Abstract
Kindlin-1 is an integrin tail binding protein that controls integrin activation. Mutations in the FERMT-1 gene lead to Kindler Syndrome in man, which is characterized by skin blistering, premature skin ageing and skin cancer of unknown etiology. Here we show that loss of Kindlin-1 in mouse keratinocytes recapitulates Kindler Syndrome, and in addition produces enlarged and hyperactive stem cell compartments, which lead to hyperthickened epidermis, ectopic hair follicle development and increased skin tumor susceptibility. Mechanistically, Kindlin-1 controls keratinocyte adhesion through β1-class integrins and proliferation and differentiation of cutaneous epithelial stem cells by promoting αvβ6 integrin-mediated TGFβ activation and by inhibiting Wnt-β-catenin signaling through an integrin-independent regulation of Wnt ligand expression. Our findings assign Kindlin-1 the novel and essential task to control cutaneous epithelial stem cell homeostasis by balancing TGFβ mediated growth inhibitory and Wnt-β-catenin mediated growth-promoting signals.
INTRODUCTION
Kindler Syndrome (KS) is an autosomal-recessive disease caused by loss of function mutations in the FERMT-1 gene, which encodes Kindlin-1. The skin is the principal affected organ of individuals with KS and displays trauma-induced blisters, photosensitivity, pigmentation defects, and increased risk for malignancies1,2.
The Kindlins belong to a family of evolutionary conserved proteins, which are primarily found at cell-matrix adhesion sites where they bind β integrin tails and increase integrin affinity for ligand (also called integrin activation)3–5. In addition, they are also present at cell-cell adhesion sites, in the cytoplasm and in the nucleus where their functions are unknown6,7. Epidermal and hair follicle (HF) keratinocytes express Kindlin-1 and Kindlin-2. However, despite the striking sequence similarity, Kindlin-1 and -2 cannot compensate for each other indicating that they have specialized functions3,8.
Epidermal keratinocytes express several integrins, most notably members of the β1 integrin subfamily9. Keratinocytes of the HF bulge express high levels of β1 integrins and αvβ6 integrin10. The HF bulge harbors dormant stem cells (SCs) that periodically become activated to sustain the hair cycle (HC)11,12. The alternation of bulge SC activation and dormancy is regulated by a tight interplay of antagonistic signaling pathways. SC dormancy is achieved by BMP and TGFβ signaling, while SC activation is elicited by shutting down BMP and TGFβ signaling and activating canonical Wnt-β-catenin signaling. Perturbations of these cell growth regulating signaling pathways or of integrin signaling can profoundly alter SC homeostasis and tumor incidence13–16. It has been shown for example, that increased integrin expression or activity is associated with an increased risk for squamous cell carcinoma16–18. Conversely, loss of β1 integrin expression in skin (M. Sibilia, manuscript in preparation) or other tissues such as mammary gland markedly reduces tumor susceptibility19. Moreover, it has recently been shown that Kindlin-2 can stabilize β-catenin and induce Wnt signaling in certain tumor cell lines20. It is therefore enigmatic why KS patients suffer from an increased tumor risk2,21,22 despite Kindlin-1 loss and compromised integrin functions in their keratinocytes3,23,24. This discrepancy suggests that Kindlin-1 harbors potent tumor suppressor function(s) in keratinocytes that operate independently of the abundant and oncogenic β1 integrins. In this paper we identified oncogenic signaling pathways that are tightly controlled by Kindlin-1.
RESULTS
Kindlin-1 loss in epidermis and HFs leads to KS-like defects
To circumvent the lethal ulcerative colitis of a constitutive3 Fermt-1 gene ablation, we efficiently deleted the Fermt-1 gene in keratinocytes by breeding floxed Kindlin-1 mice with Keratin-5 (K5)-Cre transgenics25 (Kind1-K5; Fig. 1a–c and Supplementary Fig. 1a–c). Kindlin-1 loss persisted and did not affect Kindlin-2 expression (Supplementary Fig. 1b). Heterozygous Kind1-K5 or homozygous floxed Kindlin-1 mice used as control strains (Control) had no apparent phenotype. Homozygous Kind1-K5 mice were born within the expected Mendelian ratio, were fertile and gained weight normally (Supplementary Fig. 1d).
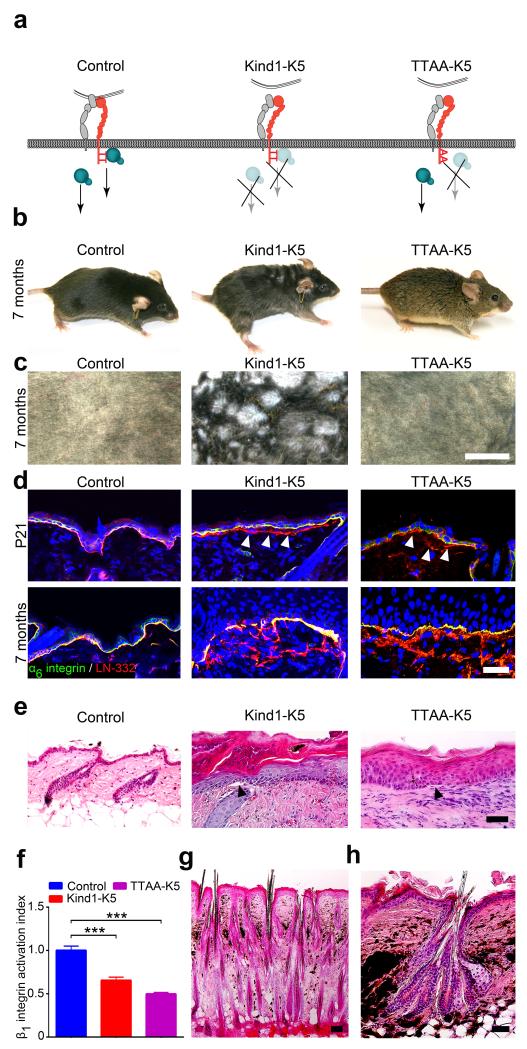
Figure 1.
Kindlin-1 controls HF growth. (a) Model of Kindlin-1 and β1 integrin functions in mice; Kind1-K5 mice lack Kindlin-1 in keratinocytes and TTAA-K5 mice express a Kindlin-binding deficient β1 integrin in which threonines-788/789 are substituted with alanines. (b) Appearance of 7 months old control, Kind1-K5 and TTAA-K5 mice. (c) View of shaved back skin of 7 months old control, Kind1-K5 and TTAA-K5 mice. (d) Immunofluorescence staining of back skin from P21 and 7 months old mice for LN-332 (red) and α6 integrin (green). Arrowheads indicate BM splitting. Nuclei are stained with DAPI (blue). (e) H/E staining of back skin from 6 months old mice. Kind1-K5 and TTAA-K5 skin shows hyperthickened skin, hyperkeratosis and blisters (arrowhead) at the epidermal-dermal junction. (f) Integrin activation index on primary control, Kind1-K5 and TTAA-K5 keratinocytes reported as mean 9EG7 binding normalized to total β1 integrin level ± SD (n=3 technical replicates). (g,h) H/E stained back skin sections of 7 months old Kind1-K5 mice. Scale bar indicates 5 mm in (c), 50 μm in (d,e,g) and 100 μm in (h).
The first histologic phenotype emerged around P21 in Kind1-K5 back skin with basement membrane (BM) splitting, small blisters at the dermal-epidermal junction and aberrant accumulation of F-actin and cell-cell adhesion proteins at the basal side of basal keratinocytes (Fig. 1d,e and Supplementary Fig. 1e). The same defects were also observed in mice expressing Kindlin-binding deficient β1 integrins in keratinocytes (Fig. 1a) due to threonine-788/789 to alanine substitutions in the β1 cytoplasmic domain (TTAA-K5)26 indicating that they are caused by malfunctioning β1 integrins (Fig. 1f). The blisters and BM defects triggered a regenerative response with granulocyte, monocyte and T cell infiltrates in the dermis of the back skin of Kind1-K5 as well as TTAA-K5 mice (Supplementary Fig. 1f–h). At postnatal age 60 (P60) Kind1-K5 mice showed progressive melanin deposits, first in the tail and then in the entire back skin (Fig. 1g,h and Supplementary Fig. 1i) resembling poikiloderma in KS. At around 3 months Kind1-K5 mice developed an irregular hair coat with small patches of densely clustered hair (Fig. 1b) that increased in size with age and appeared as meanders on the shaved back skin (Fig. 1c). At 6–8 months of age Kind1-K5 mice began to lose hair and patches with dense hair and alopecia alternated on their hair coat (Supplementary Fig. 1j). TTAA-K5 mice developed neither pigmentation defects nor densely packed hairs (Fig. 1b,c,e). Kind1-K5 mice also developed areas of atrophy next to areas of hyperkeratosis in the tail and back skin, while TTAA-K5 mice displayed hyperkeratotic areas only (Fig. 1e and Supplementary Fig. 2a). While the number of Ki67-positive cells was higher in the Kind1-K5 and TTAA-K5 epidermis, especially in hyperkeratotic areas (Supplementary Fig. 2b–d), keratinocyte differentiation and apoptosis was unaffected in both mouse strains (Supplementary Fig. 2e–i).
These results show that Kindlin-1 deletion in mouse epidermis recapitulates human KS and additionally induces an aberrant hair coat, which does not develop in TTAA-K5 mice indicating that the aberrant hair coat is caused by Kindlin-1 specific and β1 integrin- and inflammation-independent mechanisms.
Kindlin-1 loss disturbs HC and induces ectopic HFs
Back skin histology of 6 months old Kind1-K5 mice revealed two different HF abnormalities; (i) areas with isolated and densely packed HFs (Fig. 1g) separated by 1–15 interfollicullar epithelium (IFE) cells compared to 37 ±10 (mean ± SD, n = 4 mice per genotype) IFE cells between control HFs, and (ii) HFs with multiple hair shafts and bulges that clustered together and contained hair strands exiting the skin through a single hair canal (Fig. 1h).
A detailed HC analysis revealed that HF morphogenesis and the first HC proceeded normally in Kind1-K5 mouse skin (Supplementary Fig. 3a). At P50, HFs from control and Kind1-K5 mice entered telogen (resting phase of the HC). While HFs from control mice remained in a long telogen until P80 before a new HC was initiated (Supplementary Fig. 3a), Kind1-K5 HFs immediately re-entered anagen (growth phase of the HC) (Fig. 2a–c) visible by the formation of a hair germ (HG) and the expression of the transcription factor CCAAT displacement protein (CDP)27. We never observed premature anagen induction in TTAA-K5 mice (Supplementary Fig. 3b).
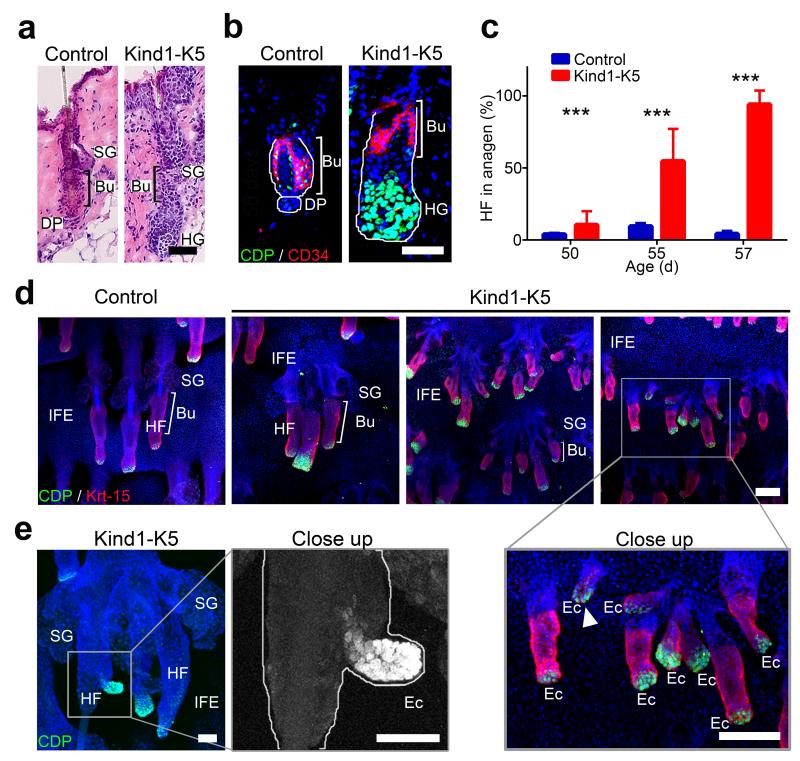
Figure 2.
Premature anagen induction and ectopic HF development in Kind1-K5 skin. (a) H/E stained back skin sections of P56 mice. (b) Immunostaining of HFs from P56 mice for CD34 (red) and CDP (green). Nuclei are stained with DAPI (blue). (c) The percentage of anagen HFs at indicated time points (mean ± SD, n=4 mice per genotype, ≥8 10× objective fields were counted). (d) Immunostaining of tail epidermal whole mounts for CDP (green), Krt-15 (red) and DAPI (blue) of P80 mice. Note ectopic HF growth in IFE of Kind1-K5 mice. (e) Ectopic HF outgrowth from a preexisting HF of the tail whole mount skin in a 6 months old Kind1-K5 mouse stained for CDP (green) and DAPI (blue). Scale bar indicates 50 μm in (a,b) and 100 μm in (d,e). Bu, bulge; SG, sebaceous gland; HG, hair germ; HF, hair follicle; DP, dermal papilla; IFE, interfollicular epidermis; Ec, ectopic HF.
Whole mount staining of P80 tail epidermis showed control mouse HFs with prominent sebaceous glands (SGs), normal sized keratin-15 (Krt-15)-positive bulges and small HGs with few CDP+ cells (Fig. 2d). The tail skin of Kind1-K5 mice contained small ectopic HFs originating from pre-existing HFs and the IFE (Fig. 2d,e). Ectopic HF- and IFE-derived Kind1-K5 HFs were also present in the back skin and expressed the dermal papilla marker alkaline phosphatase (Supplementary Fig. 3c,d). Of note, while HF numbers increased after P50 in the murine Kind1-K5 back skin their number began declining at older age (Supplementary Fig. 3i).
These findings show that Kindlin-1 deficiency leads to a premature onset of anagen, the formation of ectopic HFs in the IFE and from pre-existing HFs and a decline in HF numbers at old age.
Kindlin-1 regulates cutaneous epithelial SC homeostasis
The ectopic HFs in Kind1-K5 mice pointed to a perturbed SC homeostasis. The epidermis contains different SC populations that reside in different niches11. While bulge SCs express CD34, Krt-15, nephronectin (Npnt)28 and high levels of α6 integrin (Fig. 3a), reliable murine IFE SC markers are still missing11,29. Immunostaining revealed higher numbers of CD34+ bulge cells in back skin (Supplementary Fig. 3e) and enlarged Krt-15+ bulges in tail whole mounts of P80 old Kind1-K5 mice compared to controls (Fig. 3b and Supplementary Fig. 3f). Krt-15+ cells were also present in the Kind1-K5 infundibulum and IFE (arrowheads in Fig. 3b), but never in control mice. Npnt expression expanded into the SG, the infundibulum and the outer root sheath (Fig. 3b,c) and Npnt mRNA levels were higher in FACS-sorted bulge (Bu) and infundibulum-junctional zone (IJ) cells of Kind1-K5 mice compared to controls (Supplementary Fig. 3h). Also the Lrig1-positive SC population of the IJ zone30 extended towards the infundibulum and lower hair shaft of back and tail skin HFs of Kind1-K5 mice (Fig. 3d and Supplementary Fig. 3g).
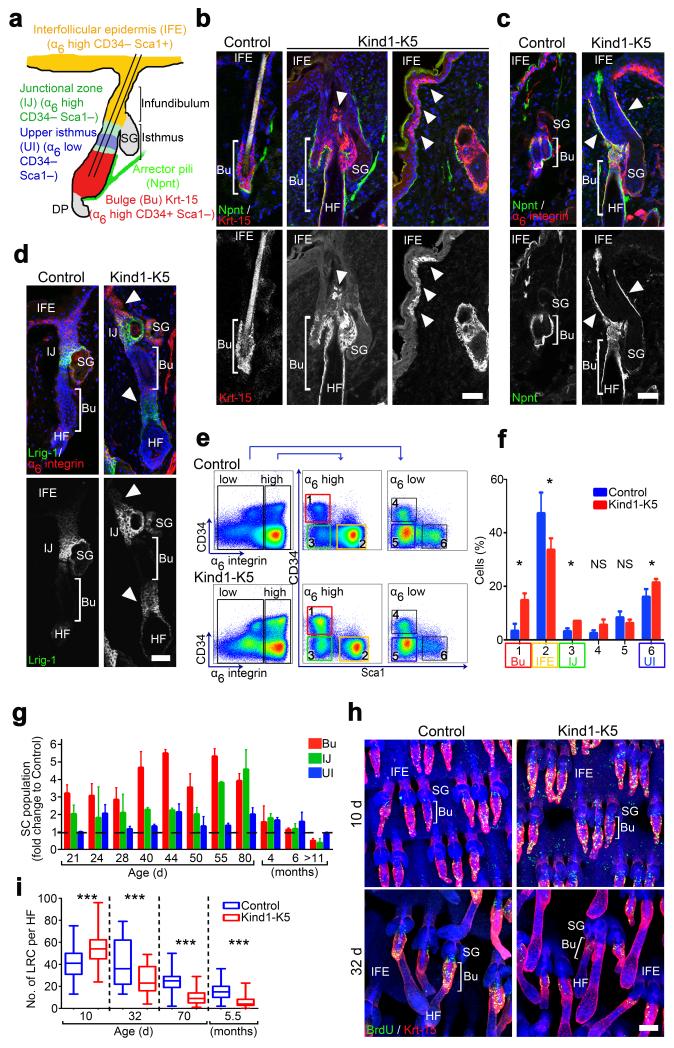
Figure 3.
Kindlin-1 regulates cutaneous epithelial SC homeostasis. (a) Distribution and expression of marker genes in SC subpopulations of murine HFs. (b–d) Back skin immunostaining from 6 months old mice for Krt-15 (red) and Npnt (green) (b), Npnt (green) and α6 integrin (red) (c), and for Lrig1 (green) and α6 integrin (red) (d). Arrowheads indicate Krt-15 positive cells (b), Npnt deposition (c) and Lrig1 expression (d) in Kind1-K5 mice. (e) Keratinocytes from P40 old mice were separated by FACS into α6 integrin-high and -low and further analyzed for CD34 and Sca1 expression. (f) Gated populations (1–6) were quantified and represented as mean ± SD (n=5 Control and 3 Kind1-K5 mice). Color code in (e–f) corresponds to the cell population denoted in (a). (g) The relative amounts of SC subpopulations over time analyzed by FACS. Data show average fold increase (± SD) relative to control mice (n and P-values are listed in Supplementary Table 1). (h) BrdU (green) retaining cells in Krt-15 (red) stained tail whole mounts after indicated chase times. (i) Numbers of LRC per HF after indicated chasing periods were quantified as boxplots (10 d n=5 Control and 3 Kind1-K5; 32 d n=3 Control and 4 Kind1-K5; 80 d n=4 Control and 3 Kind1-K5). Boxplot whisker ends show Min/Max distribution and middle line reports the median. Nuclei are stained with DAPI (blue) (b–d,h). Scale bar indicates 50 μm (b–d) and 100 μm in (h). Bu, bulge; SG, sebaceous gland; HF, hair follicle; IFE, interfollicular epidermis; UI, upper isthmus; IJ, infundibulum junctional zone.
Despite normal HCs and hair counts until P50 (Supplementary Figs. 3a,i), FACS quantifications of keratinocytes from P40 mouse skin revealed significantly more SCs in bulge, upper isthmus (UI) and IJ of Kind1-K5 mice compared to controls (Fig. 3e,f). A time course analysis showed that the expansion of SC compartments was already visible at P21, peaked at P50–80 and then began declining to become significantly smaller in 12 months old Kind1-K5 mice (Fig. 3g). In line with a normal hair coat development SC subpopulations were unaffected in TTAA-K5 mice, at least until one year of age (Supplementary Fig. 3j).
To test if elevated proliferation caused the enlarged bulges, we performed 5-bromo-2′-deoxyuridine (BrdU) label-retaining cell (LRC) assays. After 10 days of chase significantly more bulge cells were BrdU+ in tail whole mounts of Kind1-K5 mice compared to controls, while after 32, 70 days and 5.5 months significantly fewer bulge cells were BrdU+ in Kind1-K5 mice (Fig. 3h,i).
These findings demonstrate that Kindlin-1 deficiency elevates cutaneous epithelial SC proliferation, numbers and compartments in a β1 integrin-independent manner.
Kindlin-1 triggers αvβ6 integrin-mediated TGFβ release in skin
To determine whether Kindlin-1 loss alters integrin surface levels we analyzed integrin profiles on primary keratinocytes using FACS. Levels of β1 and β4 integrins were slightly lower and levels of β5, β8 slightly higher in Kind1-K5 cells compared to controls. β3 integrin was not detectable in either genotype. β6 integrin levels were significantly elevated in Kindlin-1 deficient keratinocytes (Fig. 4a), as was the β6 mRNA in FACS-sorted cutaneous SCs (Supplementary Fig. 4a). Immunostaining showed β6 integrin expression extending from the bulge and outer root sheath to the infundibulum and IFE of Kind1-K5 mice (Fig. 4b).
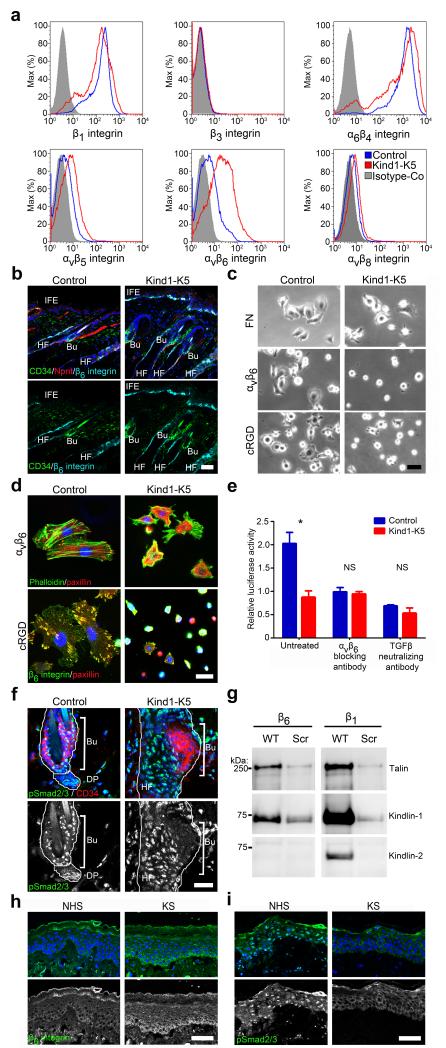
Figure 4.
Kindlin-1 promotes αvβ6 integrin induced TGFβ release in vitro and in vivo. (a) Representative histogram of integrin levels on freshly isolated keratinocytes analyzed by FACS (n=3 biological replicates). (b) Immunofluorescence analysis of back skin from P44 mice for β6 integrin (cyan), CD34 (green) and Npnt (red). (c) Brightfield images of keratinocytes plated on FN-, αvβ6- and cRGD-coated surfaces. (d) Immunostaining of keratinocytes plated on αvβ6 and cRGD surfaces for paxillin (red) and F-actin or β6 integrin (green). (e) TGFβ release from extracellular matrix by keratinocytes from control or Kind1-K5 mice in the absence or presence of indicated blocking antibodies monitored with the tMLEC reporter cell line. Luciferase activity is reported as mean ± SEM (n=3 biological replicates). (f) Immunostaining of a HF from 4 months back skin for pSmad2/3 (green) and CD34 (red). (g) Streptavidin-bead pull-down assay with biotinylated wild type (WT) integrin tails or scrambled (Scr) peptides from keratinocyte lysates (representative blot from three independent experiments). (h,i) Immunofluorescence staining of skin sections from control and individuals with KS for β6 integrin (h) and pSmad2/3 (i). Nuclei are stained with DAPI (blue) (b,d,f,h,i). Scale bar indicates 50 μm (b,c,h,i) and 25 μm (d,f). NHS, normal human skin; KS, Kindler syndrome; Bu, bulge; DP, dermal papilla; HF, hair follicle; IFE, interfollicular epidermis.
Next, we investigated β6 integrin dependent cell spreading and focal adhesion formation. While keratinocytes from Kind1-K5 mice spread normally on fibronectin (FN), they displayed severely impaired adhesion, spreading, clustering of paxillin in focal adhesion-like structures and assembly of F-actin stress fibers when plated on surfaces coated with anti-αvβ6 antibodies or αv-specific peptidomimetic cyclic RGD (cRGD) (Fig. 4c,d and Supplementary Fig. 4b,c) indicating that αvβ6 on Kind1-K5 keratinocytes is non-functional. We confirmed αvβ6 specificity of these defects by treating keratinocytes from control mice with the β6 blocking antibody 10D5, which phenocopied the spreading defects of Kindlin-1 deficient cells on cRGD surfaces (Supplementary Fig. 4d).
Active αvβ6 integrins can release TGFβ from the latency associated protein (LAP)31, which in turn suppresses proliferation of bulge SCs10,12,32,33. We tested the αvβ6-dependent TGFβ release by seeding keratinocytes together with transformed mink lung epithelial cells (tMLEC) able to report TGFβ-induced luciferase on a latent TGFβ binding protein-1-LAP-TGFβ-1 rich matrix34 (Fig. 4e). Keratinocytes from control mice efficiently induced the luciferase reporter, which could be inhibited with the αvβ6 blocking antibody 10D5 or the TGFβ neutralizing antibody 1D11. In sharp contrast, keratinocytes from Kind1-K5 mice were unable to release TGFβ confirming that their αvβ6 integrins are non-functional (Fig. 4e). In line with this in vitro result, TGFβ signaling was also severely impaired in bulge cells of Kind1-K5 mice in vivo, in which pSmad2/3 was detectable neither before nor after premature anagen induction (Fig. 4f and Supplementary Fig. 4g). Signaling by BMP via pSmad1/5/8, which also suppresses bulge cell proliferation35,36, was unaffected (Supplementary Fig. 4h). Treatment of Kindlin-1 deficient keratinocytes with soluble TGFβ-1 induced robust Smad2 phosphorylation and reduced cell proliferation (Supplementary Fig. 4e,f) indicating that TGFβ-1 signals are efficiently transduced. The severe dysfunction of αvβ6 integrins was not compensated by Kindlin-2 due to the inability of Kindlin-2 to bind β6 tails (Fig. 4g). Most importantly, subjects with KS showed a similar TGFβ defect as Kind1-K5 mice with elevated expression of β6 integrin and reduced nuclear pSmad2/3 in large areas of basal epidermal keratinocytes (Fig. 4h,i), while pSmad1/5/8 was unaffected (Supplementary Fig. 4i).
These findings indicate that Kindlin-1 is essential for β6 integrin-mediated cell adhesion, cell spreading and TGFβ release required for SC quiescence.
Kindlin-1 curbs Wnt-β-catenin signaling in skin
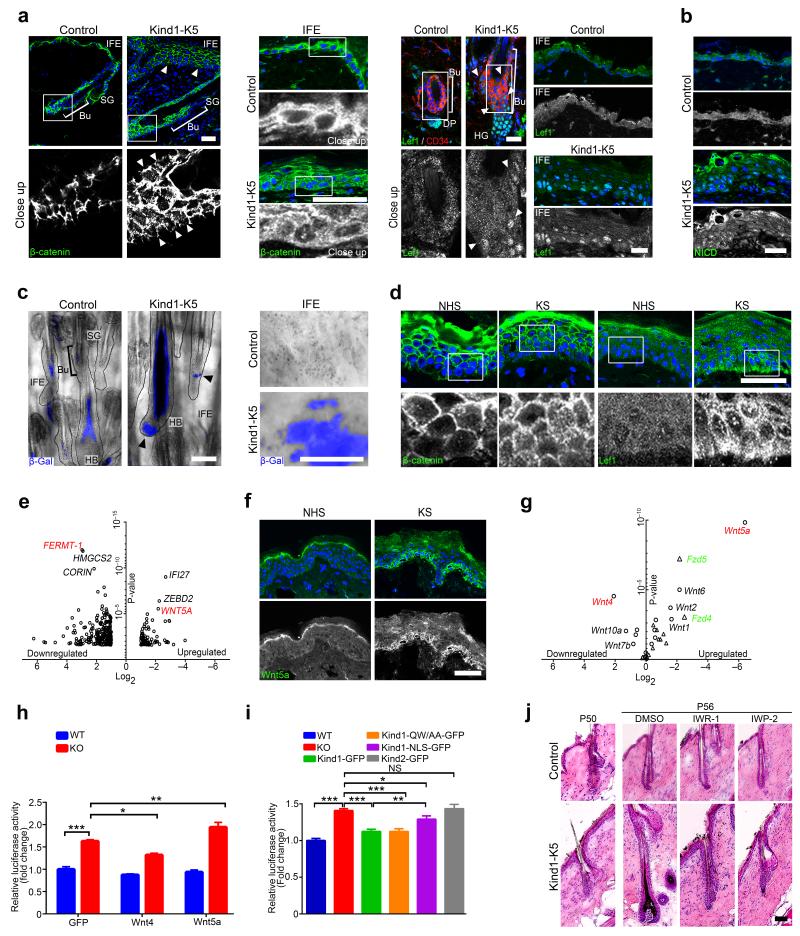
Premature anagen induction, ectopic HF development and expansion of SC compartments also develop in mice with elevated Wnt-β-catenin signaling in keratinocytes37–40, while reduced HF spacing and aberrant hair orientation occur in mice overexpressing the β-catenin cofactor Lef141 or mice with increased Notch signaling42. In skin of control mice β-catenin levels were high at cell-cell junctions, low in the nucleus and absent at the basal side of basal keratinocytes. In contrast, in epidermis of Kind1-K5 mice β-catenin also accumulated at basal sides of basal keratinocytes (Fig. 5a, arrowheads top left panel), in the nucleus of a large number of IFE cells (Fig. 5a, middle panel) and catagen HFs (Fig. 5a, lower left panel and Supplementary Fig. 5d, left). Furthermore, Kind1-K5 mice showed β-catenin in nuclei of developing HGs during premature anagen induction (Supplementary Fig. 5a upper panel, 5b), and an extended Lef1 expression from the normal site in HGs to the bulge and IFE at all stages analyzed (Fig. 5a right panel and Supplementary Figs. 5a,c,d left panel). Notably, at P50 shortly before bulges from Kind1-K5 mice induced premature anagen they showed high nuclear Lef1 levels indicating a premature onset of Wnt signaling. In control mice Lef1 was absent in telogen bulges and became highly expressed in nuclei of HG cells and weakly in the cytoplasm of IFE cells (Fig. 5a right panel and Supplementary Fig. 5a lower panel). Numbers of nuclear Notch effector Notch intracellular domain (NICD)-containing cells and NICD-induced Hes1 mRNA were unaltered in HG and pericortex, but higher in the IFE of Kind1-K5 mice compared to controls at all time points analyzed (Fig. 5b and Supplementary Fig. 5e–g). Next we determined Wnt-β-catenin signaling by intercrossing Kind1-K5 and control mice with TOPgal reporter mice, which express β-galactosidase under a TCF-Lef controlled minimal cfos promotor43. In HFs from control mice TOPgal activities were high in the pericortex during anagen, low during catagen and lost in telogen with some residual activity in club hairs. In Kind1-K5 mice, the high TOPgal activity in the pericortex and bulb of anagen HFs persisted throughout catagen, decreased in telogen (Fig. 5c left panel and Supplementary Fig. 5i) and was strongly re-induced in the following anagen stage (Supplementary Fig. 5h). Furthermore, we observed a patch-like distribution of TOPgal in the IFE of Kind1-K5 mice, which was absent in controls (Fig. 5c right panel). Importantly, the IFE of humans with KS showed similar abnormalities in β-catenin-LEF1 expression with weak nuclear β-catenin in the basal cell layer and a patch-like distribution of nuclear LEF1, whereas normal human skin contained β-catenin exclusively in cell-cell contacts and LEF1 in the cytoplasm (Fig. 5d and Supplementary Fig. 5j).
Figure 5.
Kindlin-1 controls Wnt-β-catenin signaling. (a) Immunofluorescence staining for β-catenin (green) of HF (P44) (left panel) and IFE (P55) (middle panel) and for Lef1 and CD34 (red) of HF and IFE (4 months) (right panel) from control and Kind1-K5 mice. Arrowheads indicate aberrant β-catenin (left panel) and Lef1 (right panel) localization. (b) IFE immunofluorescence staining for NICD (green) from P44 control and Kind1-K5 mice. (c) TOPgal reporter activity in tail HFs (left panel) and IFE (right panel) from 3 months old control and Kind1-K5 mice. Arrowheads indicate HF with abnormal TOPgal activity. (d) Immunofluorescence staining of human skin from normal and individuals with KS for β-catenin (left panel) and LEF1 (right panel). (e) Skin gene expression profile of NHS (n=3) versus KS (n=3) assessed with microarray and shown as volcano plot. Genes with ≥2 fold change in KS were plotted according to the Log2 fold change (x-axis) and Log10 P-value (y-axis). (f) Immunofluorescence staining for WNT5A (green) in skin of normal and individuals with KS. (g) Volcano plot of qPCR gene expression profile of keratinocytes from control versus Kind1-K5 mice. Mean expression relative to Gapdh of Wnt ligands (circle, red) and receptors (triangle, green) were plotted according to the Log2 fold change (x-axis) and log10 P-value (y-axis). (n=3 biological replicates, for mean ± SEM values see Supplementary Table 3). (h) Transient overexpression of GFP, Wnt4 and Wnt5a in floxed (WT) and Adeno-Cre treated Kindlin-1 deficient (KO) keratinocytes expressing the TOPFlash reporter. Values are corrected for the renilla control and represented as mean ± SEM fold increase relative to WT cells (n=5 biological replicates). (i) TOPFlash reporter activity in cells stably re-expressing Kindlin-1-GFP, integrin-binding deficient Kindlin-1-GFP, NLS-tagged Kindlin-1-GFP and Kindlin-2-GFP, respectively. Values are corrected for the renilla control, represented as fold increase relative to WT cells and reported as mean ± SEM (n=21 WT, KO; 13 Kind1-GFP; 9 Kind1-QW/AA-GFP, 5 Kind1-NLS-GFP, Kind2-GFP; all biological replicates). (j) H/E staining of control and Kind1-K5 mice one day (P50) and after treatment (P56) with indicated Wnt inhibitor. Nuclei are stained with DAPI (blue) (a,b,d,f). All scale bars indicate 50 μm. NHS, normal human skin; KS, Kindler syndrome, Bu, bulge; SG, sebaceous gland; HG, hair germ; HF, hair follicle; DP, dermal papilla; HB, hair bulb.
These findings demonstrate that loss of Kindlin-1 induces nuclear translocation of β-catenin-Lef1 in KS and mouse skin leading to elevated Wnt-β-catenin signaling and premature anagen onset of Kind1-K5 mouse HFs.
Kindlin-1 regulates Wnt ligand expression
To find an explanation for the increased Wnt-β-catenin signaling in the absence of Kindlin-1 we compared the skin transcriptome between healthy individuals and subjects with KS using microarray analyses. As expected, FERMT-1 was absent in skin from subjects with KS and mRNAs encoding inflammatory proteins were high (Fig. 5e, for complete list see Supplementary Table 2). Unexpectedly, several Wnt signaling components, most notably WNT5A were high in individuals with KS (Fig. 5e). Immunostaining confirmed the aberrant expression of WNT5A in basal keratinocytes of individuals with KS (Fig. 5f). qPCR and in situ hybridization of tail whole mounts from Kind1-K5 mice corroborated high Wnt5a mRNAs levels in hair bulbs (HBs), ectopic HFs and patches of IFE, which overlapped with high TOPgal activities (Fig. 5g and Supplementary Fig. 6a–c). qPCR of all known Wnt ligands and receptors revealed that several canonical (Wnt1, 2b, 3a, 9b) and non-canonical Wnts (Wnt6 and 2) were also significantly higher in keratinocytes from Kind1-K5 mice compared to controls, while Wnt4, 7a, 9a, 10a and 16 were significantly lower (Fig. 5g and Supplementary Table 3). Expression of several Frizzled (Fzd) receptors was higher, most notably Fzd4 and Fzd5, and to a lesser extent the Fzd co-receptor Lrp6. qPCR of FACS sorted bulge cells and in situ hybridization of tail whole mounts from Kind1-K5 mice confirmed the high Fzd4 mRNA levels and observed a patch-like expression of Fzd4 in their IFE (Supplementary Fig. 6d,e).
Transfection of Kindlin-1 deficient keratinocytes with SuperTOPFlash reporter revealed that the elevated Wnt signaling was cell autonomous (Fig. 5h,i) and that Lef1 was limiting the extent of Wnt signaling (Supplementary Fig. 6f). Overexpression of Wnt4, which triggers translocation of β-catenin to cell-cell junctions, thereby preventing nuclear accumulation of β-catenin and Wnt-β-catenin signaling44, decreased SuperTOPFlash activity in Kindlin-1 deficient keratinocytes compared to GFP-transfected controls (Fig. 5h). Overexpression of Wnt5a, which together with Lrp5 and Fzd4 efficiently activates the canonical Wnt-β-catenin signaling pathway45, further increased the SuperTOPFlash reporter in Kindlin-1 deficient cells but not in control cells where Fzd4 expression was low (Fig. 5h and Supplementary Fig. 6d,e).
Re-expression of similar quantities of Kindlin-1-GFP or an integrin binding-deficient Kindlin-1-QW611/612AA-GFP, but not Kindlin-2 attenuated the increased SuperTOPFlash activities and levels of Wnt4 and Wnt5a in Kindlin-1 deficient keratinocytes (Fig. 5i and Supplementary Fig. 6g–j) indicating that inhibition of Wnt-β-catenin signaling is Kindlin-1 specific and does not require integrin binding. To exclude a role for nuclear Kindlin-1, we expressed Kindlin-1-GFP fused to two SV40 nuclear localization signals (NLS) in Kind1-null cells. Although most of the protein localized to the nucleus the elevated SuperTOPFlash activity was only partially rescued (Fig. 5i), likely due to Kind1-NLS-GFP spillovers into the cytoplasm (Supplementary Fig. 6j).
To corroborate that elevated Wnt protein secretion and high Wnt-β-catenin signaling underlies premature HF anagen induction in Kind1-K5 mice, we treated them at P49, shortly before telogen onset (Fig. 5j and Supplementary Fig. 7a) with IWP-2, which blocks Wnt protein secretion by inhibiting porcupine-mediated palmitylation of Wnts46, or with IWR-1, which inhibits tankyrases thereby stabilizing the β-catenin destruction complex46,47. Both compounds efficiently blocked premature anagen induction in Kind1-K5 mice and decreased SuperTOPFlash activities in vitro (Fig. 5j and Supplementary Fig. 7a,b). In line with previous reports42, inhibition of Wnt signaling also reduced aberrant Notch activities in Kind1-K5 keratinocytes in vitro and in vivo, which in turn normalized Wnt4 levels48 (Supplementary Fig. 7c–g).
Together these findings demonstrate that Kindlin-1 inhibits Wnt-β-catenin signaling by regulating transcription of Wnt ligands and receptors in a cell autonomous and integrin-independent manner.
Loss of Kindlin-1 increases skin tumor susceptibility
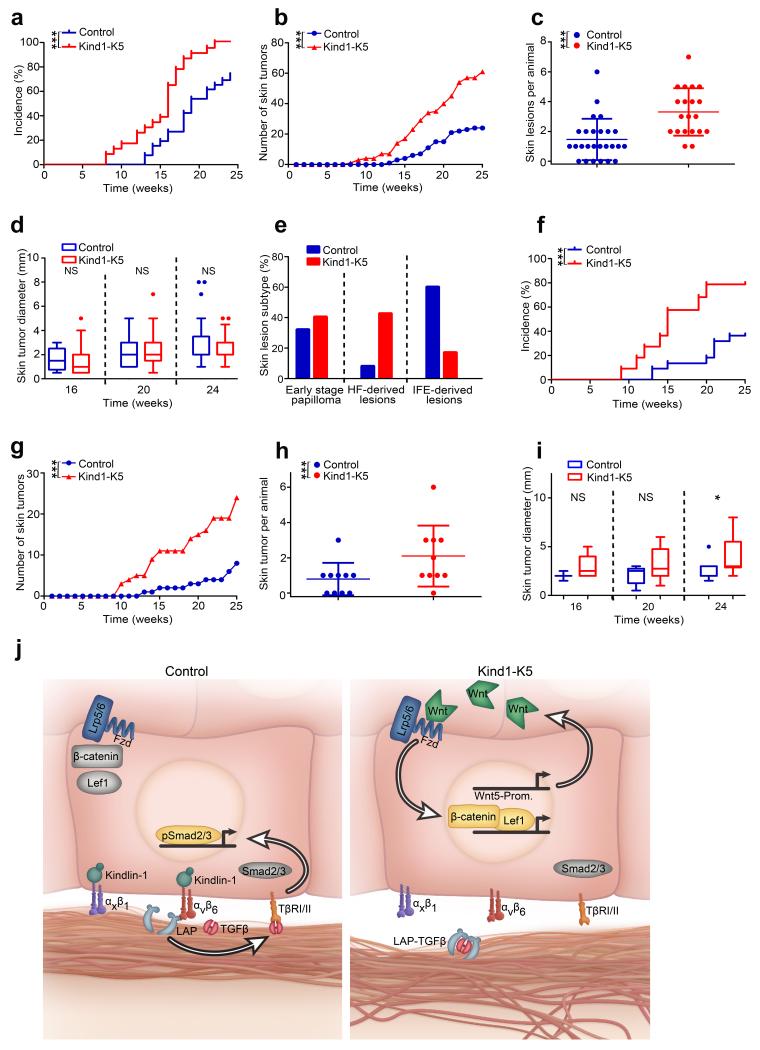
The low TGFβ and augmented Wnt signaling in humans with KS and Kind1-K5 mice represent oncogenic threats49,50. To test this hypothesis, we used the two-stage carcinogenesis protocol and treated mice with 7,12-dimethylbenz[a]anthracene (DMBA) and 12-O-tetradecanoylphorbol-13-acetate (TPA) for 25 weeks. Kind1-K5 mice developed tumors earlier and had more tumors than control mice (Fig. 6a–c). Of note, the tumor size was similar between Kind1-K5 and control mice (Fig. 6d), which could be due to different skin lesion subtypes. Indeed, histology of the tumor lesions revealed that hyperkeratotic and exophytic papillomas dominated in control mice, while HF-derived trichofolliculoma-like lesions, sebaceous papillomas, mixed papillomas and basal cell carcinoma (BCC)-like lesions dominated in Kind1-K5 mice (Fig. 6e and Supplementary Fig. 8a). In line with uncurbed Wnt signaling, we observed more cells with nuclear β-catenin and Lef1 in tumors from Kind1-K5 mice (Supplementary Fig. 8b–d).
Figure 6.
Loss of Kindlin-1 increases skin tumor incidence. (a–e) Two stage carcinogenesis (n=26 Control and 23 Kind1-K5 mice). (a) Tumor incidence (P-value by log-rank test), (b) burden and (c) skin lesions per animal (reported as mean ± SD) after 25 weeks of treatment. (d) Tumor growth reported as diameter is shown in a boxplot where whisker ends are at 1.5 interquartile ranges and middle lines represent the median. (e) The percentage of skin lesion subtypes from control (n=25 lesions) and Kind1-K5 mice (n=47 lesions) were staged by histology and immunofluorescence analysis (see Supplementary Fig. 8a). (f–i) One stage carcinogenesis with DMBA. (f) Tumor incidence, (g) skin lesion number, (h) frequency and (i) size of control (n=10) and Kind1-K5 (n=10) mice monitored as in (a–d). (j) Molecular functions of Kindlin-1; in normal cells (left panel) Kindlin-1 activates β1-class integrins and αvβ6 integrin to facilitate adhesion and TGFβ liberation from LAP, respectively. Free TGFβ activates TGFβ receptors (TβRI/II) leading to nuclear translocation of phosphorylated Smad2/3, which promotes SC quiescence. In Kindlin-1 deficient cells (right panel) activation of β1-class integrins and αvβ6 is impaired leading to adhesion defects and loss of TGFβ-mediated SC quiescence. In addition, dysregulated Wnt ligand expression leading to elevation of Wnt5a leads to canonical Wnt-β-catenin signaling via the Lrp5/6-Fzd4 complex. Wnt5-Prom, Wnt5 promotor.
A single DMBA treatment induced more and slightly larger tumors in Kind1-K5 mice compared to controls (Fig. 6f–i) indicating that the hyperproliferative state of keratinocytes in Kind1-K5 mice was sufficient to promote tumor development. Furthermore, their tumors primarily originated from HFs and SGs containing hyperactive SCs.
DISCUSSION
Loss-of-function mutations in the FERMT-1 gene cause KS, which is characterized by skin blistering at birth, premature skin aging, pigmentation defects and increased incidence of skin cancer. The ability of Kindlin-1 to activate integrins and link them to the F-actin cytoskeleton explains the development of skin blisters and BM splitting followed by an inflammatory response. However, the notion that skin tumor development is promoted by hyperactive integrins and inhibited by compromised integrins indicates that Kindlin-1 loss activates oncogenic and/or inhibits tumor suppressor functions. The task of this study was to identify such pathway(s).
Deletion of Kindlin-1 in skin epithelial cells with the K5-Cre driver line leads to small skin blistering, BM defects, chronic skin inflammation, progressive pigmentation defects and skin atrophy and hence is alike KS. Unexpectedly, the Kind1-K5 mice also develop enlarged cutaneous epithelial SC compartments, hyperactive bulge SCs, distorted HF cycles and ectopic HF (Supplementary Fig. 8e). In line with the high SC proliferation, DMBA-TPA as well as DMBA only treatment induces a significantly higher tumor incidence and tumor burden in Kind1-K5 mice. Moreover, the majority of the tumors are BCC- and trichofolliculoma-like lesions suggesting that the tumors predominantly originate from HF bulge SCs.
In search for a mechanistic explanation for the dysregulated SC and tumor cell proliferation in Kind1-K5 mice, we identified two Kindlin-1 regulated signaling pathways with opposing functions on bulge SC quiescence (Fig. 6j). Kindlin-1 binding to the cytoplasmic tail of β6 integrin51 triggers αvβ6 integrin binding to the RGD motif of LAP thereby liberating TGFβ and inducing TGFβ receptor signaling and cutaneous epithelial SC quiescence. These findings are supported by report showing that loss of β6 integrin impairs TGFβ signaling and elevates bulge SC activity in vivo32.
Despite the sequence similarities of Kindlin-1 and -252 and their expression in control and Kind1-K5 keratinocytes, Kindlin-2 can only partially compensate for Kindlin-1 at β1 integrin adhesion sites. Furthermore, Kindlin-2 is unable to bind β6 tails, which prevents it to release TGFβ-1 and to suppress SC proliferation. Interestingly, other TGFβ isoforms such as TGFβ-2 activate HF stem cells by antagonizing BMP in the bulge53. In contrast to LAP1 however, LAP2 lacks the αvβ6-binding RGD motif54 and hence the release of TGFβ-2 and BMP signaling are not affected by Kindlin-1 loss.
It is particularly interesting that high Kindlin-1 levels have also been associated with high TGFβ-1 signaling in metastatic breast cancers55. Although it was not investigated how Kindlin-1 regulates TGFβ-1 signaling, this observation together with our findings suggest that different tumor stages may benefit from different Kindlin-1 levels; in early stage tumors low TGFβ signaling supports tumor growth, while in late stage tumors high TGFβ signaling promotes epithelial to mesenchymal transition and metastatic progression56. If so, subjects with KS suffering from an increased tumor risk may be protected from metastasis.
We also found elevated Wnt-β-catenin signaling in Kind1-K5 mice and KS epidermis, which is inevitably associated with a high tumor risk and severe SC and HC defects13,50. The defect leads to ectopic HF formation and aberrant HCs, and is due to aberrant expression of several Wnt ligands, most notably Wnt5a, which probably act in an autocrine manner. Importantly, chemical compounds that inhibit Wnt protein secretion or Wnt-β-catenin signaling prevent the HC and Wnt signaling defects in keratinocytes from Kind1-K5 mice.
Wnt5a is best known for the ability to activate the non-canonical Wnt signaling pathway. However, Wnt5a can also elicit canonical Wnt-β-catenin signaling. The decision which signaling pathway is induced, is dictated by the type of cell surface receptor to which Wnt5a is binding; Wnt5a binding to the Ror2 receptor tyrosine kinase inhibits the transcriptional activity of β-catenin and thus canonical signaling, whereas Wnt5a binding to Fzd4 and the Lrp5/6 co-receptors leads to β-catenin stabilization and canonical Wnt signaling45. Interestingly, Wnt-β-catenin signaling can activate Notch42, which is also elevated in keratinocytes from Kind1-K5 mice and may, at least in part contribute to the Kind1-K5 mouse phenotype, e.g. by suppressing Wnt4 expression48 and the recruitment of β-catenin to cell-cell junctions44. Clearly, the dysregulated expression of Wnt proteins and their receptors in Kind1-K5 mice strongly suggests that Kindlin-1 safeguards the skin and HFs against an overshooting activation of the important and at the same time potentially harmful canonical Wnt-β-catenin signals.
In summary our results show that Kindlin-1 loss results in a combination of defects caused by the dysregulation of distinct pathways; the compromised β1 integrins impair adhesion and induce an inflammatory response; the impaired TGFβ liberation from LAP leads to HF SC hyperproliferation; and the increased Wnt signaling abrogates the resting phase of the HC, expands the SC compartments and together with the impaired TGFβ signaling and inflammation contributes to the increased tumor risk. The high epithelial SC activity eventually leads to SC exhaustion and skin atrophy later in life, which is another hallmark of KS. It is also conceivable that the ability of Kindlin-1 to shuttle between integrin sites where it mediates adhesion, migration and TGFβ release, and the cytoplasm where it curbs Wnt-β-catenin signaling may also contribute to physiologic SC homeostasis and hair development (Fig. 6j); mobilization of bulge SCs is accompanied by little or no integrin-ligand engagement and therefore, use Kindlin-1 to prevent Wnt-β-catenin signaling and their activation. Once activated bulge SCs move to the HG they engage integrins to ensure downward growth and therefore, have less Kindlin-1 in the cytoplasm to inhibit Wnt-β-catenin signaling.
METHODS
Mouse Strains
The conditional Kind1fl/fl mice carry a loxP-flanked ATG-containing exon 2 (Supplementary Fig. 1a) and were generated via electroporation of R1 embryonic stem (ES) cells using standard procedures57. Homologous recombination was verified with southern blots and positive ES clones were used to generate chimeric mice. Mice were crossed with transgenics carrying a deletor-flp recombinase to remove the neomycin cassette and with mice carrying deleter-Cre (to confirm the null phenotype) or K5-Cre transgenes, respectively58. For all animal studies transgenic mice were backcrossed eight times to C57BL/6. The TTAA-K5 transgenic mice were obtained through an intercross of mice carrying alanine substitutions of TT788/789 in the cytoplasmic domain of β1 integrin26 with β1 floxed mice and K5-Cre transgenics. The TOPgal Wnt reporter mice43 were intercrossed with Kind1-K5 mice. Mice were housed in a special pathogen free mouse facility and all animal experiments were carried out according to the rules of and approved by the government of Upper Bavaria.
DMBA/TPA tumor experiment
Cutaneous two stage chemical carcinogenesis was performed as previously described59 using topical applications of 100 nmol (25 μg) DMBA (Sigma) in 100 μl of acetone and twice weekly applications of 10 nmol (6.1 μg) TPA (Sigma) in 200 μl of acetone or only acetone for 25 weeks. The tumor model was terminated after 25 weeks of TPA promotion due to critical skin health condition of Kind1-K5 mice. For tumor experiment control animals were treated with acetone or TPA only. For all animal studies seven week old mice were randomized after genotyping and sample size was estimated by non-parametric Wilcoxon Test (U-Test). Number and size of tumors were recorded once per week after the start of promotion (week 0). All mice were sacrificed at the end of the experiment. Skin lesions were analyzed by histology and graded as described previously59–61.
Subjects with KS
Individuals with KS gave their informed written consent under protocols approved by St Thomas’ Hospital Ethics Committee (COREC number 06 /Q0702 /154) and the study was conducted according to the Declaration of Helsinki Principles. For IF analysis biopsies were taken from age and site matched controls (NHS) and subjects with KS with following mutations: KS patient 1 (3-year-old Indian girl; homozygous nonsense mutation +/+ p.Glu516X) and KS patient 2 (22 year old Panamanian woman; homozygous nonsense mutation +/+ p.Arg271X).
RNA for microarray analysis was isolated from upper arm biopsies taken from three KS subjects; KS patient A, 22 year old Omani woman, homozygous nonsense mutation +/+ p.Arg271X; KS patient B, 7 year old Indian girl, homozygous nonsense mutation +/+ p.Try616X and KS patient C, 19 year old Omani woman, nonsense mutation +/+ p.Try616X. Control skin RNA was isolated from four age-, site - and sex-matched biopsies.
Gene expression microarray
Total RNA was extracted using the RNeasy Fibrous Tissue Mini kit (Qiagen) and quantity and quality were measured on a Nanodrop spectrophotometer (Nanodrop, Wilmington, DE, U.S.A.). For whole-genome Illumina expression analysis, total RNA from each skin biopsy was hybridized to an Illumina Human-Ref-6 v2 BeadChip expression array (Illumina, San Diego, CA, U.S.A.). The Illumina HumanRef-6 v2 BeadChips were scanned with an Illumina Bead Array Reader confocal scanner.
The microarray data analysis was performed using Illumina’s BeadStudio Data Analysis Software (Illumina). The expression signal for all genes from each individual were grouped in KS and NHS and averaged. In order to identify statistically significant differentially regulated genes, a prefiltering set was determined for significantly higher (≥ 2-fold change) and lower expression (≤ 0.5-fold change) intensity between KS and NHS skin samples. Bonferroni’s correction was applied to each P-value to obtain an adjusted P-value to identify differentially expressed mRNAs with high statistical significance (see Supplementary Table 2). The microarray has been submitted to NCBI under the GEO accession number GSE47642.
FACS analysis
FACS analysis and sorting was performed as previously described62. A suspension of primary keratinocyte in FACS-PBS (PBS with 1% BSA) were incubated for 1 h with primary antibodies on ice and then washed twice with FACS-PBS. Cell viability was assessed by 7-aminoactinomycin (7-AAD) labeling (BD Biosciences) or ethidium monoazide (EMA) staining (Invitrogen). FACS analysis was carried out using a FACSCantoTMII Cytometer (BD Biosciences) and cell sorting with an AriaFACSII high-speed sorter (BD Biosciences), both equipped with FACS DiVa software (BD Biosciences). Purity of sorted cells was determined by post sort FACS analysis and typically exceeded 95%. Integrin surface FACS analysis of primary keratinocytes was carried out as previously described63. Data analysis was conducted using the FlowJo program (Version 9.4.10).
Real Time PCR and Notch target gene inhibition
Total RNA from total skin or FACS sorted keratinocytes was extracted with RNeasy Mini extraction kit (Qiagen) following manufacturer’s instructions. For Notch target gene inhibition analysis cells were plated on FN/collagen I coated 6-well plates (1.2 × 106 cells per well) and the next day treated with 2.5 μM N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT; Selleckchem; CatNo: S2215) in full keratinocyte growth medium (KGM) for 24 h before total RNA isolation. cDNA was prepared with a iScript cDNA Synthesis Kit (Biorad). Real Time PCR was performed with an iCycler (Biorad). Each sample was measured in triplicates and values were normalized to Gapdh. PCR primers are listed in Supplementary Table 4.
Antibodies and inhibitors
The following antibodies or molecular probes were used at indicated concentration for Western blot (W), immunofluorescence (IF), immunohistochemistry (IHC) or flow cytometry (FACS): Kindlin-13 (W: 1:5000, IF of tissue: 1:1000), Kindlin-2 (Sigma; CatNo: K3269; W: 1:1000), Talin (Sigma; clone 8D4; W: 1:1000), GAPDH (Calbiochem; clone 6C5; W: 1:10000), Phalloidin-Alexa488 (Invitrogen; CatNo: A12379; IF tissue: 1:500; IF of cells: 1:800), integrin αM (Mac-1) (eBioscience; clone M1/70; IF of tissue: 1:200), GR1 (Ly6g) (eBioscience; clone RB6-8C5; IF of tissue: 1:200), CD4 (PharMing; clone H129.19; IF of tissue: 1:200), CD19 (PharMing; clone 1D3; IF of tissue: 1:200), integrin αx (CD11c) (BD Bioscience; clone HL3; IF of tissue: 1:200), Desmoplakin (Fritzgerald Industries International; CatNo: 20R-DP002; IF tissue 1:500), α6 integrin (Itga6) (PharMingen; clone GoH3; IF of tissue: 1:200, FACS: 1:500), Laminin-332 (a gift from Monique Aumailley, University of Cologne, Germany; IF of tissue: 1:500), Keratin10 (Krt-10) (Covance; CatNo: PRB-159P; IHC: 1:600), Keratin 15 (Krt-15) (a gift from Rudolf Grosschedl; MPI Immunobiology, Freiburg, Germany), IF: 1:500), Lef1 (a gift from Rudolf Grosschedl, MPI Immunobiology, Freiburg, Germany; IF of tissue: 1:500), Loricrin (Lor) (Covance; CatNo: PRB-145P; IF: 1:500), Ki67 (Dianova; CatNo: M7249; IHC: 1:100), cleaved caspase-3 (Asp175, cCaspase3) (Cell Signaling; CatNo: 9661; IF of tissue: 1:200), CD34 (BD Bioscience; clone RAM34; IF of tissue: 1:200, FACS: 1:100), Sca1 (BioLegend; clone D7; IF of tissue 1:200, FACS: 1:200), Lrig1 (R&D Systems, CatNo: AF3688; IF: 1:500), BrdU (Abcam; CatNo: ab6326; IF: 1:500), Keratin5 (Krt-5) (Covance; CatNo: PRB-160P; IF: 1:1000), Keratin6 (Krt-6) (Covance; CatNo: PRB-169; IF: 1:500), CDP (Santa Cruz; CatNo: sc-13024; IF: 1:500), β1 integrin (BD Biosciences; clone Ha2/5; FACS: 1:200); β3 integrin (BD Bioscience; clone 2C3.G2; FACS: 1:200), β4 integrin (BD Biosciences; clone 346-11A; FACS: 1:200), β6 integrin CH2A1 (Itgb6) (a gift from Shelia Violette, Biogen Idec; IF of mouse tissue: 1:500); β6 integrin (Itgb6) (Chemicon; clone 10D5; IF of human tissue: 1:200, FACS: 1:200, IF of cells: 1:500), β5 integrin (Itgb5) (a gift from Dean Sheppard, University of California, USA ; FACS: 1:200), β8 integrin (Itgb8) (Santa Cruz; CatNo: sc-25714; FACS 1:200), paxillin (BD Bioscience; CatNo: 610051; IF: 500); pSMAD2/3 (Santa Cruz, CatNo: sc11769; IF: 1:200), pSmad2 (Millipore; W: 1:2000); total Smad2/3 (Santa Cruz; catNo: sc-8332; W: 1:1000), pSMAD1/5/8 (Millipore; CatNo: AB3848; IF of tissue: 1:200), Sox9 (a gift from Michael Wegner, University of Erlangen, Germany; IF of tissue: 1:500), Trichohyalin (AE15) (Abcam; CatNo: ab58755; IF of tissue: 1:500), activated Notch1 (NICD) (Abcam, CatNo: ab8925; IF of tissue: 1:200), and Pan Laminin (Sigma; CatNo: L9393; IF of tissue: 1:500). The following secondary antibodies were used: goat anti–rabbit Alexa488 (CatNo: A11008), goat anti–human Alexa488 (CatNo: A11013), goat anti–mouse Alexa488 (CatNo: A11029), donkey anti–goat Alexa488 (CatNo: A11055), goat anti-guinea pig Alexa 594 (CatNo: A11076), goat anti-mouse Alexa546 (CatNo: A11003), goat anti–rabbit Alexa546 (CatNo: A11010) (all from Invitrogen, FACS: 1:500, IF: 1:500), streptavidin-Cy5 (Dianova; CatNo: 016170084; FACS: 1:400), goat anti–rat HRP (Dianova; CatNo: 712035150; W: 1:10000), goat anti–mouse HRP (CatNo: 172-1011) and goat anti–rabbit HRP (CatNo: 172-1019) (all from Biorad; W: 1:10000). For mouse monoclonal primary antibodies M.O.M kit (Vector Labs) was used according to the manufacturer’s protocol. Mast cells were stained for mast cell heparin with avidin-FITC (Invitrogen; CatNo: 43-4411; IF of tissue: 1:100) as previously described64. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma).
Notch signaling inhibitor DAPT (Selleckchem; CatNo: S2215) was dissolved in ethanol at 25 μg μl−1. Wnt inhibitor IWP-2 (Calbiochem; CatNo: 681671) and IWR-1 (Sigma; CatNo: I0161) were dissolved in DMSO at 2.5 μg μl−1 (IWP-2) or 10 μg μl−1 (IWR-1).
BrdU labeling
LRC assay was performed as previously described27. Briefly, 10 d old mice were injected 4 times with 50 mg kg−1 bodyweight BrdU every 12 h to label mitotic cells and then mice were maintained for indicated chase periods. To determine LRCs per HF bulge, tail epidermal whole mounts were prepared, z projections were acquired using a Leica SP5 confocal microscope (objective 20×) and BrdU-positive cells were counted from ≥30 HFs per animal from different whole mounts.
Cell culture
TGFβ reporter cell line tMLEC and LTBP1-TGFβ matrix producing CHO-LTBP1 cells were used as described previously34. Primary keratinocytes were isolated at P21 or at indicated time points as described previously65. To generate clonal keratinocyte cell lines, primary cells from Kindlin-1fl/fl mice were spontaneously immortalized, single clones were picked and Kindlin-1 ablation was induced by transient transfection with an adenovirus expressing Cre recombinase. Keratinocytes were cultured in KGM containing 8% chelated FCS (Invitrogen) and 45 μM Ca2+ in a 5% CO2 humidified atmosphere on plastic dishes coated with a mixture of 30 μg ml−1 collagen I (Advanced BioMatrix) and 10 μg ml−1 FN (Invitrogen).
Inhibition of premature anagen induction and increased Notch signaling
At telogen onset (P49) back skin of control and Kind1-K5 mice was shaved and 100 μl of 200 μM IWR-1 or IWP-2 (diluted in PBS) was subcutaneously injected every 24 h into a marked skin region. 100 μl DMSO diluted in PBS was used for control experiments. At P56 the treated back skin was isolated, sectioned and stained with H/E. Ten serially sectioned HFs per animal were counted. To analyze Notch signaling back skin section were stained for NICD and NICD-positive nuclei were quantified.
Constructs and transfections
Wnt4 expression constructs (gift from Andreas Kispert, University of Hannover, Germany) and the human β-catenin expression construct (gift from Jürgen Behrens, University of Erlangen, Germany). Human WNT5A (ID18032), LEF1 (ID16709), pHes1-luc (ID43806), SuperTOPFlash (ID17165) and FOPFlash (ID12457) reporter plasmids were purchased from Addgene. pEGFP-C1 expression vector was acquired from Clontech.
GFP-tagged Kindlin-1, -2 and Kindlin-1-QWAA expression constructs were described3 and the Kindlin1-NLS construct was generated by fusing SV40 NLS motifs to the N-terminus of Kindlin-1. The expression cassettes were driven by a CAG promotor and flanked by two ITR sites recognized by sleeping beauty transposase 100× (SB100×)66 and transiently cotransfected with SB100× vector in a 1:1 ratio using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. After two passages cells stably expressing the GFP-fusion protein were FACS sorted for equal expression levels, which was further confirmed by western blotting.
Histology and immunostainings
Small pieces from back skin were either PFA fixed and embedded in paraffin or frozen on dry-ice in cryo-matrix (Thermo) and sectioned. Tail skin whole-mounts were prepared and immunostained as described27. Immunohistochemistry (H/E, DAB, OilRedO, AP and Xgal) of skin sections and tail whole mounts and immunofluorescence staining of tissue sections were carried out as described25. To better visualize the blue β-Gal stain the epidermal whole mount was overlaid with a grayscale image. Immunostaining of human skin sections for β6 integrin followed published protocols67.
HF bulge sizes were quantified in epidermal tail whole mounts stained for Krt-15 and z-projections that were collected with a confocal microscope using a 20× objective. Length and diameter of Krt-15 positive bulge area was measured with the ImageJ software (Version 1.41n) and bulge volumes were calculated using the circular cylinder formula (v=π×r2×h; v, volume; r, radius; h, height). The HF numbers were quantified in serial sections of comparable back skin regions from at least three mice per genotypes and indicated ages. HFs per 10× objective field of H/E stained sections were counted. HFs in anagen and early HF development were staged as described previously68,69.
Images were collected by confocal microscopy (DMIRE2 or SP5; Leica) with a 10× , 20× NA 1.4 or 40× oil objective using the Leica Confocal Software (version 2.5 Build 1227) or by bright field microscopy (Axioskop; Carl Zeiss with a 10× NA 0.3, 20× NA 0.5 or 40× NA 0.75 objective and camera DC500 with IM50 software (Leica).
In situ hybridization
Antisense riboprobes for mouse Fzd4 (SpeI-EcoRI fragment of mouse FZD4 3′-UTR), Wnt5a (gift from Irma Thesleff, University of Helsinki, Finland) and Hes1 (gift from Ryoichiro Kageyama, Kyoto University, Japan) were synthesized with T7 RNA polymerase (New England Biolabs) and DIG RNA Labeling Mix according to the manufacturer’s instructions (Roche). Isolated tail skin epidermis was fixed in 4% PFA in PBS, washed in PBS for 5 min twice and then twice for 15 min in PBS with 0.1% active diethylpyrocarbonate followed by 15 min equilibration in 5× saline-sodium citrate (SSC). Samples were first pre-hybridized for 1 h at 58 °C, in 50% formamide, 5× SSC, 40 μg/ml salmon sperm DNA, then hybridized for 16 h at 58 °C with 400 ng ml−1 of DIG-labeled probe in pre-hybridization mix, washed twice for 30 min in 2× SSC at RT, followed by 60 min wash at 65 °C in 2× SSC and then 60 min wash at 65 °C in 0.2× SSC. The tissues were washed in TBS for 5 min three times, blocked in 0.5% blocking reagent (Roche) in 0.1% Tween20/TBS (TBST) for 90 min at RT, then incubated in 1:5000 diluted anti-DIG AP (Roche) in 0.5% blocking reagent in TBST at 4 °C overnight. Following three 60 min washes in TBST, tissues were incubated in NTMT (100 mM NaCl, 100 mM Tris pH 9.5 50 mM MgCl2, 0.1% Tween 20) for 10 min at RT. Color was developed using BM Purple AP substrate precipitating reagent (Roche) at 37 °C. The reaction was stopped for 15 min with TE buffer (Tris 10 mM, EDTA 1 mM, pH 8.0) and samples were mounted with 70% glycerol.
Peptide pull-downs
Pull-downs were performed with β1 wt tail peptide (HDRREFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKYEGK-OH), β1 scrambled peptide (EYEFEPDKVDTGAKGTKMAKNEKKFRNYTVHNIWESRKVAP-OH), β6 wt tail peptide (HDRKEVAKFEAERSKAKWQTGTNPLYRGSTSTFKNVTYKHREKHKAGLSSDG-OH) and β6 scrambled peptide (KTDHAVQGDKKLSHKKNRGTSKATFPKVRHYETEEWAALESGYGSRTFKNSR-OH). All peptides were desthiobiotinylated. Prior to use, peptides were immobilized on 35 μl Dynabeads MyOne Streptavidine C1 (10 mg ml−1, Invitrogen) for 3 h at 4 °C.
Keratinocytes were lysed on ice in Mammalian Protein Extraction Reagent (Thermo Scientific) and 1 mg of cell lysate was incubated with the indicated peptides for 4 h at 4 °C. After three washes with lysis buffer the beads were boiled in SDS-PAGE sample buffer and the supernatant loaded on 8% SDS-PAGE gels.
Preparation of αvβ6 antibody-coated cover slips
Antibody coating was carried out as described70. Briefly, glass slides (24 mm × 24 mm; Menzel) were sterilized, coated with collagen I (Advanced BioMatrix) for 1 h at 37 °C, air dried, covered with nitrocellulose dissolved in methanol and air dried again. Slides were incubated with αvβ6 antibody (10D5; 10 μg ml−1; Chemicon) diluted in PBS over night at 4 °C in a humidity chamber, washed once with PBS and blocked with 1% BSA for 1 h at RT.
Generation of dense gold nanoarrays functionalized with cyclic-RGD
In a typical synthesis, 7 mg ml−1 of polystyrene(154)-block-poly(2-vinylpyridine)(33) (PS154-b-P2VP33, Polymer Source) was dissolved at RT in p-xylene (Sigma) and stirred for 2 d. Hydrogen tetrachloroaurate (III) trihydrate (HAuCl4·3H2O; Sigma) was added to the block copolymer solution (1 HAuCl4 per 2 P2VP units) and stirred for 2 d in a sealed glass vessel. Glass cover slips (Carl Roth) were cleaned in a piranha solution for at least 2 h and extensively rinsed with MilliQ water and dried under a stream of nitrogen. Micellar monolayers were prepared by dip-coating a glass cover slip into previously prepared solutions with a constant velocity equal to 24 mm min−1. The dip-coated glass slides were exposed to oxygen plasma (150 W, 0.15 mbar, 45 min, PVA TEPLA 100 Plasma System). To prevent nonspecific protein adsorption or cell binding, the glass background was covalently modified with silane-terminated polyethylene glycol (PEG; molecular weight 2000)71. Thiol-terminated cyclic-RGD was synthesized as described before72. The gold nanoparticles were then functionalized with this ligand by incubating the PEG-functionalized substrates in 100 μl of a 50 μM aqueous solution. The substrates were then thoroughly rinsed and incubated overnight with water and finally dried with nitrogen.
Spreading assay
A single cell suspension with 1.0 ×105 cells per well in a 6-well plate was plated in serum free KGM (1% BSA; 1% Penicillin-Streptomycin) on FN-coated glass cover slips (10 μg ml−1 FN in PBS for 1 h at RT) or indicated substrates for 3 h at 37 °C. Cells were imaged by bright field Axiovert 40 CFL microscope (20× objective; Carl Zeiss) and CV640 camera (Prosilica) before fixation in 4% PFA-PBS and immunostaining with indicated antibodies. Spreading area was quantified after staining with phalloidin Alexa488 (Invitrogen) with an AxioImager Z1 microscope (20× objective, Carl Zeiss) using the ImageJ software (Version 1.41n).
TGFβ stimulation assays
Keratinocytes were starved for several hours and stimulated with 5 ng ml-1 recombinant human TGFβ-1 (PEROTech) in serum free KGM for indicated time points, lysed and analyzed by western blotting. TGFβ modulated proliferation was determined in 70% confluent keratinocytes cultured in 6-well plates, incubated for 8 h with 10 μM 5-ethynyl-2′-deoxyuridine (EdU; Invitrogen) and indicated concentrations of TGFβ-1 and analyzed with Click-iT EdU Alexa Fluor488 Flow Cytometry Assay Kit (Invitrogen).
TGFβ release assay
TGFβ release was determined as described34. Briefly, CHO LTBP1 cell were seeded (5.0 × 104 cells per well) for 48 h in a 96 well plate in triplicates to deposit an LTBP1-TGFβ rich matrix. Cells were then detached with PBS/15 mM EDTA, the remaining LTBP1-TGFβ rich matrix washed twice with PBS and incubated with keratinocytes (2.0 × 104 cells per well) and tMLEC cells (1.5 × 104 cells per well) in a 100 μl final volume either in the absence or presence of αvβ6 integrin blocking antibody (10D5; 20 μg ml−1; Chemicon) or TGFβ neutralizing antibody (1D11; 15 μg ml−1; BD Biosciences). The amount of released TGFβ was measured after 16–24 h using Bright Glo luciferase Kit (Promega).
Wnt and Notch reporter assay
Cells were plated on FN and collagen I coated 12-well plates (6.0 × 105 cells per well) before transient transfection with 0.5 μg of pHes1-luc, SuperTOPFLASH or SuperFOPFLASH reporter, indicated expression plasmid and 50 ng thymidine kinase-driven renilla (Promega) for controlling transfection efficiency. The total amount of transfected plasmid DNA was kept constant at 1.5 μg per well by using pEGFP-C1 expression vector as transfection control (Clontech). After 24 h (Wnt reporter) or 48 h (Notch reporter) luciferase activity was analyzed with a Dual Luciferase reporter assay system (Promega). To determine effect of Wnt or Notch signaling cells were treated with indicated inhibitor 18 h after transfection and luciferase activity was measured 24 h later.
Statistical Analysis
Analyses were performed with GraphPad Prism software (version 5.00, GraphPad Software). Experiments were routinely repeated at least 3 times and repeat number was increased according to effect size or sample variation. If not mentioned otherwise in the figure legend, statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant) was determined by the unpaired t-test (for biological effects with assumed normal distribution) or Mann-Whitney U-test. In boxplot panels the middle line represents the median, the box ends the 25th and 75th percentile and the whisker ends show Min/Max distribution or are at 1.5 interquartile range while outliers are shown as dots, as indicated in the figure legend. Unbiased and reproducible identification of cells with positive nuclear staining for β-catenin, Lef1 and NICD was ensured by quantification of nuclear staining intensities with ImageJ tool (version1.41n).
Supplementary Material
Acknowledgments
We thank Dr. Julien Polleux for generating gold nanoarrays, Simone Bach for expert technical assistance, Dr. Charles Mein (Barts and the London Genome Centre) for generating the human microarray data and Drs. Roy Zent and Ralf Paus for carefully reading the manuscript. This work was funded by NIH (CA034282) to D.B.R., the Wellcome Trust (PhD studentship to J.E.L-C) and the UK National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London to J.A.M, the Advanced ERC Grant (ERC Grant Agreement no. 322652) and the Max Planck Society to R.F. .
References
- 1.Meves A, Stremmel C, Gottschalk K, Fässler R. The Kindlin protein family: new members to the club of focal adhesion proteins. Trends Cell Biol. 2009;19(10):504–513. doi: 10.1016/j.tcb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Lai-Cheong JE, et al. Kindler syndrome: a focal adhesion genodermatosis. Br J Dermatol. 2009;160(2):233–242. doi: 10.1111/j.1365-2133.2008.08976.x. [DOI] [PubMed] [Google Scholar]
- 3.Ussar S, et al. Loss of Kindlin-1 causes skin atrophy and lethal neonatal intestinal epithelial dysfunction. PLoS Genet. 2008;4(12):e1000289. doi: 10.1371/journal.pgen.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montanez E, et al. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 2008;22(10):1325–1330. doi: 10.1101/gad.469408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moser M, Nieswandt B, Ussar S, Pozgajova M, Fässler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med. 2008;14(3):325–330. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- 6.Dowling JJ, Vreede AP, Kim S, Golden J, Feldman EL. Kindlin-2 is required for myocyte elongation and is essential for myogenesis. BMC Cell Biol. 2008;9:36. doi: 10.1186/1471-2121-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai-Cheong JE, Ussar S, Arita K, Hart IR, McGrath JA. Colocalization of kindlin-1, kindlin-2, and migfilin at keratinocyte focal adhesion and relevance to the pathophysiology of Kindler syndrome. J Invest Dermatol. 2008;128:2156–2165. doi: 10.1038/jid.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y, Esser P, Heinemann A, Bruckner-Tuderman L, Has C. Kindlin-1 and -2 have overlapping functions in epithelial cells implications for phenotype modification. Am J Pathol. 2011;178(3):975–982. doi: 10.1016/j.ajpath.2010.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watt FM. Role of integrins in regulating epidermal adhesion, growth and differentiation. EMBO J. 2002;21:3919–3926. doi: 10.1093/emboj/cdf399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303(5656):359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watt FM, Jensen KB. Epidermal stem cell diversity and quiescence. EMBO Mol. Med. 2009;1:260–267. doi: 10.1002/emmm.200900033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo WM, Oro AE. SnapShot: hair follicle stem cells. Cell. 2011;146:334–334. doi: 10.1016/j.cell.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer. 2012;12:170–180. doi: 10.1038/nrc3217. [DOI] [PubMed] [Google Scholar]
- 14.Alonso L, Fuchs E. Stem cells in the skin: waste not, Wnt not. Genes Dev. 2003;17:1189–1200. doi: 10.1101/gad.1086903. [DOI] [PubMed] [Google Scholar]
- 15.Watt FM, Fujiwara H. Cell-extracellular matrix interactions in normal and diseased skin. Cold Spring Harb Perspect Biol. 2011;3(4) doi: 10.1101/cshperspect.a005124. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janes SM, Watt FM. New roles for integrins in squamous-cell carcinoma. Nat Rev Cancer. 2006;6(3):175–183. doi: 10.1038/nrc1817. [DOI] [PubMed] [Google Scholar]
- 17.Evans RD, et al. A tumor-associated beta 1 integrin mutation that abrogates epithelial differentiation control. J Cell Biol. 2003;160(4):589–596. doi: 10.1083/jcb.200209016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira M, Fujiwara H, Morita K, Watt FM. An activating beta1 integrin mutation increases the conversion of benign to malignant skin tumors. Cancer Res. 2009;69(4):1334–1342. doi: 10.1158/0008-5472.CAN-08-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White DE, et al. Targeted disruption of beta1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell. 2004;6(2):159–170. doi: 10.1016/j.ccr.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 20.Yu Y, et al. Kindlin 2 forms a transcriptional complex with β-catenin and TCF4 to enhance Wnt signalling. EMBO Rep. 2012;13(8):750–758. doi: 10.1038/embor.2012.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arita K, et al. Unusual molecular findings in Kindler syndrome. Br J Dermatol. 2007;157:1252–1256. doi: 10.1111/j.1365-2133.2007.08159.x. [DOI] [PubMed] [Google Scholar]
- 22.Emanuel PO, Rudikoff D, Phelps RG. Aggressive squamous cell carcinoma in Kindler syndrome. Skinmed. 2006;5:305–307. doi: 10.1111/j.1540-9740.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 23.Lai-Cheong JE, et al. Loss-of-function FERMT1 mutations in kindler syndrome implicate a role for fermitin family homolog-1 in integrin activation. Am J Pathol. 2009;175(4):1431–1441. doi: 10.2353/ajpath.2009.081154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Has C, et al. Kindlin-1 Is required for RhoGTPase-mediated lamellipodia formation in keratinocytes. Am J Pathol. 2009;175:1442–1452. doi: 10.2353/ajpath.2009.090203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brakebusch C, et al. Skin and hair follicle integrity is crucially dependent on beta 1 integrin expression on keratinocytes. EMBO J. 2000;19:3990–4003. doi: 10.1093/emboj/19.15.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Böttcher RT, et al. Sorting nexin 17 prevents lysosomal degradation of β1 integrins by binding to the β1-integrin tail. Nat Cell Biol. 2012;14:584–592. doi: 10.1038/ncb2501. [DOI] [PubMed] [Google Scholar]
- 27.Braun KM, et al. Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in wholemounts of mouse epidermis. Development. 2003;130:5241–5255. doi: 10.1242/dev.00703. [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara H, et al. The basement membrane of hair follicle stem cells is a muscle cell niche. Cell. 2011;144:577–589. doi: 10.1016/j.cell.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alonso L, Fuchs E. Stem cells of the skin epithelium. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11830–5. doi: 10.1073/pnas.1734203100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen KB, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munger JS, et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96(3):319–328. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 32.Xie Y, McElwee KJ, Owen GR, Häkkinen L, Larjava HS. Integrin β6-deficient mice show enhanced keratinocyte proliferation and retarded hair follicle regression after depilation. J Invest Dermatol. 2012;132:547–555. doi: 10.1038/jid.2011.381. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Bhatia R. Stem cell quiescence. Clin Cancer Res. 2011;17:4936–4941. doi: 10.1158/1078-0432.CCR-10-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Annes JP, Chen Y, Munger JS, Rifkin DB. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol. 2004;165(5):723–734. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proc Natl Acad Sci U S A. 2007;104(24):10063–10068. doi: 10.1073/pnas.0703004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, et al. Bone morphogenetic protein signaling inhibits hair follicle anagen induction by restricting epithelial stem/progenitor cell activation and expansion. Stem Cells. 2006;24(12):2826–2839. doi: 10.1634/stemcells.2005-0544. [DOI] [PubMed] [Google Scholar]
- 37.Gat U, DasGupta R, Degenstein L, Fuchs E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95(5):605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 38.Lo Celso C, Prowse DM, Watt FM. Transient activation of beta-catenin signaling in adult mouse epidermis is sufficient to induce new hair follicles but continuous activation is required to maintain hair follicle tumours. Development. 2004;131:1787–1799. doi: 10.1242/dev.01052. [DOI] [PubMed] [Google Scholar]
- 39.Lowry WE, et al. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19:1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva-Vargas V, et al. Beta-catenin and Hedgehog signal strength can specify number and location of hair follicles in adult epidermis without recruitment of bulge stem cells. Dev Cell. 2005;9:121–131. doi: 10.1016/j.devcel.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9(6):700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]
- 42.Estrach S, et al. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development. 2006;133(22):4427–4438. doi: 10.1242/dev.02644. [DOI] [PubMed] [Google Scholar]
- 43.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 44.Bernard P, Fleming A, Lacombe A, Harley VR, Vilain E. Wnt4 inhibits beta-catenin/TCF signalling by redirecting beta-catenin to the cell membrane. Biol Cell. 2008;100(3):167–177. doi: 10.1042/BC20070072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4(4):e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen B, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5(2):100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang SM, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461(7264):614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 48.Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 2005;19(12):1485–1495. doi: 10.1101/gad.341405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guasch G, et al. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 2007;12(4):313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beronja S, et al. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 2013;501(7466):185–190. doi: 10.1038/nature12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bandyopadhyay A, Rothschild G, Kim S, Calderwood DA, Raghavan S. Functional differences between kindlin-1 and kindlin-2 in keratinocytes. J Cell Sci. 2012;125:2172–2184. doi: 10.1242/jcs.096214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ussar S, Wang HV, Linder S, Fässler R, Moser M. The Kindlins: subcellular localization and expression during murine development. Exp Cell Res. 2006;312(16):3142–3151. doi: 10.1016/j.yexcr.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 53.Oshimori N, Fuchs E. Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell. 2012;10(1):63–75. doi: 10.1016/j.stem.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Annes JP, et al. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116(Pt 2):217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 55.Sin S, et al. Role of the focal adhesion protein kindlin-1 in breast cancer growth and lung metastasis. J Natl Cancer Inst. 2011;103(17):1323–1337. doi: 10.1093/jnci/djr290. [DOI] [PubMed] [Google Scholar]
- 56.Chaudhury A, Howe PH. The tale of transforming growth factor-beta (TGFbeta) signaling: a soigné enigma. IUBMB Life. 2009;61(10):929–939. doi: 10.1002/iub.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
References method section
- 57.Fässler R, Meyer M. Consequences of lack of beta 1 integrin gene expression in mice. Genes Dev. 1995;9(15):1896–1908. doi: 10.1101/gad.9.15.1896. [DOI] [PubMed] [Google Scholar]
- 58.Ramirez A, et al. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis. 2004;39(1):52–57. doi: 10.1002/gene.20025. [DOI] [PubMed] [Google Scholar]
- 59.Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat Protoc. 2009;4:1350–1362. doi: 10.1038/nprot.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kasper M, et al. Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes. Proc Natl Acad Sci U S A. 2011;108:4099–4104. doi: 10.1073/pnas.1014489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sundberg JP, Sundberg BA, Beamer WG. Comparison of chemical carcinogen skin tumor induction efficacy in inbred, mutant, and hybrid strains of mice: morphologic variations of induced tumors and absence of a papillomavirus cocarcinogen. Mol Carcinog. 1997;20:19–32. doi: 10.1002/(sici)1098-2744(199709)20:1<19::aid-mc4>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 62.Jensen KB, Driskell RR, Watt FM. Assaying proliferation and differentiation capacity of stem cells using disaggregated adult mouse epidermis. Nat Protoc. 2010;5:898–911. doi: 10.1038/nprot.2010.39. [DOI] [PubMed] [Google Scholar]
- 63.Lorenz K, et al. Integrin-linked kinase is required for epidermal and hair follicle morphogenesis. J Cell Biol. 2007;177(3):501–513. doi: 10.1083/jcb.200608125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kunder CA, et al. Mast cell-derived particles deliver peripheral signals to remote lymph nodes. J Exp Med. 2009;206(11):2455–2467. doi: 10.1084/jem.20090805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montanez E, et al. Analysis of integrin functions in peri-implantation embryos, hematopoietic system, and skin. Methods Enzymol. 2007;426:239–289. doi: 10.1016/S0076-6879(07)26012-4. [DOI] [PubMed] [Google Scholar]
- 66.Mátés L, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41(6):753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 67.Brown JK, et al. Integrin-alphavbeta6, a putative receptor for foot-and-mouth disease virus, is constitutively expressed in ruminant airways. J Histochem Cytochem. 2006;54(7):807–816. doi: 10.1369/jhc.5A6854.2006. [DOI] [PubMed] [Google Scholar]
- 68.Müller-Röver S, et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117(1):3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- 69.Paus R, et al. A comprehensive guide for the recognition and classification of distinct stages of hair follicle morphogenesis. J Invest Dermatol. 1999;113(4):523–532. doi: 10.1046/j.1523-1747.1999.00740.x. [DOI] [PubMed] [Google Scholar]
- 70.Shi Q, Boettiger D. A novel mode for integrin-mediated signaling: tethering is required for phosphorylation of FAK Y397. Mol Biol Cell. 2003;14(10):4306–4315. doi: 10.1091/mbc.E03-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blümmel J, et al. Protein repellent properties of covalently attached PEG coatings on nanostructured SiO(2)-based interfaces. Biomaterials. 2007;28(32):4739–4747. doi: 10.1016/j.biomaterials.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 72.Morales-Avila E, et al. Multimeric system of 99mTc-labeled gold nanoparticles conjugated to c[RGDfK(C)] for molecular imaging of tumor α(v)β(3) expression. Bioconjug Chem. 2011;22(5):913–922. doi: 10.1021/bc100551s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.