. Biol. 2009;277:103–135. doi: 10.1016/S1937-6448(09)77003-0. [DOI] [PubMed] [Google Scholar]
- Ovečka M, Lang I, Baluska F, Ismail A, Illes P, Lichtscheidl IK. Endocytosis and vesicle trafficking during tip growth of root hairs. Protoplasma. 2005;226:39–54. doi: 10.1007/s00709-005-0103-9. [DOI] [PubMed] [Google Scholar]
- Reisen D, Marty F, Leborgne-Castel N. New insights into the tonoplast architecture of plant vacuoles and vacuolar dynamics during osmotic stress. BMC Plant Biol. 2005;5:13. doi: 10.1186/1471-2229-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack FD. Plastids and gravitropic sensing. Planta. 1997;203:S63–S568. doi: 10.1007/pl00008116. [DOI] [PubMed] [Google Scholar]
- Saito C, Ueda T. Functions of RAB and SNARE proteins in plant life. Int. Rev. Cell Mol. Biol. 2009;274:183–233. doi: 10.1016/S1937-6448(08)02004-2. [DOI] [PubMed] [Google Scholar]
- Saito C, Morita MT, Kato T, Tasaka M. Amyloplasts and vacuolar membrane dynamics in the living graviperceptive cell of the Arabidopsis inflorescence stem. Plant Cell. 2005;17:548–558. doi: 10.1105/tpc.104.026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito C, Ueda T, Abe H, Wada Y, Kuroiwa T, Hisada A, et al. A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis. Plant J. 2002;29:245–255. doi: 10.1046/j.0960-7412.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- Sanderfoot AA, Kovaleva V, Zheng HY, Raikhel NV. The t-SNARE AtVAM3p resides on the prevacuolar compartment in Arabidopsis root cells. Plant Physiol. 1999;121:929–938. doi: 10.1104/pp.121.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato MH, Nakamura N, Ohsumi Y, Kouchi H, Kondo M, Hara-Nishimura I, et al. The AtVAM3 encodes a syntaxin-related molecule implicated in the vacuolar assembly in Arabidopsis thaliana. J. Biol. Chem. 1997;272:24530–24535. doi: 10.1074/jbc.272.39.24530. [DOI] [PubMed] [Google Scholar]
- Shi A, Sun L, Banerjee R, Tobin M, Zhang Y, Grant BD. Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 2009;28:3290–3302. doi: 10.1038/emboj.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silady RA, Kato T, Lukowitz W, Sieber P, Tasaka M, Somerville CR. The gravitropism defective 2 mutants of Arabidopsis are deficient in a protein implicated in endocytosis in Caenorhabditis elegans. Plant Physiol. 2004;136:3095–3103. doi: 10.1104/pp.104.050583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Morita MT, Ebine K, Okatani Y, Yano D, Saito C, et al. Vacuolar/prevacuolar compartment Qa-SNAREs, VAM3/SYP22 and PEP12/SYP21 have interchangeable functions in Arabidopsis. Plant J. 2010;64:864–873. doi: 10.1111/j.1365-313X.2010.04372.x. [DOI] [PubMed] [Google Scholar]
- Uemura T, Yoshimura SH, Takeyasu K, Sato MH. Vacuolar membrane dynamics revealed by GFP–AtVam3 fusion protein. Genes Cells. 2002;7:743–753. doi: 10.1046/j.1365-2443.2002.00550.x. [DOI] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, Bohnert HJ, Pantoja O. Novel regulation of aquaporins during osmotic stress. Plant Physiol. 2004;135:2318–2329. doi: 10.1104/pp.104.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Fukaki H, Fujisawa H, Tasaka M. Mutations in the SGR4, SGR5 and SGR6 loci of Arabidopsis thaliana alter the shoot gravitropism. Plant Cell Physiol. 1997;38:530–535. doi: 10.1093/oxfordjournals.pcp.a029201. [DOI] [PubMed] [Google Scholar]
- Yano D, Sato M, Saito C, Sato MH, Morita MT, Tasaka M. A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc. Natl Acad. Sci. USA. 2003;100:8589–8594. doi: 10.1073/pnas.1430749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Grant B, Hirsh D. RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol. Biol. Cell. 2001;12:2011–2021. doi: 10.1091/mbc.12.7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.