Abstract
The elusive etiology of germline bias of the T cell receptor (TCR) for major histocompatibility complex (MHC) has been clarified by recent ‘proof-of-concept’ structural results demonstrating the conservation of specific TCR-MHC interfacial contacts in complexes bearing common variable segments and MHC allotypes. We suggest that each TCR variable-region gene product engages each type of MHC through a ‘menu’ of structurally coded recognition motifs that have arisen through coevolution. The requirement for MHC-restricted T cell recognition during thymic selection and peripheral surveillance has necessitated the existence of such a coded recognition system. Given these findings, a reconsideration of the TCR–peptide-MHC structural database shows that not only have the answers been there all along but also they were predictable by the first principles of physical chemistry.
By focusing the molecular ‘lens’ through which immunologists look at experimental questions, structural biology has had a considerable effect on immunology. There is a relatively high degree of structural sophistication among immunologists, as molecular recognition lies at the heart of immunology. However, although many complexes of T cell antigen receptor (TCR) and peptide–major histocompatibility complex (pMHC) have been published in the past 10 years1, there is far from uniform agreement among structural biologists and immunologists about how to interpret this structural information2–12. We believe the cause of most of this confusion is rooted in misunderstandings about very basic physicochemical aspects of molecular interactions between proteins. The intention of this perspective is to clarify some of these principles and to illustrate that both recent and previous structural data are actually very much in accord with functional biological observations. The main issue addressed here is often referred to as ‘TCR-MHC bias’ or ‘germline-encoded recognition’ of MHC. These terms are used to describe the original idea that TCR and MHC molecules coevolved a ‘preference’ to interact with one another13. Although this hypothesis has been supported by several cellular, biochemical and functional studies14–17, the molecular basis of this phenomenon, and even its existence, remains controversial3,10,12,18,19. Here we attempt to provide a clear structural explanation of both the questions and the answers.
Unexplained features of TCR-pMHC interactions
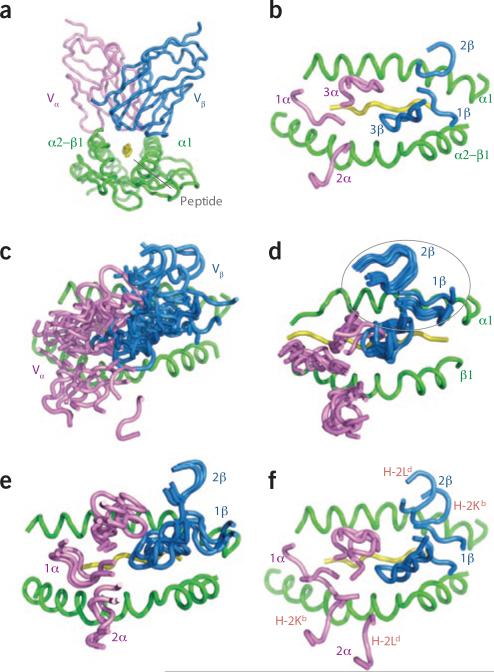
Ten years after the first complex structures were reported20,21, TCR-pMHC interactions still present one of the most interesting and enduring structural puzzles in biology, mainly because the binding interface is functionally and structurally segregated into four distinct components that collectively form composite surfaces22 (Fig. 1a,b). The pMHC-binding site of the TCR is segregated into invariant germline variable (V) gene–encoded components (complementarity-determining region 1 (CDR1) and CDR2) and components encoded by junctions between somatically rearranged variable-(diversity)-joining regions (CDR3). The MHC-binding site ‘visible’ to the TCR is also segregated into a mostly conserved helical scaffold, which presents within it a diverse array of peptides to the TCR. The TCR-pMHC recognition event ‘pairs’ invariant and variant structural components of the TCR and pMHC, in that the most variable regions of the TCR (CDR3) are positioned in the center of the binding interface where they contact the peptide, whereas the more conserved elements of the TCR (CDR1 and CDR2) and the tops of the MHC helices engage in contacts that surround the central CDR3-peptide region like a gasket. Thus, the structures of the individual molecules, together with their mode of interaction (the TCR ‘footprint’ on MHC), beautifully mirror the genetic makeup of the proteins involved, as originally predicted22. The idea that most of the binding interface (75–80%) involves contact between the germline-encoded CDR1 and CDR2 TCR regions and the MHC helices (Fig. 1b) seems logical, given that the TCR V-gene repertoire is specific for MHC molecules regardless of the peptide presented16. This ‘focus’ on the MHC may facilitate the rapid, TCR-mediated ‘scanning’ of MHC molecules15,23 to identify those presented peptides that stabilize the half-life of the TCR-pMHC complex sufficiently for signaling to occur.
Figure 1.
Divergence and convergence of TCR footprints on MHC molecules. (a) Interaction of VαVβ with peptide-MHC, viewed down the MHC groove (Protein Data Bank accession number, 2CKB). (b) ’Footprint‘ view of a showing the stereotyped polarity of the Vα and Vβ CDR loops on pMHC. (c) Convergent footprint polarity but diverse CDR loop positions in nine different TCR-pMHC complexes (Protein Data Bank accession numbers, 1AO7, 1FO0, 1J8H, 1KJ2, 1ZG1, 2NX5, 1MI5, 1OGA and 1U3H). (d) Close superpositions (in circle) of the contacts of Vβ8 CDR1 and CDR2 with the I-A MHC α1 helix in six different TCR-pMHC complexes (Protein Data Bank accession numbers, 1U3H, 2Z31, 2PXY, 1D9K, 3C60 and 3C61). (e) Retention of similar germline-mediated contacts by the BM3.3 TCR with H-2Kb in three different peptide complexes (Protein Data Bank accession numbers, 1FO0, 2OL3 and 1NAM). (f) Use of alternative ‘codons’ for interaction of the 2C TCR Vα and Vβ with H-2Kb versus H-2Ld (Protein Data Bank accession numbers, 2CKB and 2OI9). H-2Kb and H-2Ld (in red) adjacent to the respective loops indicate the positions of CDR2α and CDR2β in the structures.
Intriguing to biophysicists is the use of the term ‘bias’ to describe the apparent ‘preference’ of the αβ TCR repertoire for engaging MHC molecules. In contrast, antibodies show no bias for a particular antigen and can be polyspecific24. The ‘jury is still out’ for the γδ TCR repertoire25,26. Also fascinating is the idea that an entire repertoire of V genes encodes proteins that interact, at least in the context of an intact immune system, only with a particular structural class of ligands (MHC), which are highly diverse themselves. There are no other analogous protein-protein interaction systems in the mammalian genome that present such intriguing structural questions. What could be the structural solution to such a conundrum? Bias is a ‘fuzzy’ term for a biophysicist and is an idea about which immunologists tend to have highly polarized opinions. This need not be the case. ‘Bias’ is simply a euphemism for ‘specificity’, and specificity in protein-protein interactions is something that is well understood from the standpoint of structure and chemistry, although understanding of the relationship between structure and binding energetics is still a long way off. Thus, ‘bias’ can more correctly be referred to as TCR ‘specificity’ for MHC. That is, the natural TCR repertoire seems to physically react with MHC proteins. Given that the TCR is ‘specific’ for the MHC, then the most logical prediction, from first principles of physical chemistry, is that TCR V-domain CDR1 and CDR2 loops have evolved specific sets of contacts with residues found exclusively on MHC helices. Thus, the basis of MHC restriction is simply the coevolution of specific, conserved constellations of contacts between TCR V-gene products and MHC gene products.
However, it is not quite as simple as that. The TCR is faced with a considerable challenge, as it is specific for the MHC but is also ‘cross-reactive’ with many MHC molecules16. That is, each αβ TCR can in principle recognize any MHC allele; there does not seem to be class (I or II) or allotype specificity by particular TCR germline segments as there is, for example, in natural killer cell receptors27. This cross-reactivity is essential in that it enables the TCR to briefly dock and ‘scan’ the peptide contents of many different MHC molecules. Although there do seem to be TCR Vα and Vβ TCR chain usage ‘preferences’ in certain immune responses28, in general, the αβ chains pair randomly, and practically any αβ combination can be used to recognize most MHC molecules during an immune response. Given the combinatorial diversity of the αβ chain repertoire (approximately 2,000–3,500 possible pairs), how is the TCR specific for only MHC and many MHC molecules at the same time? This question has captured the interest of structural biologists and immunologists alike.
Until recently, the database of crystal structures was apparently not very informative about an answer to this problem1,12,29 (Fig. 1c). Although they vary considerably in the details, the known TCR-pMHC structures share a roughly diagonal docking mode (±75°) with a uniformly stereotyped binding polarity, in which the Vα domain lies mainly over the amino-terminal end of the peptide and the α2 helix (MHC class I) or β1 helix (MHC class II), and the Vβ domain lies mainly over the carboxy-terminal region of the peptide and the respective α1 MHC helices. Notably, there have not seemed to be obviously conserved contacts between TCR V regions and MHC helices among the TCR-pMHC complexes; such a broadly conserved structural determinant might have constituted the ‘smoking gun’ of MHC ‘bias’. Such a lack of conservation supports the idea that the prethymic TCR repertoire has unbiased, random specificity19 and that ‘extrinsic influences’, such as the antigenic peptides, constraints encountered during thymic selection30, or coreceptors2,18 focus the docking orientations of the V-domain interactions on the MHC helices. This calls into question the idea that there is indeed a specific germline-derived structural origin of the TCR bias for MHC. The implication is that the V-domain contacts with MHC helices are an energetically continuous or flat landscape that forms as a secondary consequence of CDR3-peptide interactions and/or coreceptor-imposed steric influences.
Nevertheless, although the existing collection of TCR-pMHC structures has not shown conserved contacts, the conservation of the docking polarity (that is, Vα always lies over the α2-β1 helix, and Vβ always lies over the α1 helix) is a clear indication, dating back to the first structures of complexes, of germline specificity between the Vα and Vβ domains and particular helices of the MHC (Fig. 1c). Given such specific docking polarity, the answer to this problem, consequently, seems predictable on the basis of basic structural principles: as interatomic contacts in protein interfaces are specific and persistent, the contacts of CDR1 and CDR2 with the MHC helices represent germline-encoded energetic minima (‘clicks’, if thought of as analogous to a socket wrench) rather than slippery surfaces devoid of innate specificity. Thus, despite the ‘blurred’ structural data, from first principles, a TCR-MHC recognition ‘code’ seems likely to orchestrate germline bias9. For a better understanding of this, clarification of some basic ideas is in order.
Protein-protein interactions are governed mainly by van der Waals interactions, hydrogen bonds and charges (salt bridges)31,32. Interactions between TCRs and MHC molecules are no different from any other protein-protein interactions in that their engagement is governed by the same first principles of molecular recognition. Protein-protein interactions generally bury several thousand square angstroms of surface area in the interfaces. As with all protein-protein interactions, most contacts in TCR-pMHC interfaces are mediated by van der Waals interactions1,7, which are individually weak but, when present in large numbers across broad interfaces, ‘sum’ to substantial binding energies. Hydrogen bonds are important in specificity because of their exquisite energetic dependence on the stereochemistry and geometry of the bond, as is also true for salt bridges33. Proteins associate mainly because water is expelled from the interacting surfaces (the hydrophobic effect), which generates a gain in entropy34. These ‘desolvated’, interacting surfaces then engage in many van der Waals contacts, whose specificity is enhanced by hydrogen bonds and salt bridges. Protein interfaces with good ‘shape complementarity’ (such as ‘knob-in-hole’ interfaces) maximize the number of van der Waals contacts and generally have higher affinity35. Although TCR-pMHC interactions do bury large amounts of surface area, they seem to have relatively less optimal shape complementarity (dissociation constants in the micromolar range)36 than do cytokine-receptor interactions (dissociation constants in the nanomolar range); this low shape complementarity no doubt contributes to the low average affinity of TCR-pMHC interactions. Of importance to the argument about germline bias is the fact that interatomic contacts in low-affinity protein-protein interactions are no less specific than those in high-affinity interactions32. Thus, the collection of interatomic contacts mediating the several thousand square angstroms of contact between TCR germline–derived regions and the MHC helices are probably not formed passively or secondarily as a result of ‘steering’ or ‘steric exclusion’ by a coreceptor or MHC-bound peptide, respectively.
While this may explain TCR specificity for one MHC molecule, how can the TCR cross-reactivity for so many different MHC molecules be explained? Is this not evidence of a lack of germline MHC specificity, given how diverse MHC alleles are? Again, no matter how cross-reactive, protein complexes bind each other only with highly specific, evolutionarily refined docking chemistries. Cases of true cross-reactivity are more correctly called ‘polyspecificity’37. There are many examples in the immune system of specific recognition by one protein of a spectrum of diverse ligands24,38–40. The fact that the TCR has specificity for many MHC molecules does not indicate that this polyreactivity is due to a flat energetic landscape mediated by ‘promiscuous’ interactions between the V segments and the MHC helices. Structurally, the idea of ‘promiscuity’ is a complete misnomer and runs counter to the basic first principles of physical chemistry: there is no such thing as a ‘promiscuous interface’ from a structural chemistry standpoint. If there is a measurable binding event between two macromolecules, even very weak binding, it is mediated by specific interatomic contacts. In addition to mutational data that have identified focused energetic interaction ‘hot spots’ on MHC helices and CDR1 and CDR2 domains6,9,15,17,41, evidence for the specificity of the interatomic contacts is that low-affinity protein complexes can be crystallized and structures can be determined to high resolution. Crystal structure determination requires that hundreds of millions of different complexes in the crystal lattice be in the exact same structural arrangement to within fractions of an angstrom. Nonspecific complex interfaces simply cannot be crystallized because of the requirement that each complex in the lattice be an exact duplicate of every other molecule. Thus, were germline-encoded interactions between TCR and MHC nonspecific and merely a consequence of a loosely constrained shape complementarity, degenerate main-chain contacts or ‘extrinsic factors’, TCR-pMHC complexes simply could not be crystallized.
Clarification provided by recent observations
The fact that the database of approximately 20 TCR-pMHC crystal structures consists of complexes containing many different αβ chains, peptides and MHC molecules has blurred and obscured any structural conservation (Fig. 1c). Recently, however, some clarity has emerged for this problem (Fig. 1d). One study focusing on the interaction of one particular germline Vβ segment (Vβ8.2) with a particular MHC molecule (I-Au) has determined crystal structures of three Vβ8.2 TCR–I-Au MHC complexes9. Comparisons among these structures has revealed a notably coincident pair-wise interaction motif, which was termed a ‘codon’, between the Vβ8.2 CDR1 and CDR2 loops, involving mainly Glu54β, Tyr48β and Asn31β, contacting residues on the top of the I-Au α1 helix that are conserved across I-A alleles: Lys39, Gln57 and Gln61, respectively. This constellation of amino acids engages in hydrogen bonds, van der Waals interactions and charged contacts; notably, these interactions occur between amino acid side chains and are therefore specific contacts. Another study determined the structures of three additional Vβ8 TCR–I-Ab complexes (two with Vβ8.2 and one with Vβ8.1) that, notably, were derived from mice with impaired negative selection42. Thus, these TCRs might represent ‘preselection’ TCRs and could demonstrate whether thymic selection ‘filters’ an unbiased TCR repertoire into one that binds MHC exclusively with the typical geometry. These complexes have shown binding geometries similar to ‘conventional’ TCRs, which suggests that these interfacial contacts are indeed ‘engrafted’ in the TCR germline before selection42. Even more notably, the complexes crystallized in these studies, together with a published D10 TCR (Vβ8.2)–I-Ak complex, constitute a set of six different Vβ8.2 TCRs and one Vβ8.1 TCR in complex with I-A (Fig. 1d). Superposition of these complexes shows close convergence of the CDR1β and CDR2β contacts with MHC, which presents a very different picture than that of previous superpositions (Fig. 1c). Although there is nearly exact correspondence of the Vβ contacts in four of the complexes, in two of them, the CDR1 and CDR2 interactions with MHC show some ‘wobble’ in the positions of side chains, as would be expected given the pairing of different peptides and Vβ with different Vα chains (as well as differing resolutions of the structures). Nevertheless, all of the complexes show a similar ‘knob-in-hole’ complementarity in which Tyr48 of CDR2β occupies a depression on the α1 helix of I-A; this interaction is then surrounded by a ‘halo’ of side chain–specific interactions. The coincidence is striking. The appearance of this Vβ8-interaction motif in the different structures with distinct MHC alleles (I-Au, I-Ak and I-Ab), Vα gene segments (Vα4.2, Vα4.1, Vα3.1 and Vα2.3) and peptides (MBP1-11, ConA and 3K) is the strongest evidence to date that this interaction is germline ‘encoded’ at the level of pairwise amino acid contacts.
Further evidence supporting the idea of germline-encoded specificity is provided by crystallized complexes of TCR molecules bound to allogeneic MHC class I molecules. The 2C TCR (also Vβ8.2), together with several high-affinity variants of 2C generated by randomization of the CDR3α, have been crystallized in complex with H-2Ld–QL9 (refs. 8,43). These studies have shown identical germline-encoded contacts between the CDR1 and CDR2 domains and the helices of H-2Ld in 2C and its variants. The importance of this experiment is that the selection of the high-affinity variants is done by yeast display in a cell-free system, in which the docking orientation of the TCR on H-2Ld is not influenced by coreceptors or any other extrinsic influences44. After selection for high-affinity binding to H-2Ld–QL9 by yeast display, each of the CDR3α loops of the variants has an entirely different sequence. In the complex structures, these CDR3α conformations and QL9 peptide contacts have been substantially remodeled, yet the surrounding ‘gasket’ of germline contacts with MHC remains unperturbed. A similar CDR3-randomization experiment with a series of human MHC class I–specific TCRs has produced the same end result45. Finally, a series of studies of the BM3.3 TCR bound to H-2Kb molecules presenting very different peptides has shown that the CDR1 and CDR2 docking positions with H-2Kb helices are mostly preserved, whereas the CDR3 contacts differ to accommodate the different peptides46 (Fig. 1e). Notably, the germline contacts are not exactly similar but instead seem to adjust slightly to the different peptides. As with the Vβ8–I-A MHC class II examples, these results collectively indicate that germline-encoded interactions represent energetic minima (‘clicks’) that are not super-ceded or dictated by divergent peptide-CDR3 contacts.
Returning to the issue of germline-encoded cross-reactivity for different MHC molecules, it is also clear from the structural database that each V segment will have distinct interaction codons with each MHC (Fig. 2). That is, there does not seem to be a universally shared or ‘cryptic’ structural epitope on all MHC molecules that orchestrates MHC recognition by the TCR. Structural ‘mimicry’ by different MHC molecules is not needed to explain their shared ability to engage a single TCR V segment47. Instead, a given MHC can use entirely structurally distinct codons to engage different V segments (Fig. 2b), as one V segment can engage different MHC molecules through entirely unrelated chemistries9. For example, Vβ8.2 has completely different binding modes on H-2Ld and on H-2Kb despite the high degree of amino acid conservation on the tops of the H-2Kb and H-2Ld helices8 (Fig. 1f). Furthermore, the amino acid pairwise interactions of the Vβ8.2-containing 2C TCR with H-2Kb and H-2Ld do not even remotely resemble the interactions of Vβ8.2 with I-A molecules9,42. The structural database also shows that each V-segment type can use more than one codon with a given MHC haplotype (Fig. 2b). For example, two Vβ2-containing TCRs with H-2Kb presenting different peptides have been crystallized, showing that the Vβ2 docking footprints on the α1 helix differ considerably48,49; therefore, we interpret this as showing that the Vβ2 uses alternative codons for H-2Kb recognition. The peptide is probably involved in ‘editing’ germline-encoded TCR docking codons (‘clicks’) by selecting the most energetically and/or functionally optimal for that particular peptide. In this way, the peptide is certainly involved in the final docking geometry in that it selects from a limited ‘menu’ of defined codon options (Fig. 2b).
Figure 2.

The ‘codon hypothesis’ for germline TCR-MHC interactions. (a) each V-gene product (where ‘Vx’ is Vα or Vβ) interacts with diverse MHC surface residues on the tops of the helices of different MHC molecules (Y, X and Z) by distinct yet specific mechanisms. That is, each CDR1 and/or CDR2 engages different MHC surfaces in diverse ways: the pairwise interactions that form each codon need not be shared by different MHC molecules. The CDR-MHC interface is presented as ‘teeth’ on the respective interacting surfaces. The same ‘teeth’ in a particular Vα or Vβ CDR1-CDR2 engage different MHC surface structures (opposing ‘teeth’) in each complex in unique, highly specific ways. (b) An individual TCR germline V segment can engage one MHC molecule using several distinct codons (A, B and C) that are influenced by the interactions of CDR3 with the MHC-bound peptide. This is presented here as different docking geometries (‘footprints’) on the MHC that are mediated by the interaction of common residues on the TCR CDR1-CDR2 with different ‘registers’ of the MHC helix in each peptide complex. Inset, free energy diagram indicating that each footprint represents a low-energy binding solution (‘click’) rather than an energetic continuum.
The extension of the codon idea is that in principle, a table of interaction codons between each type of Vα or Vβ segment and each MHC molecule could be constructed that would allow prediction of a docking footprint for any TCR-pMHC complex for which the V-gene segment usage and MHC type is known. The difficulty in this will no doubt be in understanding the nature and extent of the peptide-mediated editing of codon ‘choice’.
The coded nature of TCR-MHC interactions is a unique feature of T cell recognition not found in any other receptor-ligand system in the mammalian genome. In contrast, antibody-antigen interactions need not have coded interactions because the antibody is not restricted to binding antigen presented in a composite structural scaffold that is partly conserved and partly variable. Thus, the purpose of the coding in the TCR repertoire is to ensure that every TCR has some residual affinity for most MHC molecules for the purposes of positive selection and peripheral scanning. In addition, by ‘hard-wiring’ the TCR germline to recognize MHC helices and straddle the peptide in the MHC groove, TCR CDR3 domains are ideally positioned to ‘read out’ the centrally located peptide contents.
If the idea that the TCR is structurally specific for MHC is accepted, what can be made of results showing that T cells from mice lacking MHC class I, MHC class II, CD4 and CD8 (quadruple-knockout mice) can be activated by non-MHC ligands, apparently in the absence of coreceptors18? Does this prove that TCRs are not biased for MHC recognition? The suggestion of that paper18 is that the monomorphic coreceptors, which associate their cytoplasmic domains with the kinase Lck on T cells and physically engage their extracellular domains with MHC in the TCR signaling complex, lead to a misleading appearance of TCR bias for MHC. However, when coreceptors are removed, they can no longer sequester Lck away from the TCR and the TCR can now signal through engagement of non-MHC ligands. Thus, that study concludes that TCRs are being focused on MHC molecules merely by coreceptors (that is, an ‘extrinsic factor’)18. Surely a chief function of coreceptors is to focus the TCR on MHC and to limit TCR ligand recognition ‘options.’ However, from a purely structural standpoint, one would never say that the TCR can bind only MHC among the ‘universe’ of potential protein ligands in an artificial system. When taken out of its normal biological context (for example, through the removal of coreceptors and MHC ligands), the TCR simply becomes a binding protein, like an antibody, with a diverse ‘library’ of central CDR3 sequences. It is very likely that if this combinatorial ‘library’ of CDR3 domains is ‘panned’ against a broad spectrum of proteins on the surface of a mammalian cell, there will be reactivity. For example, a TCR reactive with the non-MHC ligand fluorescein has been reported50. When an antigen-specific Fab fragment is displayed on phage with its CDR3 domains randomized, Fab fragments with binding specificities for practically any antigen can be recovered. Thus, the quadruple-knockout experiment is essentially the surface display of a variable antigen receptor ‘panned’ against naive antigens on mammalian cell surfaces. No detection of reactivity would have been a more unexpected result. The finding of interactions between TCRs and non-MHC molecules in this system does not indicate that the TCR repertoire is not biased in the germline toward MHC specificity.
Outstanding questions
While we now have ‘glimpses’ of codons of MHC restriction, many questions remain in order to refine this concept. For example, there are several cases of TCR-pMHC complexes that have minimal germline interactions yet still retain the stereotypical orientation and polarity51; one particularly intriguing example is the ‘superbulged’ peptide complex that forms a ‘triad’ of MHC contacts5. Is this minimal triad sufficient to ‘encode’ germline bias for MHC? Will the codon idea apply to TCR interactions with nonclassical MHC molecules and/or CD1 (refs. 52,53)? These are all open questions. The structural basis of TCR-MHC recognition, like the structural foundations of most complicated biological processes, is a continuum of more or less ‘canonical’ binding modes, and surprises and exceptions to the ‘rules’ will no doubt appear. This perspective has highlighted our views of the most parsimonious explanations for how the system works based on fundamental principles of physical chemistry, now buttressed by structural results that have allowed us to reexamine the structural database with fresh eyes.
ACKNOWLEDGMENTS
We thank J. Kuriyan and D. Kranz for discussions. Supported by the National Health and Medical Research Council (CJ Martin Fellowship to L.K.E.), the Canadian Institute of Health Research (J.J.A.), N.I.H. (D.F., K.C.G.) and the Howard Hughes Medical Institute (K.C.G.).
References
- 1.Rudolph MG, stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 2.Buslepp J, Wang H, Biddison WE, Appella E, Collins EJ. A correlation between TCR Vα docking on MHC and CD8 dependence: implications for T cell selection. Immunity. 2003;19:595–606. doi: 10.1016/s1074-7613(03)00269-3. [DOI] [PubMed] [Google Scholar]
- 3.Collins EJ, Riddle DS. TCR-MHC docking orientation: natural selection, or thymic selection? Immunol. Res. 2008;41:267–294. doi: 10.1007/s12026-008-8040-2. [DOI] [PubMed] [Google Scholar]
- 4.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 2006;6:883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 5.Tynan FE, et al. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat. Immunol. 2005;6:1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 6.Huseby ES, Crawford F, White J, Marrack P, Kappler JW. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat. Immunol. 2006;7:1191–1199. doi: 10.1038/ni1401. [DOI] [PubMed] [Google Scholar]
- 7.Marrack P, scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu. Rev. Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colf LA, et al. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 9.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat. Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 10.Gras S, Kjer-Nielsen L, Burrows SR, McCluskey J, Rossjohn J. T-cell receptor bias and immunity. Curr. Opin. Immunol. 2008;20:119–125. doi: 10.1016/j.coi.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Housset D, Malissen B. What do TCR-pMHC crystal structures teach us about MHC restriction and alloreactivity? Trends Immunol. 2003;24:429–437. doi: 10.1016/s1471-4906(03)00180-7. [DOI] [PubMed] [Google Scholar]
- 12.Wilson IA, Stanfield RL. MHC restriction: slip-sliding away. Nat. Immunol. 2005;6:434–435. doi: 10.1038/ni0505-434. [DOI] [PubMed] [Google Scholar]
- 13.Jerne NK. The somatic generation of immune recognition. Eur. J. Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- 14.Blackman M, et al. The T cell repertoire may be biased in favor of MHC recognition. Cell. 1986;47:349–357. doi: 10.1016/0092-8674(86)90591-x. [DOI] [PubMed] [Google Scholar]
- 15.Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418:552–556. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- 16.Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 17.Sim BC, Zerva L, Greene MI, Gascoigne NRJ. Control of MHC restriction by TCR Vα CDR1 and CDR2. Science. 1996;273:963–966. doi: 10.1126/science.273.5277.963. [DOI] [PubMed] [Google Scholar]
- 18.Van Laethem F, et al. Deletion of CD4 and CD8 coreceptors permits generation of αβT cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat. Rev. Immunol. 2008;8:231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- 20.Garboczi DN, et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. [PubMed] [Google Scholar]
- 21.Garcia KC, et al. An αβ T cell receptor structure at 2.5Å and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 22.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 23.Matsui K, et al. Low affinity interaction of peptide-MHC complexes with T cell receptors. Science. 1991;254:1788–1791. doi: 10.1126/science.1763329. [DOI] [PubMed] [Google Scholar]
- 24.Inman JK. In: Theoretical Immunology. Bell GI, Perelson AS, Pimbley GH, editors. Marcel Dekker; New York: 1978. pp. 243–278. [Google Scholar]
- 25.Adams EJ, Strop P, Shin S, Chien YH, Garcia KC. An autonomous CDR3δ is sufficient for recognition of the nonclassical MHC class I molecules T10 and T22 by γδ T cells. Nat. Immunol. 2008;9:777–784. doi: 10.1038/ni.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin S, et al. Antigen recognition determinants of γδ T cell receptors. Science. 2005;308:252–255. doi: 10.1126/science.1106480. [DOI] [PubMed] [Google Scholar]
- 27.Radaev S, Sun PD. Structure and function of natural killer cell surface receptors. Annu. Rev. Biophys. Biomol. Struct. 2003;32:93–114. doi: 10.1146/annurev.biophys.32.110601.142347. [DOI] [PubMed] [Google Scholar]
- 28.Acha-Orbea H, et al. Limited heterogeneity of T cell receptors from lymphocytes mediating autoimmune encephalomyelitis allows specific immune intervention. Cell. 1988;54:263–273. doi: 10.1016/0092-8674(88)90558-2. [DOI] [PubMed] [Google Scholar]
- 29.Garcia KC, Adams EJ. How the T cell receptor sees antigen–a structural view. Cell. 2005;122:333–336. doi: 10.1016/j.cell.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Fink PJ, Bevan MJ. Positive selection of thymocytes. Adv. Immunol. 1995;59:99–133. doi: 10.1016/s0065-2776(08)60630-6. [DOI] [PubMed] [Google Scholar]
- 31.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 32.Reichmann D, Rahat O, Cohen M, Neuvirth H, schreiber G. The molecular architecture of protein-protein binding sites. Curr. Opin. Struct. Biol. 2007;17:67–76. doi: 10.1016/j.sbi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 34.Richards FM, Richmond T. Solvents, interfaces and protein structure. Ciba Found. Symp. 1977;60:23–45. doi: 10.1002/9780470720424.ch2. [DOI] [PubMed] [Google Scholar]
- 35.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 36.Ysern X, Li H, Mariuzza RA. Imperfect interfaces. Nat. Struct. Biol. 1998;5:412–414. doi: 10.1038/nsb0698-412. [DOI] [PubMed] [Google Scholar]
- 37.Wucherpfennig KW, et al. Polyspecificity of T cell and B cell receptor recognition. Semin. Immunol. 2007;19:216–224. doi: 10.1016/j.smim.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol. Cell. 2003;12:577–589. doi: 10.1016/s1097-2765(03)00365-4. [DOI] [PubMed] [Google Scholar]
- 39.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 40.McFarland BJ, Strong RK. Thermodynamic analysis of degenerate recognition by the NKG2D immunoreceptor: not induced fit but rigid adaptation. Immunity. 2003;19:803–812. doi: 10.1016/s1074-7613(03)00320-0. [DOI] [PubMed] [Google Scholar]
- 41.Manning TC, et al. Alanine scanning mutagenesis of an αβ T cell receptor: mapping the energy of antigen recognition. Immunity. 1998;8:413–425. doi: 10.1016/s1074-7613(00)80547-6. [DOI] [PubMed] [Google Scholar]
- 42.Dai S, et al. Crossreactive T cells spotlight the germline rules for αβ T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones LL, Colf LA, Stone JD, Garcia KC, Kranz DM. Distinct CDR3 conformations in TCRs determine the level of cross-reactivity for diverse antigens, but not the docking orientation. J. Immunol. 2008;181:6255–6264. doi: 10.4049/jimmunol.181.9.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holler PD, Chlewicki LK, Kranz DM. TCRs with high affinity for foreign pMHC show self-reactivity. Nat. Immunol. 2003;4:55–62. doi: 10.1038/ni863. [DOI] [PubMed] [Google Scholar]
- 45.Sami M, et al. Crystal structures of high affinity human T-cell receptors bound to peptide major histocompatibility complex reveal native diagonal binding geometry. Protein Eng. Des. Sel. 2007;20:397–403. doi: 10.1093/protein/gzm033. [DOI] [PubMed] [Google Scholar]
- 46.Mazza C, et al. How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? EMBO J. 2007;26:1972–1983. doi: 10.1038/sj.emboj.7601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huseby ES, et al. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 48.Reiser JB, et al. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat. Immunol. 2003;4:241–247. doi: 10.1038/ni891. [DOI] [PubMed] [Google Scholar]
- 49.Reiser JB, et al. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat. Immunol. 2000;1:291–297. doi: 10.1038/79728. [DOI] [PubMed] [Google Scholar]
- 50.Ganju RK, Smiley ST, Bajorath J, Novotny J, Reinherz EL. Similarity between fluorescein-specific T-cell receptor and antibody in chemical details of antigen recognition. Proc. Natl. Acad. Sci. USA. 1992;89:11552–11556. doi: 10.1073/pnas.89.23.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat. Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott-Browne JP, et al. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat. Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- 53.Borg NA, Kjer-Nielsen L, McCluskey J, Rossjohn J. Structural insight into natural killer T cell receptor recognition of CD1d. Adv. Exp. Med. Biol. 2007;598:20–34. doi: 10.1007/978-0-387-71767-8_3. [DOI] [PubMed] [Google Scholar]