Abstract
A fundamental question in developmental biology is how tissue growth and patterning are coordinately regulated to generate complex organs with characteristic shapes and sizes. We showed that in the developing primordium that produces the Drosophila adult trachea, the homeobox transcription factor Cut regulates both growth and patterning, and its effects depend on its abundance. Quantification of the abundance of Cut in the developing airway progenitors during late larval stage 3 revealed that the cells of the developing trachea had different amounts of Cut, with the most proliferative region having an intermediate amount of Cut and the region lacking Cut exhibiting differentiation. By manipulating Cut abundance, we showed that Cut functioned in different regions to regulate proliferation or patterning. Transcriptional profiling of progenitor populations with different amounts of Cut revealed the Wingless (known as Wnt in vertebrates) and Notch signaling pathways as positive and negative regulators of cut expression, respectively. Furthermore, we identified the gene encoding the receptor Breathless (Btl, known as fibroblast growth factor receptor in vertebrates) as a transcriptional target of Cut. Cut inhibited btl expression and tracheal differentiation to maintain the developing airway cells in a progenitor state. Thus, Cut functions in the integration of patterning and growth in a developing epithelial tissue.
INTRODUCTION
During animal development, the rates and patterns of cell growth, cell division, and cell death ultimately determine the size of tissues and organs. Growth encompasses both cell size and cell number and is important to amass enough cellular material to form an organ, whereas patterning is necessary for the generation of specialized cell types that enable the organ to perform its functions. Growth and patterning need to be tightly coordinated for normal development, and errors can lead to developmental defects and disease. Each organ uses different ways to achieve coordinated growth and differentiation. In some cases, patterning precedes growth (for example, in fly wing disc development), whereas in others, growth precedes patterning (for example, during segmentation in short-germ band arthropods, such as the red flour beetle Tribolium castaneum, and during segmentation in vertebrates) (1–3). In addition, uniform (the cells all divide at the same rate and at a similar time during development of the organ), localized (cells in one region of the developing organ divide, the remainder do not), and nonuniform (cells divide at different rates at a given time during development of the organ) mitosis are alternately used as driving forces of organogenesis (1, 3–5). Thus, to understand organogenesis, we need to identify the signals that regulate patterning and growth and identify the spatiotemporal activation of signaling pathways and their effectors during morphogenesis.
A model for quantitative and mechanistic analysis of integrated patterning and growth is the Drosophila wing imaginal disc, the morphogenesis of which is achieved through balanced proliferation and differentiation driven by the morphogen gradients of Decapentaplegic (Dpp), a member of the transforming growth factor (TGF) family, and Wingless (Wg) along the anterior-posterior (A/P) and dorsal-ventral (D/V) axes, respectively (2, 4). Although in the Drosophila wing disc, Dpp functions as an integrator molecule for both patterning and growth and exhibits a graded abundance, it promotes uniform proliferation in the disc (4, 6). This makes the wing disc an inadequate system for understanding how proliferation and differentiation are coordinated during development of other organs, such as the vertebrate limb or spinal cord, that develop by nonuniform proliferation in space and time (7–11). In addition, in flies and vertebrates, secreted morphogens, such as Sonic hedgehog (Shh), fibroblast growth factor (FGF), and Wnt, or cell-cell communication pathways, such as Notch and Hippo, regulate proliferation in various contexts (4, 12–16), but little is known about their potential effects and crosstalk on proliferation rates across the same tissue during organogenesis. A well-documented example for the role of a morphogen that regulates mitosis is in the development of the chick central nervous system (CNS) in which Wnt acts in a concentration-dependent manner both as a morphogen to specify distinct neuronal cell fates and as a mitogen to stimulate progenitor proliferation. This dual action of Wnt coordinates proliferation and differentiation in the developing CNS (8), and therefore, Wnt represents an integrator of patterning and growth in this system. An emerging model system to study coordination of patterning and growth during organogenesis is the remodeling of the Drosophila airways during metamorphosis. Specifically, the pupal and adult airways are formed by small numbers of progenitor cells, called tracheoblasts, that are specified in the embryo but remain quiescent until later in development (17–22). We and others have characterized the morphogenesis of the Drosophila spiracular branch (SB) tracheoblasts (23–26), which are multipotent progenitors that generate pupal tracheoles (specialized pupal airways) (26), as well as adult tracheal tubes, spiracles, and epidermal cells (24). The homeobox transcription factor Cut is necessary in the embryo for the specification of the SB tracheoblasts and their survival, whereas at the larval 3 (L3) stage, Cut coincides with dividing SB cells, suggesting that at these later times, it functions to promote proliferation. Unlike the extensively studied Drosophila wing disc epithelium, the SB does not exhibit uniform proliferation during growth. Instead, a growth zone is established during development that produces all the different cell types of the adult trachea (24).
Here, we investigated the patterning and growth of the SB and the role of Cut in their coordination. First, we developed an assay to precisely measure Cut protein abundance in the developing SB and found that Cut was nonuniformly distributed at L3; the most proliferative region had an intermediate amount of Cut. Using gene expression arrays, we investigated the signals that control the development of the SB and identified Wg and Notch as antagonistic regulators of the Cut gradient. We found that positive Wg and negative Notch signals established the proliferative growth zone that generates the different parts of the adult airways. By carefully manipulating Cut abundance, we affected organ growth: (i) Elimination of Cut resulted in cell death, whereas reduction of its amount increased mitosis; and (ii) ectopic expression of cut in cells destined to enter the endocycle (a modified G1-S cell cycle that promotes cellular growth) and differentiate into trachea was sufficient to induce mitosis and blocked tracheal differentiation. We identified the FGF receptor (FGFR), called Breathless (Btl) in flies, as a transcriptional target of Cut. Thus, our study revealed the importance of a transcription factor gradient in both patterning and nonuniform growth of a tubular organ, as well as identified the spatial deployment of signaling pathways in the regulation of this gradient.
RESULTS
Graded amounts of Cut in the SB correlate with differential expression of cell cycle and signaling genes
At the embryonic stage, Cut specifies the multipotent abdominal airway progenitors in Drosophila (24). In addition, at the L3 stage, a later stage during larval development, when the different regions of the SB exhibit differential proliferation rates, Cut is present in a subset of SB cells in an apparent graded pattern and its abundance may influence cell proliferation (24) (Fig. 1A). To test whether Cut abundance controlled cell proliferation, we developed tools to precisely measure and manipulate Cut abundance in the SB.
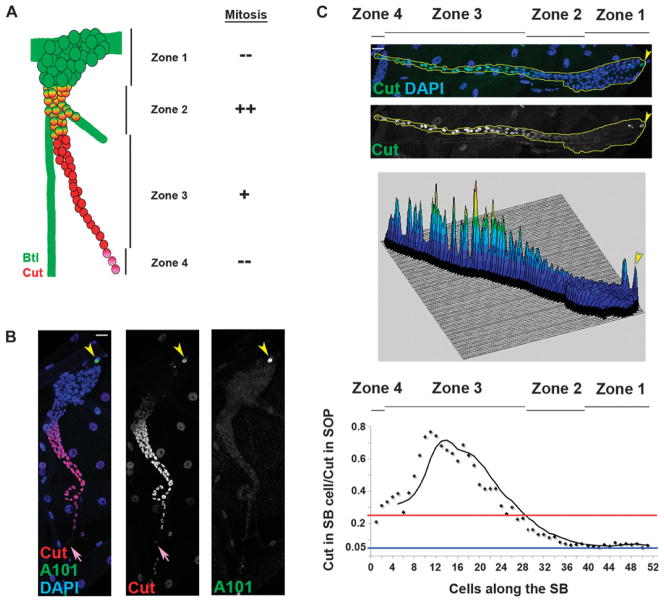
Fig. 1.
Cut abundance is graded in the developing airway progenitors. (A) Model of the four zones of SB cells during morphogenesis and their corresponding mitotic activity. The solid green areas lacking cells (circles) represent gas-transporting tracheal tubes. (B) neurA101-lacZ late L3 larva stained for Cut (red), β-galactosidase (green), and 4′,6-diamidino-2-phenylindole (DAPI) (blue). Yellow arrowheads indicate the SOP staining for both Cut and β-galactosidase, and purple arrows indicate the distal part of the SB connecting to the epidermis. (C) Top: Wild-type (WT) SB stained for Cut (green) and DAPI (blue) rotated 90° clockwise compared to (A) and (B) so that the proximal part (toward the dorsal trunk) is to the right and the distal part (toward the epidermis) is to the left. Yellow line outlines the entire SB, and yellow arrowhead indicates the Cut-positive SOP. Middle: Surface plot of the intensity of the Cut signal along the length of the SB. Bottom: Graph shows normalized Cut abundance along the length of the SB. Blue line indicates a y ratio of 0.05 (Cut is absent), and red line, 0.25 (moderate Cut amount). SB zones are indicated in the graph. Scale bar in (B) applies to all images: 20 μm.
To measure the amount of Cut in SBs, we developed a quantitative immunofluorescence assay that takes advantage of the presence of Cut in the nucleus of a sensory organ precursor (SOP) cell as a normalization control. This Cut-positive SOP is closely associated with the trachea and is easily identified in all samples due to its shape, location, and expression of the SOP marker neuralized (neur) (Fig. 1B). We used the SOP reference cell to calculate the normalized Cut abundance per cell as a ratio of the amount of Cut in each SB cell versus the SOP. Cut abundance in this SOP was not affected by any of the manipulations that we performed using cut-Gal4 (expressed in zones 2 to 4), btl-Gal4 (expressed in zones 1 and 2 and in the differentiated tracheal cells), or esg-Gal4 (weak expression in zones 2 to 4) to drive Gal4 expression in different subsets of SB cells (fig. S1, A to C). Quantification of the wild-type SB showed that Cut abundance is nonuniform in the tissue. Zone 1, which is closest to the dorsal trunk (DT) and has no detectable Cut, had a normalized Cut abundance of 0.05; zone 2, with intermediate Cut amounts, had a normalized Cut abundance more than 0.05 and less than 0.25; zone 3, with the highest abundance of Cut, had a normalized Cut abundance more than 0.25; and zone 4, located at the most distal tip of the SB with moderate Cut amounts, had a normalized Cut abundance less than 0.4 (Fig. 1C). The two regions in the SB with low but detectable Cut abundance exhibited extensive proliferation: Zone 2 coincided with the zone of highest proliferation in the L3 stage (24), and zone 4 encompassed the spiracular abdominal histoblasts, which proliferate extensively later during the pupal stage (27). Nonuniform Cut abundance was also present in SBs from younger larvae at the early L2 stage (fig. S1D), which precedes proliferation of SB cells that occurs during the L3 stage (24).
To understand how the Cut gradient is generated, we looked for factors expressed in spatially restricted patterns in the SB by transcriptional profiling. To identify these factors, we microdissected wild-type SBs from late L3 larvae, isolated two groups of cells, zone 1 cells (in which Cut was not detected) and zone 3 cells (in which Cut was abundant), and performed transcriptional profiling using Affymetrix Drosophila 2.0 arrays (Fig. 2A, see details in Materials and Methods). Gene Ontology (GO) enrichment of the 482 annotated differentially expressed genes in the two samples indicated that genes involved in the cell cycle and, more specifically, mitosis are highly expressed in cells in zone 3, whereas zone 1 cells highly express genes involved in tracheal differentiation (Fig. 2B and table S1). The more enriched categories in the Cut-positive zone 3 include genes involved in “spindle,” “mitosis,” “mitotic sister chromatid segregation,” “cell division,” “cytokinesis,” and “kinetochore” (Fig. 2B, blue bars), consistent with the mitotic ability of Cut-positive cells. In contrast, zone 1 cells, which lack Cut, express genes involved in “open tracheal system development,” “primary branching,” “secondary branching,” “epithelial cell migration,” and “fibroblast growth factor receptor signaling” (Fig. 2B, red bars), consistent with zone 1 cells undergoing tracheal differentiation.
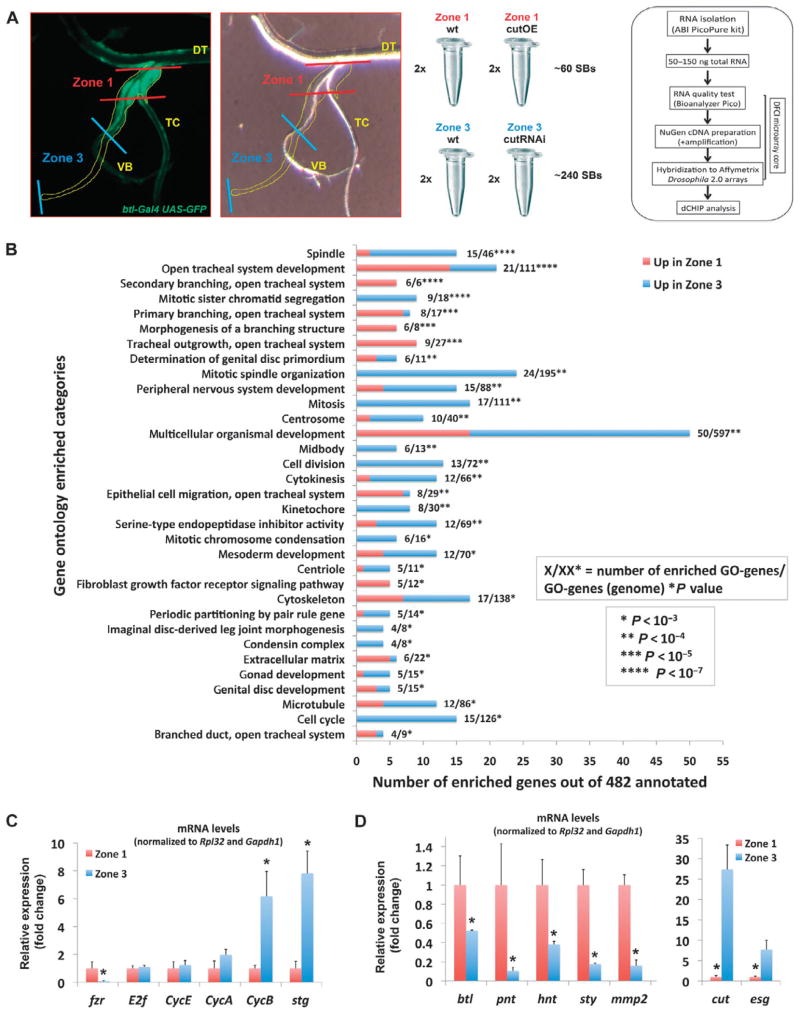
Fig. 2.
Zones 1 and 3 express different sets of genes. (A) Microarray strategy used to assess differential gene expression in different populations of SB cells. Fluorescent btl-Gal4 UAS-actGFP and bright-field images of the anterior abdominal part of the L3 tracheal system under a dissecting microscope, indicating the boundaries of different populations of SB cells (zone 1, red; zone 3, blue) used for genomic analyses. DT, dorsal trunk; TC, transverse connective; VB, visceral branch. Duplicate zone 1 samples from WT and cut-overexpressing (cutOE) flies, as well as zone 3 samples from WT and cut-knockdown (cutRNAi) flies, were collected and their transcriptome was analyzed using Affymetrix arrays as shown in the box. (B) GO enrichment analysis of filtered genes differentially expressed in WT zones 1 and 3 (fold change ≥1.5). Each bar corresponds to the total number of enriched genes in each category. The ratio next to each bar corresponds to the enriched genes over the total number of genes in the genome in each GO category (P values were calculated by Student’s t test in dChip). The red and blue parts of each bar represent the number of genes up-regulated in zone 1 (positive FC, up in zone 1) and down-regulated in zone 1 (negative FC, up in zone 3), respectively. (C) qRT-PCR validation of cell cycle genes. (D) qRT-PCR validation of tracheal genes. Error bars in (C) and (D) correspond to the SD from the mean of biological triplicates. *P ≤ 0.05 by Student’s t test.
To validate the microarray experiment, we performed quantitative reverse transcription polymerase chain reaction (qRT-PCR) experiments and found that, indeed, genes involved in mitosis, such as Cyclin B (CycB) and the Drosophila homolog of cdc25, string (stg), were highly expressed in zone 3. Cyclin A (CycA) also showed a trend of higher expression in zone 3, whereas E2f and Cyclin E (CycE), genes involved in G1-S phase progression, were not differentially expressed (Fig. 2C). In addition, fizzy-related ( fzr), a regulator of the endocycle, was enriched in zone 1 (Fig. 2C). qRT-PCR analysis of genes involved in tracheal cell differentiation and migration confirmed that members of the Btl pathway, such as the receptor encoded by btl, the Ets transcription factor encoded by pointed ( pnt), the negative regulator of receptor tyrosine kinases encoded by sprouty (sty), Matrix metalloproteinase 2 (Mmp2), and hindsight (hnt), were enriched in zone 1 (Fig. 2D). Finally, genes previously shown to be expressed in zone 3 (24), such as cut and escargot (esg), showed the expected pattern (Fig. 2D). These data agree with our previous results (24), provided further evidence that cut expression coincides with dividing SB cells and inversely correlates with tracheal differentiation, and identified additional genes that are specifically expressed in different populations of the SB.
To assess which signaling pathways are activated in different SB populations and regulate Cut gradient formation, we compiled a list of all the genes encoding components of conserved developmental signaling pathways (table S2) and performed hierarchical clustering of their expression in zones 1 and 3 (fig. S2). Because ligands seemed likely candidates to reveal information about spatial activity of signaling pathways, we focused on genes encoding ligands and limited our analysis to those genes that met the criteria for confidence scoring (transcripts were detected in more than 20% of the samples with an expression of at least 65 in at least 25% of the samples; see Materials and Methods for details) in the micro-array data (Fig. 3A). We found that transcription of the genes encoding the Notch ligand Delta (Dl), the FGFR ligand Branchless (bnl), and the platelet-derived growth factor (PDGF) and vascular endothelial growth factor (VEGF) receptor (PVR) ligand Pvf1 was enriched in zone 1, and Wg pathway ligands and others were enriched in zone 3 (Fig. 3A).
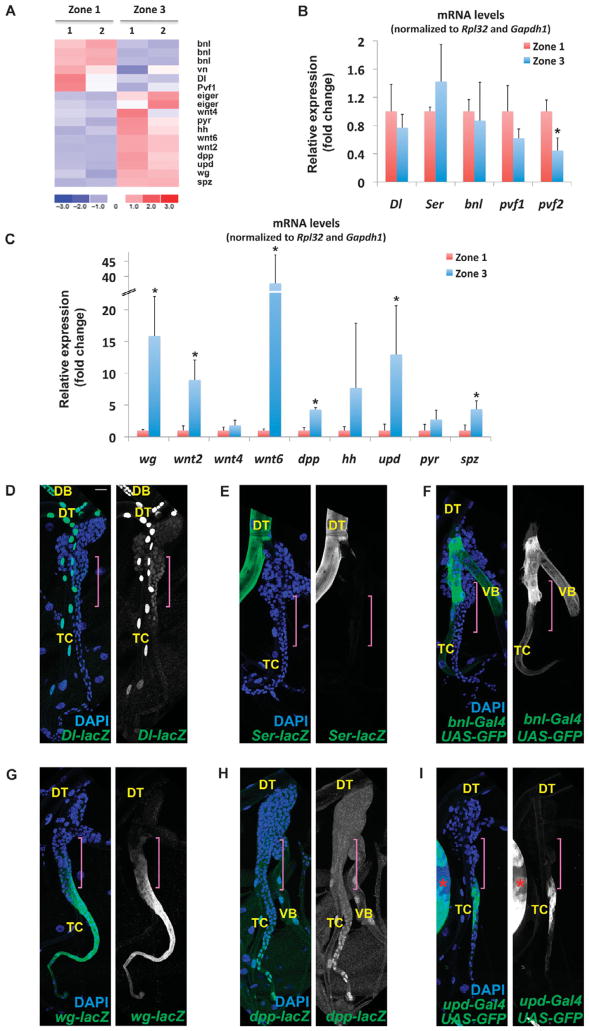
Fig. 3.
Differential expression of signaling pathway ligands in the SB. (A) Heat map of conserved ligand gene expression in zone 1 and zone 3 samples in duplicates (samples 1 and 2). (B) qRT-PCR validation of genes encoding ligands with higher expression inzone1. (C) qRT-PCR validation of genes encoding ligands with higher expression in zone 3. Error bars in (B) and (C) correspond to the SD from the mean of biological triplicates. *P < 0.05 by Student’s t test. (D) Dl-lacZ. (E) Ser-lacZ. (F) bnl-Gal4 UAS-GFP. (G) wg-lacZ. (H) dpp-lacZ. (I) upd-Gal4;UAS-GFP. Reporter expression in (D) to (I) is labeled in green by either green fluorescent protein (GFP) or an antibody against β-galactosidase. DAPI is blue in all colored panels. Gray panels show only reporter expression. The red asterisk in (I) indicates part of a wing disc strongly expressing the reporter. Purple brackets in all panels indicate zone 2 identified on the basis of its position at the TC-SB junction. Scale bar, 20 μm. Tracheal branches other than the SB are indicated. DT, dorsal trunk; DB, dorsal branch; TC, transverse connective; VB, visceral branch.
We performed qRT-PCR from zone 1 and zone 3 RNA extracts and analyzed the transcripts for relative abundance to that in the zone 1 samples (Fig. 3, B and C) and also assessed the activity of known reporters of these genes in the SB in vivo (Fig. 3, D to I). Of the genes encoding ligands tested by qRT-PCR, only Pvf 2 was significantly enriched in zone 1 by qRT-PCR (Fig. 3B). Because the microarray data suggested that expression of Dl was enriched in zone 1, we also tested another Notch ligand–encoding gene, Serrate (Ser), but neither of these two Notch ligand–encoding genes exhibited significant enrichment in expression in zone 1 by qRT-PCR (Fig. 3B). In vivo reporters for Dl (Dl-lacZ), Ser (Ser-lacZ), and bnl (bnl-Gal4) showed that Dl was expressed in differentiated tracheal cells of the DT, dorsal branch (DB), and transverse connective (TC), as well as zones 1 and 2 of the SB (Fig. 3D); Ser was expressed only in the gas-transporting DT region (Fig. 3E); and bnl was expressed in differentiated tracheal cells of the TC and visceral branch (VB) but not in the SB (Fig. 3F).
We found a robust signature of the Wg pathway in zone 3, where expression of wg, wnt2, and wnt6 was significantly enriched (Fig. 3C). In addition, the qRT-PCR validation indicated that mRNAs of ligands of other conserved signaling pathways were also enriched in zone 3: decapentaplegic (dpp) for the TGF-β pathway, unpaired [upd, also known as outstretched (os)] for the Janus kinase and signal transducer and activator of transcription (Jak/Stat) pathway, and spaetzle (spz) for the Toll pathway. In contrast, genes encoding the ligands of the Hedgehog pathway [hedgehog (hh)] and the FGFR Heartless (Htl) pathway [pyramus ( pyr)] did not show significant enrichment in this assay (Fig. 3C). In vivo reporters for some of these genes showed that wg (wg-lacZ) was strongly expressed in zone 3 but was weakly expressed in zones 2 and 4 (Fig. 3G), dpp (dpp-lacZ) was expressed in zones 3 and 4 (Fig. 3H), and upd (upd-Gal4) was expressed non-uniformly in zones 2 and 3 (Fig. 3I). The analysis of ligand gene expression in the SB indicated the spatially restricted activation of conserved developmental signaling pathways in different zones: the Notch pathway in differentiated tracheal cells, zone 1, and zone 2, and the Wg, Dpp, and Jak/Stat pathways in zone 3 cells.
Notch and Wg establish a growth zone through their antagonistic actions on Cut
Because transcriptional profiling and in vivo analysis indicated that the genes encoding the Notch pathway ligand, Dl, and the Wg pathway ligand, wg (as well as other wnt genes), were active in spatially restricted populations of the SB, we hypothesized that the signaling pathways themselves might be active in distinct populations of SB cells. In addition, the Notch and Wg signaling pathways have been shown to activate cut expression at the wing DV boundary (28, 29), and Notch has also been shown to inhibit cut in the follicular epithelium (30, 31).
To assess whether Notch and Wg modulated the amount of Cut in SB tracheoblasts and trachea during larval stages, we tested transcriptional reporters and antibodies that reflect the activity of the Notch and Wg pathways. We found that the Notch pathway transcriptional reporter Su(H)-GBE-lacZ (32) was active in zones 1 and 2 of the L3 SB, as well as in the mature tracheal cells (Fig. 4A), and its expression starts at the junction of the SB and TC early in development (fig. S3A). This pattern of Su(H)-GBE-lacZ expression was similar to that observed for Dl-lacZ (Fig. 3D), consistent with Dl functioning as a ligand of Notch in the SB. In addition, Wg, which coincided with cells expressing the wg-lacZ reporter (Fig. 3G), was present in SB cells positive for a Cut reporter during early development (fig. S3B) and labeled zones 2, 3, and 4 in late L3 larvae (Fig. 4A). These results indicated that the Notch and Wg pathways are activated in complementary overlapping patterns in the SB (Fig. 4A), and their activity relative to Cut suggested that they may regulate Cut in opposite ways, with Wg activating and Notch repressing Cut.
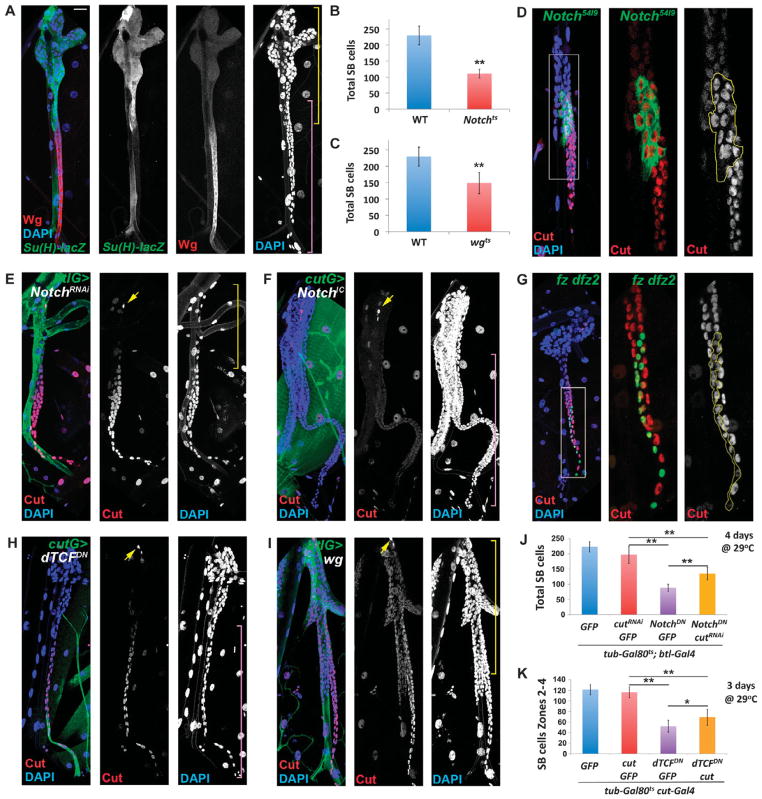
Fig. 4.
Notch represses and Wingless promotes the appearance of Cut in the SB. (A) The Notch activity reporter Su(H)-GBE-lacZ (green) is expressed in a complementary pattern to that of Wg protein (red). Gray panels show Su(H)-GBE-lacZ, Wg, and DAPI separately. Yellow bracket denotes btl-Gal4 expression domain; purple bracket denotes cut-Gal4 expression domain. (B) Reduction in the number of SB tracheoblast cells in Notchts1 late L3 larvae compared to WT (n = 15 SBs). (C) Reduction in the number of SB tracheoblast cells in wgts late L3 larvae compared to WT (n = 9 SBs). (D) Notch54l9 clone labeled by GFP (green) stained for Cut (red) and DAPI (blue). Boxed area in (D) is shown to the right. Yellow line delineates the clone boundaries. (E) tub-Gal80ts; btl-Gal4 UAS-srcGFP;UAS-NotchRNAi (btlG>NotchRNAi) stained for Cut (red) and DAPI (blue). Yellow bracket denotes btl-Gal4 expression domain. (F) cut-Gal4 tub-Gal80ts;UAS-srcGFP/UAS-NotchICD (cutG>NotchIC) stained for Cut (red) and DAPI (blue). Purple bracket denotes cut-Gal4 expression domain. (G) fz Dfz2 double-mutant clone labeled with GFP (green) and stained for Cut (red) and DAPI (blue). Boxed area in (G) is shown to the right. Yellow line delineates clone boundaries. (H) cut-Gal4 tub-Gal80ts;UAS-srcGFP/UAS-dTCFDN (cutG>dTCFDN) stained for Cut (red) and DAPI (blue). Purple bracket denotes cut-Gal4 expression domain. (I) tub-Gal80ts; btl-Gal4 UAS-srcGFP/UAS-wg (btlG>wg) stained for Cut (red) and DAPI (blue). Yellow bracket denotes btl-Gal4 expression domain. (J) Rescue of NotchDN-induced proliferation defect by UAS-cutRNAi(n=10SBs). (K) Rescue of dTCFDN-induced proliferation defect by UAS-cut (n = 10 SBs). Scale bar in (A) is representative of all fluorescent images: 20 μm, except for boxed areas in (D) and (G), where the scale bar is 8 μm. Yellow arrows in (E), (F), (H), and (I) indicate the Cut-positive SOP. **P < 0.001; *P < 0.05 by Student’s t test.
If the Notch and Wg pathways regulate Cut and Cut controls cell proliferation, then these two pathways would be predicted to affect SB cell number. Consistent with this hypothesis, when Notchts or wgts larvae were reared at the permissive temperature (18°C) and then shifted to the restrictive temperature (29°C) 24 to 48 hours before dissection, the number of SB cells was significantly reduced (Fig. 4, B and C).
To test whether Notch restricts the amount of Cut, we generated Notch54l9 mutant clones and found that Cut protein abundance was increased in clones abutting zone 2 where Cut amount is normally low (Fig. 4D). Thus, Notch activity is necessary to reduce Cut abundance in zone 2. In addition, when we lowered Notch pathway activity in zones 1 and 2 of the SB by overexpressing UAS-NotchRNAi using the btl-Gal4 driver, the number of SB tracheoblasts was reduced (Fig. 4E). Conversely, when Notch signaling was ectopically activated in zone 3 using a cut-Gal4 driver to overexpress a constitutively active form of the Notch receptor (NotchIC), the abundance of Cut was reduced and excessive proliferation was observed (Fig. 4F). These findings indicate that Notch suppressed cut, reduced the amount of Cut protein, and limited SB cell number.
To assess whether the Wg pathway contributed to the induction of Cut in the SB, we generated clones mutant for the two Wg receptors, Frizzled (Fz) and Frizzled 2 (dFz2), and thus the clones cannot respond to Wg (33). The double-mutant fz dfz2 cells showed reduced amounts of Cut (Fig. 4G). However, Cut was not entirely eliminated, indicating that additional pathways likely participate in Cut induction (Fig. 4G). In addition, Cut abundance and the SB cell number were reduced when a dominant-negative form of TCF (UAS-dTCFDN), which blocks the β-catenin transcriptional pathway activated by Wg (34), was expressed using the cut-Gal4 driver (Fig. 4H). To test whether the Wg pathway was sufficient to induce cut expression and promote SB cell proliferation, we ectopically activated the Wg pathway in zones 1 and 2 by overexpressing the Wg ligand (UAS-wg) in the btl-Gal4 domain (Fig. 4I). This led to proliferation of the SB cells and expansion of zone 2. The Wg-expressing zone 1 cells resembled zone 2 cells; they did not undergo endoreplication as indicated by their small nuclei, but instead remained in an undifferentiated state with low amounts of Cut (Fig. 4I). Therefore, we concluded that Wg pathway activity stimulated Cut induction and SB cell proliferation in zones 2 to 4, and forced activation of the Wg pathway was sufficient to stimulate Cut induction and SB cell proliferation and inhibit endoreplication and differentiation in zone 1.
To further examine the functional relationship between Notch, Wg, and Cut in SB cell proliferation, we performed genetic epistasis experiments to assess whether both pathways exert their effects through Cut. To show whether Cut functioned as a downstream effector of the Notch pathway in the SB, we coexpressed UAS-NotchDN and UAS-cutRNAi in the btl-Gal4 domain, representing zones 1 and 2, in the presence of the temperature-sensitive Gal4 repressor Gal80ts to ensure careful control of transgene overexpression in time and viability of the larvae to the late L3 stage. As expected, if Cut acts downstream of Notch, the cell number defect caused by loss of Notch was significantly rescued by loss of Cut (Fig. 4J).
Furthermore, if Cut is a downstream effector of the Wg pathway in the SB, we predicted that we could rescue the UAS-dTCFDN–induced reduction of Cut-positive SB cells by simultaneously expressing a UAS-cut transgene. For these experiments, we used cut-Gal4, representing zones 2 to 4, in the presence of Gal80ts to overexpress the transgenes in the SB temporally and monitored the number of cells in the overexpression domain. Indeed, we found a significant rescue of the cell number phenotype in the SBs of larvae overexpressing both UAS-dTCFDN and UAS-cut compared to those over-expressing UAS-dTCFDN and UAS-GFP (Fig. 4K). Thus, the spatially restricted antagonistic actions of the Notch and Wg pathways regulated the proliferation and differentiation of airway progenitors (as indicated by the effects on endoreplication), and their coordination of Cut abundance contributed to these actions.
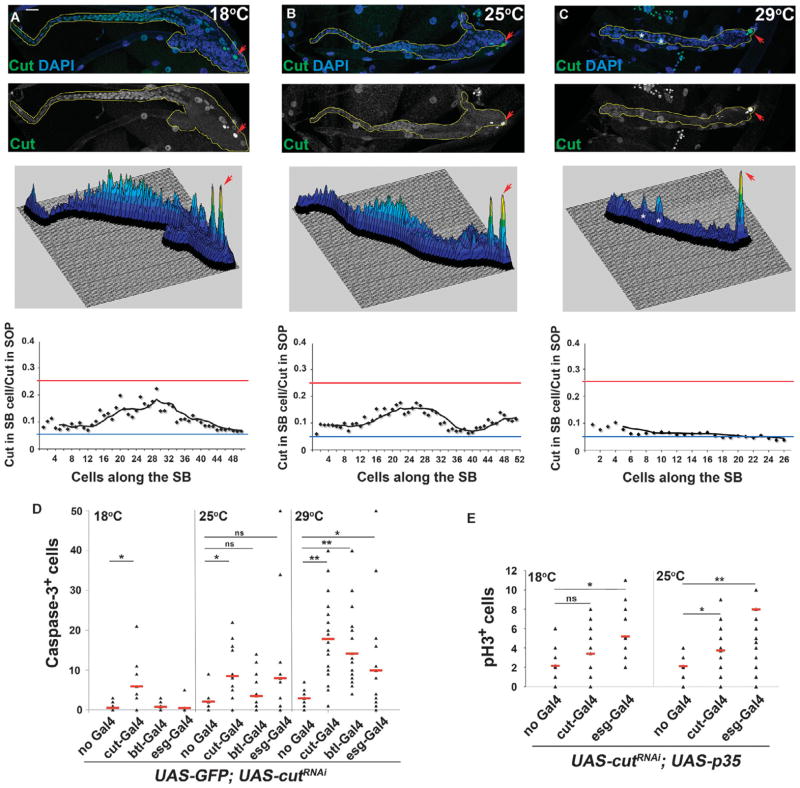
The amount of Cut is critical for SB cell survival, proliferation, and differentiation
We have shown that the Cut gradient in the SB correlated with different states and growth activities. To demonstrate that the abundance of Cut is responsible for these effects and cellular states, we manipulated the amount of Cut in the SB with tissue-specific RNA interference (RNAi) and over-expression experiments. To reduce Cut abundance, we used a UAS-cutRNAi transgene (see Materials and Methods), which, in conjunction with the tissue specificity (fig. S1, A to C) and temperature sensitivity of the Gal4-UAS system (35), allowed us to generate subsets of SB cells that expressed different amounts of Cut at different temperatures. To quantify the amount of Cut in SB cells in cut-Gal4 UAS-GFP;UAS-cutRNAi late third instar (L3) larvae reared at 18°C, 25°C, and 29°C, we calculated the normalized Cut abundance along the length of the SB. Consistent with a decrease in the amount of Cut protein in the SB at higher temperatures, the normalized Cut abundance per cell decreased with temperature (Fig. 5, A to C). We also tested the effects of Cut knockdown in specific regions of the SB using cell-specific drivers (fig. S4). We expressed UAS-cutRNAi with btl-Gal4, which is expressed in zones 1 and 2, and with esg-Gal4, which is expressed at lower levels in zones 2 to 4 (fig. S1, A and C). Although there is very little, if any, apoptotic cell death in wild-type SBs (Fig. 5D, see “no Gal4” controls at different temperatures), we found that the decrease in Cut abundance, achieved by induction of the RNAi at increasing temperatures (Fig. 5, C and D), caused increased apoptosis in the SB as measured by cleaved caspase-3 immunostaining (fig. S4). The number of apoptotic cells was greatest at higher temperatures in which the reduction in Cut abundance was greatest for all Gal4-mediated RNAi expression (Fig. 5D). This role of Cut in promoting cell survival is consistent with our previous observation that clones of SB cells mutant for cutDB7, a protein-null allele (30), were smaller than their wild-type twin spots due to apoptosis (24). Thus, elimination of Cut leads to cell death.
Fig. 5.
Reduction of Cut impairs airway progenitor survival and increases proliferation in a dose-dependent manner. (A) Top: cut-Gal4;UAS-cutRNAi SB reared at 18°C stained for Cut (green) and DAPI (blue). Middle: Surface plot of the intensity of the Cut signal along the length of the SB. Bottom: Graph showing normalized Cut abundance along the length of the SB. (B) Top: cut-Gal4;UAS-cutRNAi SB reared at 25°C stained for Cut (green) and DAPI (blue). Middle: Surface plot of the intensity of the Cut signal along the length of the SB. Bottom: Graph showing normalized Cut abundance along the length of the SB. (C) Top: cut-Gal4;UAS-cutRNAi SB reared at 29°C stained for Cut (green) and DAPI (blue). Middle: Surface plot of the intensity of the Cut signal along the length of the SB. Asterisks in (C) indicate muscle cells that are Cut-positive and produce the corresponding peaks in the surface plot. Bottom: Graph showing normalized Cut abundance along the length of the SB. Red arrowheads in (A) to (C) indicate the SOP. Blue line in (A) to (C) indicates a y ratio of 0.05, and red line, 0.25. Scale bar, 20 μm. (D) Number of apoptotic cells in SBs in which Cut was reduced with a UAS-cutRNAi transgene driven by cut-Gal4, btl-Gal4, or esg-Gal4 at 18°C, 25°C, and 29°C. (E) Number of mitotic cells in SBs in which Cut was reduced using cut-Gal4 or esg-Gal4 at 18°C and 25°C in the presence of a UAS-p35 transgene to prevent apoptosis. Data presented in (D) and (E) represent n > 10 SBs. **P < 0.001; *P < 0.05; ns, not significant (Student’s t test).
When UAS-cutRNAi was expressed such that it did not completely eliminate the Cut protein, for example, by rearing the animals at 18°C or 25°C instead of 29°C, or by using the esg-Gal4 driver, which is weaker than cut-Gal4, the number of SB cells increased compared to wild type (compare Fig. 5A to Fig. 1C). Thus, lowering the amount of Cut in SB cells, without reducing it so far that apoptosis occurred, resulted in more cells. To measure cell proliferation directly, we quantified the number of SB cells that were positive for phosphorylated histone H3 (pH3+) in the larvae grown at 18°C and 25°C. To avoid problems caused by loss of tissue integrity due to apoptosis in UAS-cutRNAi larvae (Fig. 5D and fig. S4), we coexpressed a general apoptosis inhibitor, the baculovirus protein p35 (36). This UAS-p35 transgene effectively rescued the apoptosis phenotype caused by expression of UAS-cutRNAi by cut-Gal4 (fig. S5) at both 18°C and 25°C. The number of proliferating SB cells increased in both UAS-p35/cut-Gal4;UAS-EGFP;UAS-cutRNAi and UAS-p35;esg-Gal4 UAS-EGFP;UAS-cutRNAi larvae (Fig. 5E), and this increase correlated with a reduction of Cut protein abundance with increasing temperature. The knockdown studies indicated that complete elimination of Cut protein from the SB cells resulted in cell death, whereas a moderate decrease in Cut protein abundance in zones 2 to 4 resulted in increased proliferation.
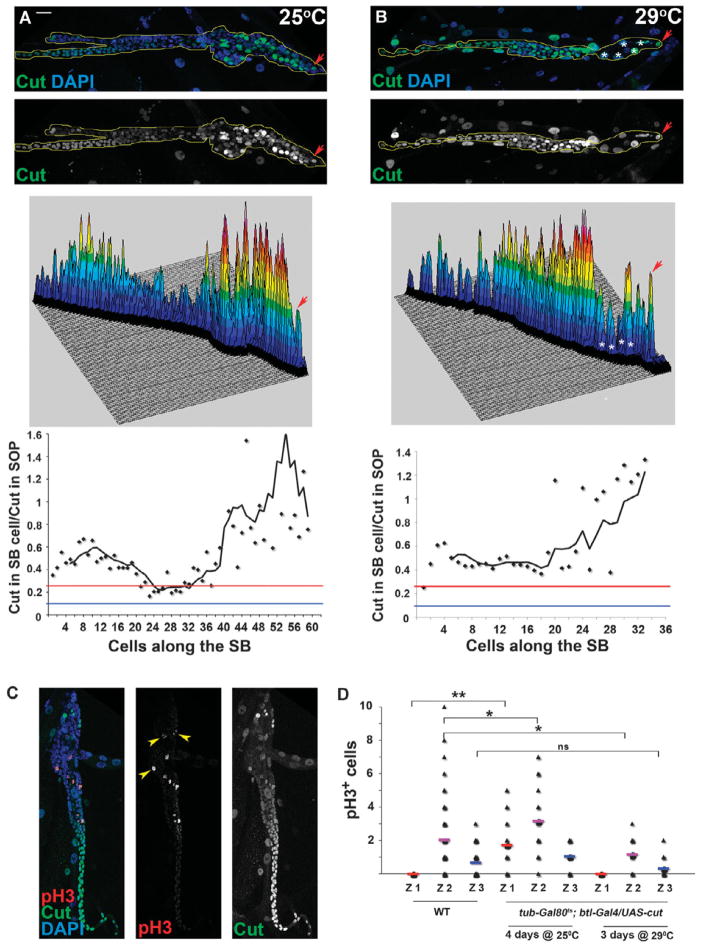
We tested whether ectopic expression of Cut had any effect(s) on the growth and patterning of the SB by ectopically expressing a UAS-cut transgene in the endoreplicating zone 1 cells and in the zone 2 cells in which Cut abundance is low (Fig. 1A), using the btl-Gal4 driver (fig. S1A) in the presence of the temperature-sensitive Gal80ts (37). This genetic configuration allowed us to activate the UAS-cut transgene to different levels at different temperatures (Fig. 6, A and B). If Cut abundance is critical for the proliferation and patterning of the SB cells, we expected to observe different results by expressing the transgene at low or high levels in a context-dependent manner: For example, zone 2 cells over-expressing cut might become less proliferative (like zone 3 cells), whereas zone 1 cells ectopically expressing cut might show proliferation rather than differentiation (like zone 2 cells). We found that low-level over-expression of UAS-cut at 25°C resulted in cells with small nuclei in zone 1 (Fig. 6, A and C), which is in agreement with the role of Cut in suppressing endoreplication, a process associated with zone 1 tracheal differentiation (24). Overexpression of UAS-cut at 25°C also stimulated proliferation in zone 1 as indicated by the presence of pH3+ cells in this area (Fig. 6, C and D). Thus, the presence of a low amount of Cut stimulated SB cell proliferation and inhibited differentiation when ectopically expressed in SB zone 1 (Fig. 6C). In addition to, and consistent with, the observation that the moderate amount of Cut in zone 2 of the SB is critical for normal development, strong overexpression of the UAS-cut transgene at 29°C prevented normal growth of SB tracheoblasts in zones 1 and 2. The cells in zones 1 and 2 were almost completely eliminated (Fig. 6B), the growth of the SB was impaired, and the number of pH3+ cells in zone 2 was significantly reduced (Fig. 6D).
Fig. 6.
Ectopic expression of cut stimulates cell proliferation. (A) Ectopic expression of Cut at 25°C (tub-Gal80ts; btl-Gal4/UAS-cut) for 4 days. SB is stained for Cut (green) and DAPI (blue). Middle: Surface plot of the intensity of the Cut signal along the length of the SB. Bottom: Graph showing normalized Cut abundance along the length of the SB. (B) Ectopic expression of UAS-cut at 29°C (tub-Gal80ts; btl-Gal4/UAS-cut) for 3 days. SB is stained for Cut (green) and DAPI (blue). Middle: Surface plot of the intensity of the Cut signal along the length of the SB. Asterisks in (B) indicate Cut-positive cells in the main trachea that produce the corresponding peaks in the surface plot and they are not part of the SB. Bottom: Graph showing normalized Cut abundance along the length of the SB. Arrowheads in (A) and (B) indicate the SOP. Blue line indicates a y ratio of 0.05, and red line, 0.25. Scale bar, 20 μm. (C) SB of tub-Gal80ts; btl-Gal4/UAS-cut genotype reared at 25°C for 4 days and stained for pH3 (red) and Cut (green). (D) Graph showing the number of pH3-positive cells per zone (Z1, zone 1; Z2, zone 2; Z3, zone 3) in the SBs of WT larvae and larvae ectopically expressing cut at 25°C and 29°C for 3 to 4 days [n > 10 SBs; **P < 0.001; *P < 0.05; ns, not significant (Student’s t test)]. Zones were identified by position in the SB.
The knockdown analysis (Fig. 5) and the ectopic expression analysis (Fig. 6) showed that there is a dose effect of Cut on the SB. Moderate excess of Cut in zone 1 impaired differentiation and promoted ectopic proliferation, whereas large excess of Cut in zone 2 decreased proliferation and impaired SB development. Moderate loss of Cut in zones 2 to 4 increased proliferation, whereas complete loss of Cut in zones 2 to 4 resulted in apoptosis and a reduction in the number of SB cells.
Cut inhibits btl expression and tracheal differentiation
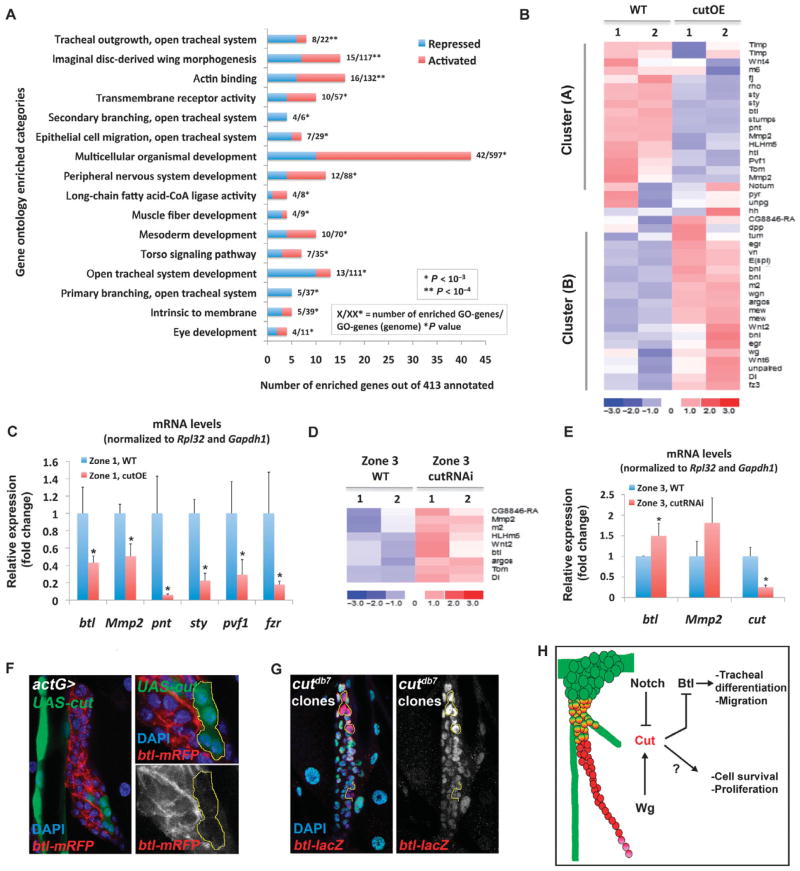
To understand potential mechanisms for how Cut affected SB development, we performed transcriptional profiling to identify Cut targets relevant to SB development. We compared wild-type zone 1 cells to zone 1 cells expressing a moderate amount of ectopic cut (“cutOE”) and compared wild-type zone 3 cells to zone 3 cells with a moderate reduction of Cut abundance (“cutRNAi”). To achieve ectopic expression of cut in zone 1 cells, we expressed a UAS-cut transgene with btl-Gal4 under the control of Gal80ts. We found that induction of the UAS-cut transgene for only 12 hours at 29°C was sufficient to induce Cut protein in the tracheal system (fig. S6, A and B) and simultaneously preserved zone 1 SB cells (it exerted the same phenotype as 3 to 4 days of induction at 25°C; fig. S6C). Conversely, to achieve reduction of cut in zone 3 with a minimal loss of tissue integrity, we expressed a UAS-cutRNAi transgene under cut-Gal4 control at 18°C (as in Fig. 5A).
We performed GO enrichment analysis of the 413 annotated genes that were differentially expressed between wild-type and cutOE zone 1 cells (Fig. 7A and table S3). The most significantly enriched category includes genes involved in “tracheal outgrowth, open tracheal system,” and the expression of six of eight of these differentially expressed genes (including btl, pnt, and sty) was reduced in the cutOE sample [Fig. 7A (blue bars) and table S3]. Other enriched categories include “secondary branching open tracheal system,” “primary branching open tracheal system,” “epithelial cell migration open tracheal system,” and “open tracheal system development,” indicating that Cut possibly targets multiple genes involved in differentiation of tracheal cells and branching morphogenesis (Fig. 7A). The expression of most of the differentially expressed genes in these categories was reduced in the cutOE zone 1 cells [Fig. 7A (blue bars) and table S3], suggesting that tracheal system genes were repressed by Cut.
Fig. 7.
Cut represses tracheal differentiation genes. (A) GO enrichment analysis of filtered genes differentially expressed in WT zone 1 and cutOE zone 1 (fold change ≥1.5). (B) Heat map of differentially expressed signaling pathway genes between WT and cutOE SB zone 1 cells in micro-array duplicates (samples 1 and 2). (C) qRT-PCR validation of tracheal gene expression upon cut overexpression in zone 1. Data are shown with the WT values normalized to 1. (D) Heat map of the most prominent cluster of up-regulated signaling pathway genes in cutRNAi zone 3 cells. (E) Relative amount of the indicated mRNAs detected by qRT-PCR. Data are shown with the WT values normalized to 1. Error bars in (C) and (E) correspond to the SD from the mean of biological triplicates. *P ≤ 0.05. (F) Clone overexpressing cut (labeled by GFP) in the background of the btl-mRFPmoe reporter (red) stained with DAPI (blue). Gray shows the btl-mRFP reporter only. (G) cutdb7 mutant clone (marked by the absence of GFP) in the background of the btl-lacZ reporter (red) stained with DAPI (blue). Gray shows btl-lacZ. Yellow line delineates clone boundaries in (F) and (G). (H) Model showing the spatial activation and the antagonistic actions of the Notch and Wg pathways that generate the nonuniform abundance of Cut, which orchestrates SB morphogenesis. Scale bars, 20 μm (F and G); 10 μm (right panels of F).
To identify which signaling pathways might be the targets of Cut during tracheal differentiation, we assessed the transcriptional changes of signaling pathway components (table S2) in wild-type and cutOE zone 1 cells (Fig. 7B). Both genes with increased expression (cluster A) and genes with decreased expression (cluster B) were observed in the cutOE samples. Because Cut functions as a transcriptional repressor (38), we expected that expression of its direct targets would be decreased in the ectopic expression experiment and therefore would be included in cluster A, whereas cluster B would include indirectly regulated genes. To validate cluster A genes, we performed qRT-PCR and found that transcripts for components of the Btl pathway, such as btl, Mmp2, pnt, and sty, were reduced when cut was ectopically expressed in zone 1 (Fig. 7C), suggesting that the Btl pathway might be targeted by Cut during normal development of the SB. In addition, expression of fzr, an endoreplication marker enriched in zone 1 (Fig. 2C), was also reduced by ectopic cut (Fig. 7C), as previously demonstrated by mosaic analysis (24).
If Cut transcriptionally represses the Btl pathway in the SB, then we expected to find the expression of genes encoding members of this pathway to be increased in the cutRNAi sample due to loss of Cut repression. Indeed, clustering of the signaling pathway components gene set (table S2) indicated that btl and Mmp2 were transcriptionally increased when cut was reduced in zone 3 (fig. S7 and Fig. 7D), and the expression of btl was validated by qRT-PCR (Fig. 7E). Therefore, Cut is likely a transcriptional repressor of btl.
To assess whether Cut repressed btl in vivo, we generated act-FLPout clones (39) overexpressing UAS-cut in the SB and observed repression of the btl-enhancer-mRFPmoe transcriptional reporter (40) in zone 1 (Fig. 7F). In addition, cutDB7 null clones in the SB showed an increase in the expression of the btl-lacZ reporter (41) (Fig. 7G). The ectopic expression and RNAi experiments indicated that Cut inhibited tracheal differentiation by targeting genes involved in the Btl pathway and that one of its targets is btl.
DISCUSSION
Our study of SB morphogenesis showed that (i) the abundance of Cut is tightly controlled to allow coordination of growth and patterning during normal development, and its loss leads to cell death; (ii) the Notch and Wg signaling pathways exert antagonistic actions on Cut to regulate its protein amount in the developing SB; and (iii) Cut represses tracheal differentiation by targeting Btl at the transcriptional level. Our results illustrate how the spatial integration of the antagonistic actions of the Notch and Wg pathways along the SB controls the gradient of the transcription factor Cut, and show that the abundance of Cut plays a key role in the patterning and differentiation of tracheal cells (Fig. 7H).
Drosophila cut is expressed in a highly complex pattern in various tissues and cell types (42, 43). Although its expression and presence of the protein in differentiated cell types (for example, neurons and Malpighian tubules) underscores its role in terminal differentiation, we find that Cut is present in undifferentiated progenitors, the imaginal SB tracheoblasts, where it controls their survival, proliferation, and differentiation. Contrary to its role in the follicular epithelium, where it regulates the switch from mitotic cycles to endoreplication (30), in the SB Cut has a pleiotropic function. Depending on its protein abundance, Cut specifies survival, proliferation, or differentiation. In zone 1, the abundance of Cut is low, which is necessary for tracheal differentiation; in zone 2, Cut abundance is moderate with amounts that stimulate proliferation; and, in zone 3, Cut abundance is high and specifies the fate of the spiracle. We showed that changing the abundance of Cut experimentally in any of these zones compromised SB organogenesis.
To understand which signals regulate the emergence of an adult epithelial organ from undifferentiated progenitors, we performed transcriptional profiling to identify genes encoding proteins in conserved signaling pathways that were differentially regulated between distinct populations of SB cells. For example, we analyzed gene expression patterns in zone 1 cells destined to become tracheal tubes and in zone 3 cells destined to divide and become the spiracle. We identified several conserved signaling pathways with spatially restricted activation, including the Notch, Btl, and PVR pathways in zone 1, and the Wg, Hh, Dpp, Jak/Stat, and Toll pathways in zone 3. These are good candidates to act as regulators of the Cut gradient that organizes morphogenesis of the SB or as potential targets of Cut activity involved in the growth and patterning of the SB.
We identified the Notch and Wg pathways as key upstream regulators of Cut. Notch and Wg were activated in a spatially complementary pattern in the SB and exerted antagonistic actions on Cut: Notch inhibited Cut in zone 2, whereas Wg induced Cut in zones 2 and 3. This resulted in the generation of a Cut gradient in the developing SB and the establishment of the highly proliferative zone 2 in which Cut abundance is low, and this zone contributes to cells of the entire SB and subsequently the adult tracheal system (24). Although we showed that Notch is necessary for reducing Cut abundance in zone 2, we found that Notch-null mosaics had no effect on Cut in zone 1, suggesting that additional pathways activated in zone 1, such as the PVR pathway, are likely involved in Cut regulation in zone 1. Finally, we cannot exclude the possibility that Notch has an additional role in zone 1 or the mature tracheal tubes where it is highly activated.
In contrast to Notch, Wg was a positive Cut regulator, and Wg over-expression stimulated Cut induction. However, overexpression of Cut only partially rescued the decrease in cell number associated with loss of Wg signaling, suggesting that Wg may have additional roles in the SB, which might include regulating proliferation in a Cut-independent manner. Finally, transcriptional profiling also identified other pathways that could potentially regulate cut expression in zone 3. In particular, the Dpp or the Jak/Stat pathway, as suggested by the spatial expression of their ligands, may play a role.
Our quantitative manipulations of Cut abundance in the SB showed that its gradient contributed to the growth differences observed in zones 1 to 3. The graded abundance of Cut was required for nonuniform proliferation of epithelial progenitors. Moderate, rather than high, amounts of Cut resulted in the highest proliferation. The genes targeted by Cut in the SB that regulate the cell cycle may be different from those targeted in other tissues in which Cut functions. In the follicular epithelium, Cut promotes the expression of genes encoding Cyclin A and the phosphatase String (also known as cdc25), which stimulate cell proliferation (30). During development of the posterior spiracle (where no cell proliferation is involved), Cut functions in cell survival by directly repressing the gene encoding the cell death–promoting protein Reaper to inhibit apoptosis (44). In mammalian cell lines, the Cut homolog Cux-1 represses the genes encoding the cell cycle inhibitors p21 and p27 to stimulate proliferation (30, 45).
Btl plays a key role in the morphogenesis and differentiation of the embryonic, as well as the adult, tracheal system (17, 18, 20, 21, 23–26, 46). Many transcription factors that induce btl expression during embryonic tracheal specification and differentiation have been identified, but we identified Cut as a transcription factor that keeps tracheal progenitor cells in an undifferentiated state and that it targeted btl. Transcriptional profiling showed that many tracheal differentiation genes were repressed by ectopic expression of cut, and in vivo experiments showed that Btl was transcriptionally regulated by Cut. Because Cut is a DNA binding factor, a detailed characterization of the promoter region of putative Cut target genes should help to understand how Cut abundance controls its various functions.
Our findings on the role of Cut are likely to be relevant to the role of the Cut vertebrate homologs Cux-1 and Cux-2 in organogenesis (38, 47). Cux-1 is detected in tubular organs, such as the lung and kidney, and Cux-1–knockout mice die shortly after birth because of lack of lung formation (48). Furthermore, in the developing kidney, Cux-1 abundance is inversely correlated with the degree of cellular differentiation (49), and transgenic Cux-1 mice consistently exhibit kidney overgrowth due to excess proliferation (50). Finally, expression of Cux-1 and Cux-2 overlaps with that of Notch and Wnt pathway components in multiple tissues during vertebrate development (48, 51, 52), suggesting that Notch and Wnt may regulate Cux-1 and Cux-2 expression in these tissues.
MATERIALS AND METHODS
Fly stocks
Description of the following stocks, unless indicated, can be found at FlyBase (http://www.flybase.org). Numbers in parentheses correspond to Bloomington stock numbers unless otherwise indicated. Notchts1 (#2533), wgts (#7000), fzP21 dfz2C1 (33), btl-Gal4 UAS-actGFP (#8807), wg-lacZ (#1672), dpp-lacZ10638 (#12379), upd-Gal4E132 (#26796), bnl-Gal4NP2211 [Drosophila Genomics Resource Center (DGRC), #112825], esg-Gal4NP6267 (DGRC, #113886), UAS-dTCFDN (#4784), UAS-srcGFP (#5432), tub-Gal80ts (#7016), UAS-wgE4 (#5919), UAS-p35 (#6298), UAS-cut (a gift from B. Mathey-Prevot), Dl-lacZ05151 (#11651), Ser-lacZ9.5 (53), btl-lacZB123 (41), btl-mRFPmoe (40), neur A101-lacZ (54), cut-Gal4PG142 (55), UAS-cutRNAi [Vienna Drosophila RNAi Center (VDRC), #5687] (56), UAS-cutRNAiJF03304 (#29625) (57), UAS-cutRNAiHMS00924 (#33967) (58), UAS-NotchRNAi (59), UAS-NotchDN, UAS-NotchIC, Su(H)-GBE-lacZ, and esg-Gal4 UAS-EGFP (60). For all experiments, we focused on the SBs of tracheal segments Tr4 and Tr5.
Generation of mitotic clones
To assess the phenotype of Notch and the Wg receptors fz and dfz2, we generated MARCM clones (61) in larvae of the following genotypes: Notch54l9 FRT19A/tub-Gal80 FRT19A; tub-Gal4/UAS-CD8GFP;hsFLP (62) and hsFLP UAS-nlsEGFP tub-Gal4; fzP21dfz2C1 FRT40A/tub-Gal80 FRT40A (33), respectively. Mutant cutDB7 clones and cut overexpression clones were generated as previously described (24) in the background of btl-lacZB123 and btl-mRFPmoe, respectively. At least five flies with mosaic tissues were analyzed.
Transgene overexpression
All UAS transgenes were overexpressed in the presence of tub-Gal80ts (37) to overcome embryonic and larval lethality. In particular, tub-Gal80ts;btl-Gal4 UAS-srcGFP and cut-Gal4 tub-Gal80ts;UAS-srcGFP females were crossed to UAS transgenic males. Crosses were reared at 18°C until early L2 and then shifted to 25°C (Fig. 6A) or 29°C (Figs. 4 and 6B) for 3 to 4 days to allow moderate and strong Gal4 activation, respectively. At least five larvae for each transgene were analyzed. For rescue experiments (Fig. 4, J and K), the same number of UAS trans-genes was used in controls to ensure that the observed phenotype is not due to titration of the Gal4. For RNAi experiments, we used btl-Gal4 UAS-srcGFP, cut-Gal4; UAS-srcGFP, and esg-Gal4 UAS-EGFP females crossed to UAS-cutRNAi males. Crosses were reared at 21°C for 1 day to allow egg laying, and then the parents were removed. The vials containing the L1 progeny of the crosses were incubated at 18°C, 25°C, or 29°C for 10, 5, or 5 days, respectively (until they reached the late L3 stage), and then the L3 larvae were dissected. To reduce Cut abundance, we characterized UAS-cutRNAi transgenes that efficiently eliminated the Cut protein, as assessed by immunostaining with the monoclonal antibody recognizing Cut, 2B10 (24, 42). To ensure that the phenotypes we observed were not due to off-target RNAi effects (63), we used three different UAS-cutRNAi lines generated in different laboratories (56–58) and obtained similar results. We present data for the weaker line (56) here because this weaker line allowed more flexibility in manipulating Cut abundance without the need for a Gal80 repressor.
Immunohistochemistry
For all quantification experiments, we assessed phenotypes at the wandering L3 stage (110 to 120 hours after egg laying), whereas for fig. S3, we dissected L2 and early L3 larvae. Larvae were filleted ventrally in dissection chambers (64), ensuring that the tracheal system remained in place and the SBs were not detached from the epidermis. After removal of the internal organs, the larval carcasses with the associated trachea were fixed and stained as previously described (62). The following antibodies were used: mouse antibody recognizing Cut [2B10; 1:50 dilution; Developmental Studies Hybridoma Bank (DSHB)], mouse antibody recognizing Wg (4D4; 1:50 dilution; DSHB), rabbit antibody recognizing β-galactosidase (1:10,000 dilution; Cappel), mouse antibody recognizing β-galactosidase (1:500 dilution; Promega), rabbit antibody recognizing pH3 (1:2000 dilution; Millipore), and rabbit antibody recognizing GFP (1:2000 dilution; Molecular Probes). Secondary antibodies recognizing mouse or rabbit and conjugated to Alexa Fluor 488, 555, 594, and 647 were used at 1:1000. For nuclear staining, DAPI was used at 0.3 μg/ml along with the secondary antibodies.
Image analysis
Fluorescent images were acquired with a Leica TCS SP2 AOBS or TCS DM-IRE2 AOBS confocal microscope or a Zeiss AxioImager. Image processing and single-cell quantification of the Cut staining intensity in the SOP and SB cells (Figs. 1C, 4, A to C, and 5, A and B, bottom panels) were performed using Photoshop CS3. Each SB cell nucleus was outlined using the DAPI channel, and the total Cut signal in this nucleus was recorded as integrated density in Photoshop. Normalized Cut abundance (the ratio Cut in SB cell/Cut in SOP) was calculated for single SB cells from distal (zone 4) to proximal (zone 1, close to the DTand SOP) positions in the SB. When more than one cell was found in a specific position along the distal-proximal axis of the SB (x axis in the graph), values of two to three cells were averaged. Thus, the total number of cells shown in the x axis of the graph is smaller than the actual number of SB cells (which is indicated by the DAPI staining in blue in the top panels). The surface plots (middle panels) were generated using ImageJ (National Institutes of Health); the surface outlined by the yellow line is included in the plot, and the peaks indicate the intensity of the Cut signal so that the same color indicates similar intensities of the fluorescence (black corresponds to background). Note that the surface plot measures the fluorescence intensity in each pixel of the image and does not account for the size (surface) of each nucleus. Due to its larger surface, the SOP nucleus produces a lower peak in the plot, even though its total Cut signal (integrated density) is larger.
RNA isolation and microarrays
Total RNA (50 to 150 ng) was prepared from zone 1 (wild type or cutOE: tub-Gal80ts/w1118; btl-Gal4 UAS-actGFP/+ or tub-Gal80ts/w1118; btl-Gal4 UAS-actGFP/UAS-cut reared at 18°C and shifted to 29°C for 12 hours before dissection) and zone 3 SB cells (wild type or cutRNAi: cut-Gal4/w1118; UAS-srcGFP/+; +/+ or cut-Gal4/w1118; UAS-srcGFP/+; UAS-cutRNAi/+ reared at 18°C). L3 larvae were filleted in magnetic chambers, and the whole trachea including the SBs was carefully dissected from the larval carcass with forceps and tungsten needles in 1× phosphate-buffered saline (PBS). The dissected tracheawas placed in one drop of 1× PBS on a Sylgard plate (VWR) and was allowed to stick to the plate. Tracheoblasts were isolated from the Tr4 and Tr5 tracheal segments. Zone 1 tracheoblasts (100 to 150 endoreplicated cells) and zone 3 tracheoblasts (30 to 40 diploid cells of the distal SB beyond the TC junction) were dissected on the Sylgard plate with a micro-knife (Fine Science Tools). To generate pure zone 1 cells, we removed the associated TC and VB from the dissected fragments by exploiting the property of air-filled trachea (TC and VB) to stick to Sylgard. Isolated zone 1 and 3 fragments were immediately frozen in plastic tubes in dry ice/ethanol and stored at −80°C until all replicates were ready. Empirically, we found that we needed about six times more zone 3 SBs compared to zone 1 SBs to isolate similar amounts of total RNA from zones 1 and 3. RNA was extracted with the ABI PicoPure RNA isolation kit following the manufacturer’s instructions. After RNA quality assessment with the Bioanalyzer Pico, one round of NuGen linear amplification followed by complementary DNA (cDNA) preparation was performed before hybridization to Affymetrix Drosophila 2.0 arrays (DFCI microarray core facility). dChip (http://www.hsph.harvard.edu/cli/complab/dchip/) was used for array normalization, filtering (we limited our analysis to genes that, according to the perfect match-mismatch probe assessment, were present in >20% of the arrays with a signal value higher than the microarray background level >65 in ≥25% of the samples), data analysis, and presentation. Comparisons were generated on filtered gene lists using fold change ≥1.5 between treatments and P ≤ 0.05 calculated from biological replicates by Student’s t test in dChip.
qRT-PCR validation was performed on a Bio-Rad CFX-96 qPCR machine from biological triplicates. Total RNA extracted from dissected SB cells with the ABI PicoPure RNA isolation kit was quantified, and 50 ng was used for reverse transcription using the Bio-Rad iScript cDNA Synthesis Kit. The Bio-Rad iQ SYBR Green Supermix kit was used for qPCR of biological triplicates. Primers were designed using the Roche Assay Design Center tool (http://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp?id=UP030000), and their sequences are available in table S4. To assess differences in gene expression between samples, we used the relative quantification (ΔΔCt ) method. Data were normalized against the expression of Gapdh1 and Rpl32 and are shown as fold-change differences between sample pairs to be compared. P values were calculated using Student’s t test on biological triplicates.
Supplementary Material
Acknowledgments
We thank Y. Apidianakis, R. Binari, N. Giagtzoglou, M. Kulkarni, and B. Mathey-Prevot for helpful comments on the manuscript and members of the Perrimon laboratory for discussions. We also thank the DSHB for antibodies; the Bloomington Stock Center, DGRC, VDRC, Transgenic RNAi Project (TRiP), M. Affolter, C. Delidakis, W.-M. Deng, A. Ghabrial, M. Krasnow, B. Mathey-Prevot, and G. Struhl for fly stocks; and P. Skouridis for help with microscopy in Cyprus.
Funding: N.P. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions: C.P. designed and performed the experiments, analyzed the data, and wrote the paper; N.P. provided guidance, materials, and help with paper writing.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Microarray data are available at the EBI ArrayExpress database under accession number E-MEXP-3831.
www.sciencesignaling.org/cgi/content/full/6/263/ra12/DC1
Fig. S1. The Cut gradient in flies of different genotypes and stages.
Fig. S2. Signaling pathway genes are differentially expressed in zone 1 and zone 3 SB cells.
Fig. S3. The Notch and Wg pathways are activated during early SB development.
Fig. S4. A temperature-sensitive UAS-cutRNAi line recapitulates the loss of cut.
Fig. S5. p35 prevents the apoptosis induced by expression of UAS-cutRNAi at 18°C and 25°C.
Fig. S6. Scheme for ectopic expression of cut in zone 1 cells.
Fig. S7. Transcriptional effects of cut knockdown on signaling pathway genes in zone 3.
Table S1. Enrichment of GO categories in zone 1 compared to zone 3.
Table S2. Signaling pathway gene list in Affymetrix Drosophila Genome 2.0 arrays.
Table S3. Enrichment of GO categories in wild-type zone 1 compared to cutOE zone 1.
Table S4. qPCR primer sequences.
REFERENCES AND NOTES
- 1.Aulehla A, Pourquié O. Signaling gradients during paraxial mesoderm development. Cold Spring Harb Perspect Biol. 2010;2:a000869. doi: 10.1101/cshperspect.a000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence PA, Struhl G. Morphogens, compartments, and pattern: Lessons from Drosophila? Cell. 1996;85:951–961. doi: 10.1016/s0092-8674(00)81297-0. [DOI] [PubMed] [Google Scholar]
- 3.Liu PZ, Kaufman TC. Short and long germ segmentation: Unanswered questions in the evolution of a developmental mode. Evol Dev. 2005;7:629–646. doi: 10.1111/j.1525-142X.2005.05066.x. [DOI] [PubMed] [Google Scholar]
- 4.Schwank G, Basler K. Regulation of organ growth by morphogen gradients. Cold Spring Harb Perspect Biol. 2010;2:a001669. doi: 10.1101/cshperspect.a001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serrano N, O’Farrell PH. Limb morphogenesis: Connections between patterning and growth. Curr Biol. 1997;7:R186–R195. doi: 10.1016/s0960-9822(97)70085-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milán M, Campuzano S, García-Bellido A. Cell cycling and patterned cell proliferation in the wing primordium of Drosophila. Proc Natl Acad Sci USA. 1996;93:640–645. doi: 10.1073/pnas.93.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hornbruch A, Wolpert L. Cell division in the early growth and morphogenesis of the chick limb. Nature. 1970;226:764–766. doi: 10.1038/226764a0. [DOI] [PubMed] [Google Scholar]
- 8.Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- 9.Towers M, Mahood R, Yin Y, Tickle C. Integration of growth and specification in chick wing digit-patterning. Nature. 2008;452:882–886. doi: 10.1038/nature06718. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y. Growth and patterning in the limb: Signaling gradients make the decision. Sci Signal. 2009;2:pe3. doi: 10.1126/scisignal.253pe3. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J, Nakamura E, Nguyen MT, Bao X, Akiyama H, Mackem S. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev Cell. 2008;14:624–632. doi: 10.1016/j.devcel.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 13.Bénazet JD, Zeller R. Vertebrate limb development: Moving from classical morphogen gradients to an integrated 4-dimensional patterning system. Cold Spring Harb Perspect Biol. 2009;1:a001339. doi: 10.1101/cshperspect.a001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuccillo M, Joyner AL, Fishell G. Morphogen to mitogen: The multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:772–783. doi: 10.1038/nrn1990. [DOI] [PubMed] [Google Scholar]
- 15.Halder G, Johnson RL. Hippo signaling: Growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ten Berge D, Brugmann SA, Helms JA, Nusse R. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development. 2008;135:3247–3257. doi: 10.1242/dev.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabernard C, Affolter M. Distinct roles for two receptor tyrosine kinases in epithelial branching morphogenesis in Drosophila. Dev Cell. 2005;9:831–842. doi: 10.1016/j.devcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Guha A, Kornberg TB. Tracheal branch repopulation precedes induction of the Drosophila dorsal air sac primordium. Dev Biol. 2005;287:192–200. doi: 10.1016/j.ydbio.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Manning G, Krasnow MA. Development of the Drosophila Tracheal System. Vol. 1 CSHL Press; New York: 1993. [Google Scholar]
- 20.Samakovlis C, Hacohen N, Manning G, Sutherland DC, Guillemin K, Krasnow MA. Development of the Drosophila tracheal system occurs by a series of morphologically distinct but genetically coupled branching events. Development. 1996;122:1395–1407. doi: 10.1242/dev.122.5.1395. [DOI] [PubMed] [Google Scholar]
- 21.Sato M, Kornberg TB. FGF is an essential mitogen and chemoattractant for the air sacs of the Drosophila tracheal system. Dev Cell. 2002;3:195–207. doi: 10.1016/s1534-5807(02)00202-2. [DOI] [PubMed] [Google Scholar]
- 22.Whitten J. In: The Post-Embryonic Development of the Tracheal System in Drosophila melanogaster. Wright T, editor. 2d Academic Press; New York: 1980. [Google Scholar]
- 23.Guha A, Lin L, Kornberg TB. Organ renewal and cell divisions by differentiated cells in Drosophila. Proc Natl Acad Sci USA. 2008;105:10832–10836. doi: 10.1073/pnas.0805111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitsouli C, Perrimon N. Embryonic multipotent progenitors remodel the Drosophila airways during metamorphosis. Development. 2010;137:3615–3624. doi: 10.1242/dev.056408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato M, Kitada Y, Tabata T. Larval cells become imaginal cells under the control of homothorax prior to metamorphosis in the Drosophila tracheal system. Dev Biol. 2008;318:247–257. doi: 10.1016/j.ydbio.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 26.Weaver M, Krasnow MA. Dual origin of tissue-specific progenitor cells in Drosophila tracheal remodeling. Science. 2008;321:1496–1499. doi: 10.1126/science.1158712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ninov N, Chiarelli DA, Martín-Blanco E. Extrinsic and intrinsic mechanisms directing epithelial cell sheet replacement during Drosophila metamorphosis. Development. 2007;134:367–379. doi: 10.1242/dev.02728. [DOI] [PubMed] [Google Scholar]
- 28.de Celis JF, Bray S. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 1997;124:3241–3251. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- 29.de Celis JF, Garcia-Bellido A, Bray SJ. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development. 1996;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- 30.Sun J, Deng WM. Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development. 2005;132:4299–4308. doi: 10.1242/dev.02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J, Deng WM. Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Dev Cell. 2007;12:431–442. doi: 10.1016/j.devcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furriols M, Bray S. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr Biol. 2001;11:60–64. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- 33.Chen CM, Struhl G. Wingless transduction by the Frizzled and Frizzled2 proteins of Drosophila. Development. 1999;126:5441–5452. doi: 10.1242/dev.126.23.5441. [DOI] [PubMed] [Google Scholar]
- 34.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 35.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 36.Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 37.McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- 38.Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- 39.Struhl G, Basler K. Organizing activity of Wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 40.Ribeiro C, Neumann M, Affolter M. Genetic control of cell intercalation during tracheal morphogenesis in Drosophila. Curr Biol. 2004;14:2197–2207. doi: 10.1016/j.cub.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 41.Ohshiro T, Saigo K. Transcriptional regulation of breathless FGF receptor gene by binding of TRACHEALESS/dARNT heterodimers to three central midline elements in Drosophila developing trachea. Development. 1997;124:3975–3986. doi: 10.1242/dev.124.20.3975. [DOI] [PubMed] [Google Scholar]
- 42.Blochlinger K, Bodmer R, Jan LY, Jan YN. Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 1990;4:1322–1331. doi: 10.1101/gad.4.8.1322. [DOI] [PubMed] [Google Scholar]
- 43.Blochlinger K, Jan LY, Jan YN. Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development. 1993;117:441–450. doi: 10.1242/dev.117.2.441. [DOI] [PubMed] [Google Scholar]
- 44.Zhai Z, Ha N, Papagiannouli F, Hamacher-Brady A, Brady N, Sorge S, Bezdan D, Lohmann I. Antagonistic regulation of apoptosis and differentiation by the Cut transcription factor represents a tumor-suppressing mechanism in Drosophila. PLoS Genet. 2012;8:e1002582. doi: 10.1371/journal.pgen.1002582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma M, Brantley JG, Alcalay NI, Zhou J, Heystek E, Maser RL, Vanden Heuvel GB. Differential expression of Cux-1 and p21 in polycystic kidneys from Pkd1 null and cpk mice. Kidney Int. 2005;67:432–442. doi: 10.1111/j.1523-1755.2005.67099.x. [DOI] [PubMed] [Google Scholar]
- 46.Ghabrial A, Luschnig S, Metzstein MM, Krasnow MA. Branching morphogenesis of the Drosophila tracheal system. Annu Rev Cell Dev Biol. 2003;19:623–647. doi: 10.1146/annurev.cellbio.19.031403.160043. [DOI] [PubMed] [Google Scholar]
- 47.Sansregret L, Nepveu A. The multiple roles of CUX1: Insights from mouse models and cell-based assays. Gene. 2008;412:84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 48.Ellis T, Gambardella L, Horcher M, Tschanz S, Capol J, Bertram P, Jochum W, Barrandon Y, Busslinger M. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 2001;15:2307–2319. doi: 10.1101/gad.200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vanden Heuvel GB, Brantley JG, Alcalay NI, Sharma M, Kemeny G, Warolin J, Ledford AW, Pinson DM. Hepatomegaly in transgenic mice expressing the homeobox gene Cux-1. Mol Carcinog. 2005;43:18–30. doi: 10.1002/mc.20091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanden Heuvel GB, Bodmer R, McConnell KR, Nagami GT, Igarashi P. Expression of a cut-related homeobox gene in developing and polycystic mouse kidney. Kidney Int. 1996;50:453–461. doi: 10.1038/ki.1996.336. [DOI] [PubMed] [Google Scholar]
- 51.Sharma M, Fopma A, Brantley JG, Vanden Heuvel GB. Coexpression of Cux-1 and Notch signaling pathway components during kidney development. Dev Dyn. 2004;231:828–838. doi: 10.1002/dvdy.20175. [DOI] [PubMed] [Google Scholar]
- 52.Tavares AT, Tsukui T, Izpisúa Belmonte JC. Evidence that members of the Cut/Cux/CDP family may be involved in AER positioning and polarizing activity during chick limb development. Development. 2000;127:5133–5144. doi: 10.1242/dev.127.23.5133. [DOI] [PubMed] [Google Scholar]
- 53.Bachmann A, Knust E. Dissection of cis-regulatory elements of the Drosophila gene Serrate. Dev Genes Evol. 1998;208:346–351. doi: 10.1007/s004270050190. [DOI] [PubMed] [Google Scholar]
- 54.Huang F, Dambly-Chaudière C, Ghysen A. The emergence of sense organs in the wing disc of Drosophila. Development. 1991;111:1087–1095. doi: 10.1242/dev.111.4.1087. [DOI] [PubMed] [Google Scholar]
- 55.Bourbon HM, Gonzy-Treboul G, Peronnet F, Alin MF, Ardourel C, Benassayag C, Cribbs D, Deutsch J, Ferrer P, Haenlin M, Lepesant JA, Noselli S, Vincent A. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev. 2002;110:71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- 56.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 57.Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, Villalta C, Laverty TR, Perkins LA, Perrimon N. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ni JQ, Markstein M, Binari R, Pfeiffer B, Liu LP, Villalta C, Booker M, Perkins L, Perrimon N. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5:49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 61.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 62.Pitsouli C, Delidakis C. The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development. 2005;132:4041–4050. doi: 10.1242/dev.01979. [DOI] [PubMed] [Google Scholar]
- 63.Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 64.Budnik V, Gorczyca M, Prokop A. Selected methods for the anatomical study of Drosophila embryonic and larval neuromuscular junctions. Int Rev Neurobiol. 2006;75:323–365. doi: 10.1016/S0074-7742(06)75015-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.