Abstract
Objective
Conduct a representative survey of Human papillomavirus (HPV) prevalence and its genotype distribution in invasive anal cancer specimens in the U.S.
Methods
Population-based archival anal cancer specimens were identified from Florida, Kentucky, Louisiana and Michigan cancer registries and SEER tissue repositories in Hawaii, Iowa and Los Angeles. Sections from one representative block per case were used for DNA extraction. All extracts were assayed first by Linear Array and re-tested with INNO-LiPA if inadequate or HPV negative.
Results
Among 146 unique invasive anal cancer cases, 93 (63.7%) were from women and 53 (36.3%) from men. HPV (any type) was detected in 133 (91.1%) cases and 129 (88.4%) contained at least one high risk type, most (80.1%) as a single genotype. HPV16 had the highest prevalence (113 cases, 77.4%); HPV6, 11, 18 and 33 were also found multiple times. Among HPV16 positive cases, 37% were identified as prototype variant Ep and 63% were non-prototypes: 33% Em, 12% E-G131G, 5% Af1, 4% AA/NA-1, 3% E-C109G, 3% E-G131T, 2% As and 1% Af2. No significant differences in the distributions of HPV (any), high-risk types, or HPV16/18 were seen between gender, race or age group.
Conclusions
The establishment of pre-vaccine HPV prevalence in the U.S. is critical to the surveillance of vaccine efficacy. Almost 80% of anal cancers were positive for the vaccine types HPV16 or HPV18 and in 70% these were the only types detected suggesting that a high proportion might be preventable by current vaccines.
Keywords: anal cancer, HPV typing, Human papillomavirus, archived tissue, cancer registry
Introduction
Anal cancers are relatively rare malignancies, most of which occur as squamous cell carcinomas (SCC) of the anal tract. According to 2009 NPCR/SEER combined data, the incident rate for invasive anal cancer in the U.S. was 1.8 per 100,000, totaling 2236 cases in males (rate 1.5) and 3692 among females (rate 2.1) annually (1). While incidence is relatively low, the number of anal malignancies has steadily increased over the past three decades (2). Many of the recognized risk factors for anal cancer, such as number of sexual partners and anal receptive intercourse, are associated with persistent human papillomavirus (HPV) infection of the anal canal (3;4). Anal histology shares common anatomic characteristics with the cervix, including a transformation zone where most HPV-associated neoplastic transformation occurs. It is therefore anticipated that available HPV vaccines will provide protection against anal cancer too and clinical trials of the quadrivalent Gardasil (Merck & Co, Inc., Whitehouse Station, NJ) showed good efficacy against anal intraepithelial neoplasia (5;6).
The proportion of anal carcinomas attributable to HPV has been estimated to be about 90% (7–9) attributing the majority to HPV16 and HPV18. However, no population-based data are available for the United States regarding the type-specific HPV prevalence in anal cancers preceding vaccine implementation. Such information will provide a baseline to monitor vaccine effectiveness against this malignancy. To meet this surveillance objective, population-based sampling of anal cancer tissue from US central state registries was conducted to determine the type-specific HPV prevalence in these cases.
HPV 16 cases were further evaluated to determine HPV16 genotype variants since some reports had indicated that certain non-prototype sequences have a disproportionally high representation in anal diseases (10;11).
Materials and Methods
Selection of anal cancer tissues
Representative tissue specimens were obtained as part of the Centers for Disease Control Central Cancer Registries (CDC CCR) study to provide a baseline prevalence of HPV types in HPV-associated cancers from a representative sample of the US population. A systematic full case selection of anatomic regions coded as anal canal was pursued depending on specimen availability. Cases were recruited from seven participating registries, including four central cancer registries (CCRs) in Florida, Kentucky, Louisiana and Michigan as well as three SEER cancer registry-based residual tissue repositories (RTRs) in Los Angeles County, Hawaii and Iowa. CCR associated pathology laboratories and RTRs were asked to select one representative archived, formalin-fixed paraffin-embedded (FFPE) tissue block from each anal cancer case that was diagnosed between 1995 and 2005. CDC and each participating state received approval from the Institutional Review Board (IRB) for the study.
Specimen preparation and DNA extraction
Six microtome sections were prepared from each block using a fresh disposable blade and applicator for each case. The first and last sections were stained with Hematoxylin and Eosin (H&E) and intervening sections - two 5-µm sections per sample - were transferred into 2 ml conical screw cap tubes with tether cap (Simport, Beloeil, Canada). H&E sections were reviewed by a study pathologist (ERU) to confirm that the sections included viable tumor. Cases that did not have representative material were excluded; otherwise samples were processed as previously described using high temperature-assisted tissue lysis (12) and automated DNA purification with a Chemagic MSM1 (PerkinElmer, Waltham, MA, USA). The resulting 100 µL DNA eluate was tested immediately or stored at −20°C. A blank sample without tissue was included in every extraction batch to monitor potential cross contamination.
HPV Genotyping
All extracts were tested with the Linear Array HPV Genotyping Test (LA, Roche Diagnostics, Indianapolis, IN), which distinguishes 37 different HPV genotypes (6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52(XR), 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73, 81, 82, 83, 84, 89, IS39). Templates for the PCR reaction were prepared with 10 µl DNA and 40 µl H2O, otherwise following the manufacturer’s protocol. The reverse line blot hybridization was performed with an automated platform (Beeblot instruments (Bee Robotics, Caernarfon, UK)). Samples with negative or inadequate (negative for HPV and cellular β-Globin control) LA results were re-tested with the INNO-LiPA HPV Genotyping Assay (LiPA, Innogenetics, Gent, Belgium) which detects 29 HPV types (6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68, 69, 70, 71, 73, 74, 81, 82). The assay was performed according to the manufacturers specifications using an Autoblot 3000 (MedTec, Buffalo Grove, IL) for the line-blot procedure.
HPV16 variant determination
All samples positive for HPV16 by either LA or LiPA genotyping assays were subjected to variant analysis with a pyrosequencing assay (13). Briefly, 10 µl of the extracted DNA was used as a template to amplify a 314 bp fragment of the E6 region. If no amplicon was visible by gel electrophoresis, the procedure was repeated with an enriched template that was first amplified by whole genome amplification with the GenomePlex WGA2 kit (Sigma-Aldrich, St Louis, MO, USA) in accordance with the manufacturer’s recommendations. Subsequently, nucleotide identities at position 109, 131, 132, 143, 145, 178, 350 were determined with a Pyromark Q96 MD (Qiagen, Valencia, CA, USA) to identify the variants AA/NA-1, Af1, Af2, As, E-C109G, E-G131G, E-G350, Em, Ep. In cases, where the pyro signals were ambiguous, the E6 amplicon was also sequenced by the traditional Sanger method as described previously (14).
Analysis
Prevalence was consistently calculated as percentage of positives from the total number of cases tested. HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68 were considered high-risk (hr) with regards to oncogenic potential (15) and all other types as low-risk. Differences in prevalence for any HPV type, hr HPV positive or HPV16 and 18 positive by gender, race, age, and disease stage were evaluated using Fisher’s Exact Test. Age was grouped into 10-year intervals. Cancer stage was crudely classified as local, regional, distant, or unknown, and termed SEER stage.
Hierarchical categories for HPV status were assigned as follows: (1) HPV16 – includes all cases positive for this type regardless of other results, (2) HPV18 - adds all case positive for HPV18, but not for HPV16, (3) other hr HPV – adds cases positive for any high risk type as listed above, but not HPV16 or 18, (4) any HPV – adds all cases positive for any HPV type not included in the previous groups.
Results
Within the time period between 1995 and 2005, a total of 401 eligible anal cancer cases were identified from the records of the seven registries and RTRs. Due to lack of availability or participation tissues were not received from 222 cases. An additional 32 cases were excluded for insufficient malignant representation and one sample was inadequate in HPV testing. Despite this reduction, the admitted tissue samples appeared to be representative of cancers diagnosed in participating registries, based on demographic variables such as age, race, and sex (data not shown).
Specimens from the remaining 146 anal cancer cases were successfully tested. The patient’s gender, race and age were distributed as shown in Table 1. HPV genotyping results were obtained by LA from 127 cases and by LiPA for the remaining 19. Any HPV was detected in 133 (91.1%) cases. A high risk-type was found in 129 cases (88.4%) and four (2.7%) contained only low risk HPV (two each with HPV6 and HPV26). In 117 (80.1%) cases a single HPV infection was detected. Multiple types (2 – 6) were observed in 16 (11%) cases averaging 1.17 types per positive sample. HPV16 was present in 113 (77.4%) cases, HPV18 in 5 (3.4%) and two of them (1.4%) had both of these types. Other frequently identified types included HPV33 (9 cases, 6.2%), as well as HPV6 (6 cases, 4.1%) and 11 (5 cases, 3.4%) (see Table 2 for further details).
Table 1.
HPV prevalence in anal cancers by gender, race, age and SEER stage
| n | HPV2 (%) | hr HPV (%) | HPV16/18 (%) | ||||
|---|---|---|---|---|---|---|---|
| Gender | |||||||
| Female | 93 | 86 | (92.5) | 84 | (90.3) | 74 | (79.6) |
| Male | 53 | 47 | (88.7) | 45 | (84.9) | 42 | (79.2) |
| P = 0.548 | P ≥ 0.422 | P = 1.000 | |||||
| Race | |||||||
| Asian Pac. Islands | 5 | 5 | (100) | 5 | (100) | 3 | (60.0) |
| Black | 15 | 14 | (93.3) | 13 | (86.7) | 12 | (80.0) |
| Hispanic | 21 | 20 | (95.2) | 19 | (90.5) | 17 | (81.0) |
| White | 105 | 94 | (89.5) | 92 | (87.6) | 84 | (80.0) |
| P ≥ 0.928 | P = 1.000 | P ≥ 0.684 | |||||
| Age Groups | |||||||
| ≤ 39 | 10 | 10 | (100) | 10 | (100) | 8 | (80.0) |
| 40 – 49 | 23 | 21 | (91.3) | 21 | (91.3) | 19 | (82.6) |
| 50 – 59 | 40 | 37 | (92.5) | 35 | (87.5) | 30 | (75.0) |
| 60 – 69 | 28 | 26 | (92.9) | 25 | (89.3) | 22 | (78.6) |
| 70 – 79 | 30 | 28 | (93.3) | 27 | (90.0) | 26 | (86.7) |
| ≥ 80 | 15 | 11 | (73.3) | 11 | (73.3) | 11 | (73.3) |
| P ≥ 0.337 | P ≥ 0.541 | P ≥ 0.846 | |||||
| SEER stage | |||||||
| Local | 83 | 75 | (90.4) | 73 | (88.0) | 67 | (80.7) |
| Regional | 30 | 27 | (90.0) | 26 | (86.7) | 23 | (76.7) |
| Distant | 9 | 9 | (100) | 9 | (100) | 8 | (88.9) |
| Unknown | 24 | 22 | (91.7) | 21 | (87.5) | 18 | (75.0) |
| P = 1.000 | P ≥ 0.899 | P ≥ 0.824 | |||||
HPV = positive for any of the types tested
hr HPV = positive for one of the high risk types HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68
HPV16/18 = positive for HPV16 or HPV18
P-values for differences between categories were calculated by Fisher’s Exact Test. Percentage from each category’s total (in n column) are stated in brackets
Table 2.
Summary of HPV detection in anal cancers
| Adequate, eligible samples | 146 | |
|---|---|---|
| HPV positive (any type) | 133 | 91.1% |
| High Riska only | 116 | 79.5% |
| Low Riskb only | 4 | 2.7% |
| High and low risk | 13 | 8.9% |
| Single type | 117 | 80.1% |
| Multiple types (2 – 6) | 16 | 11.0% |
| HPV16 | 113 | 77.4% |
| HPV33 | 9 | 6.2% |
| HPV6 | 6 | 4.1% |
| HPV11 | 5 | 3.4% |
| HPV18 | 5 | 3.4% |
| HPV26 | 2 | 1.4% |
| HPV31 | 2 | 1.4% |
| HPV51 | 2 | 1.4% |
| HPV58 | 2 | 1.4% |
| HPV62 | 2 | 1.4% |
| HPV66 | 2 | 1.4% |
| Other types (one case each) | 8 | 5.5% |
HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68
any HPV not included in the high risk HPV types
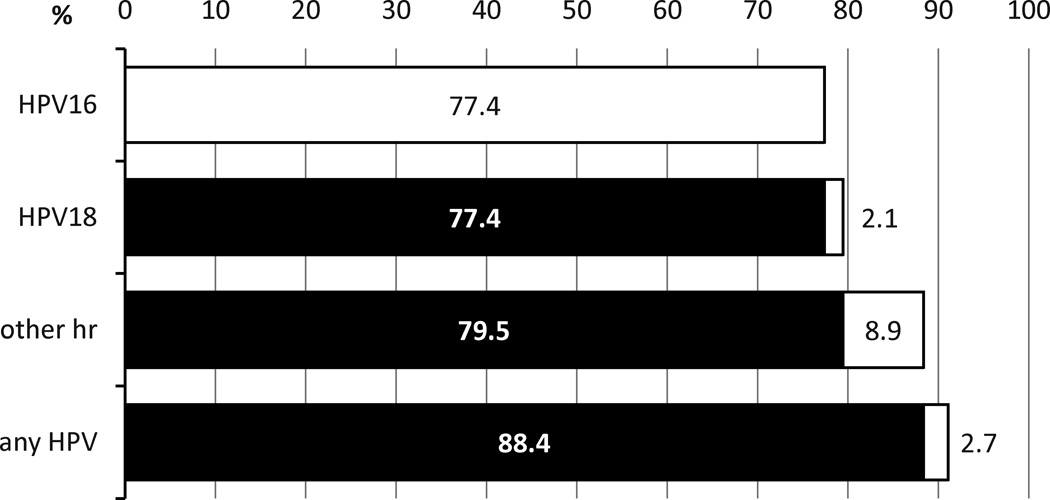
By histological type, hr HPV types were found in 126 of 133 (94.7%) squamous cell carcinomas (SCC), in 2 of 11 (18.2%) adenocarcinomas (both HPV16) and in one of the tumors with “other” histology (HPV31). Applying hierarchical assignment, 113 anal cancers (77.4%) were attributable to HPV16, an additional 3 (2.1%) to HPV18, a further 13 (8.9%) to all other high risk types, and 4 (2.7%) to all remaining HPV types (Fig. 1). Proportions of cases positive for HPV (any type tested), hr HPV or HPV16/18 were not statistically different by sex, race, age group, or SEER stage (Table 1).
Figure 1.
Hierarchical attribution of HPV types to anal cancers. White fields indicating attribution of the specific type or group and black the cumulative prevalence of types with higher hierarchy.
Pyrosequencing allowed determination of HPV 16 variant lineage in 98 HPV16 positive cases. An additional 2 were clarified by Sanger sequencing. For the remaining 13, PCR amplification or sequencing could not be achieved at a quality necessary for unambiguous variant typing. As detailed in Table 3, the majority of HPV16 variants were European (88%) followed by African and Asian variants (6% each).
Table 3.
Variants determined in 100 HPV16 positive anal cancers
| Variant | Found in Anal Cancers (= %) |
|
|---|---|---|
| African | Af1 | 5 |
| Af2 | 1 | |
| Asian American or North American | AA/NA-1 | 4 |
| Asian | As | 2 |
| European | Ep | 37 |
| Em | 33 | |
| E-C109G | 3 | |
| E-G131G | 12 | |
| E-G131T | 3 |
100 of 113 HPV16 positive anal cancer specimens were successfully sequenced, the remaining 13 did not yield data at sufficient quality.
Discussion
The tissues obtained for this study were requested from seven state cancer registries and provide the first U.S. population-based assessment of HPV prevalence and type distribution in anal cancers. As expected, HPV prevalence was high and at least one hr type was found in almost 90% of the anal cancer cases. Internationally, findings have varied substantially, likely a result of small sample sizes, as well as disparate HPV detection methodologies and sensitivity. Nonetheless, most studies based on consensus primer PCR systems also have reported HPV prevalence in anal cancer tissue of 80% or greater (3;16;17). While the combined use of the LA and LiPA HPV genotyping tests has potentially increased overall detection sensitivity in our study, the type-specific prevalence was similar to that reported in a meta-analysis including North American studies mainly among MSM (3), where over 70% were HPV16-positive and prevalence of HPV6, 18 and 33 were observed at 5 and 10% each. It is unclear if the remaining small proportion of anal cancer specimens are HPV negative as a result of sampling artifact, deficiencies in the HPV detection methodology, or if these cases were truly triggered by alternative carcinogenic agents. By histological examination, only 4 of the 13 HPV negative tumors were squamous carcinomas. The rest were adenocarcinomas that could be of colorectal origin (7), Paget’s disease (1) or undifferentiated (1).
No evidence was found for differences in HPV prevalence between any of the demographic subgroups or cancer stage included in the analysis. This was not surprising considering the high HPV prevalence, especially for HPV16, and the modest size of the study population.
Distribution of HVP16 variants was dominated by European lineages and was very similar to results found in cervical cytology samples in the United States (18). The frequency of the HPV16 G131 variants in our invasive anal tumors was 2.5-fold greater than that reported by Da Costa et al. (10) in anal neoplasia tissue from a mostly HIV positive population, which may support their hypothesis that infection with the E-G131G or E-G131T genotypes pose elevated risk for malignant progression. It is not known if HIV infection plays any role in this regard and information on HIV status was not collected in our study.
This study represents the first assessment of HPV type distribution in anal cancers derived from a random sample of population based U.S. cancer registries representing different geographical regions. HPV16 and HPV18 DNA were found in almost 80% of the anal cancers in this study, and 103 anal cancers (70%) were exclusively positive for these two types. These results will assist in the establishment of a baseline for etiologic fractions of anal cancer attributable to specific HPV genotypes. The data will be integral to the surveillance and evaluation of prophylactic HPV vaccine efficacy for anal cancer in the United States.
Acknowledgment
We thank all members of HPV typing of Cancers Workgroup for contributions made toward this study:
- CDC:
-
○Mona Saraiya, MD, MPH, CDC, Division of Cancer Prevention and Control
-
○Elizabeth R Unger, MD, PhD, CDC, CCID/NCEZID/DHCPP/CVDB
-
○Martin Steinau, PhD, CDC, CCID/NCEZID/DHCPP/CVDB
-
○Mariela Z Scarbrough, BS, CCID/NCEZID/DHCPP/CVDB
-
○Meg Watson, MPH, Division of Cancer Prevention and Control
-
○Mangalathu Rajeevan, PhD, CDC, CCID/NCEZID/DHCPP/CVDB
-
○Daisy Lee, MS, CDC, CCID/NCZVED, HPV Lab
-
○Maung Maung Khin, MS, CCID/NCEZID/DHCPP/CVDB
-
○Juanita Onyekwuluje, MS, CDC, CCID/NCEZID/DHCPP/CVDB
-
○Deblina Datta, MD, Division of STD Prevention
-
○Susan Hariri, PhD, Division of STD Prevention
-
○
- Florida:
-
○Jill MacKinnon, PhD, University of Miami, Florida Cancer Data System
-
○Youjie Huang, MD, DrPH, MPH, Florida State Department of Health
-
○Carlos Alvarez, BBA, University of Miami, Florida Cancer Data System
-
○Edward Wilkinson, MD, University of Florida
-
○Martha Campbell-Thompson, DVM, PhD, University of Florida
-
○Amy Wright, MS, University of Florida
-
○Kelley Durden, HT (ASCP), University of Florida
-
○
- Hawaii:
-
○Brenda Hernandez, PhD, University of Hawaii, Cancer Research Center of Hawaii
-
○Marc Goodman, PhD, University of Hawaii, Cancer Research Center of Hawaii
-
○Hugh Luk, BS, HTL, University of Hawaii, Cancer Research Center of Hawaii
-
○David Horio, MD, University of Hawaii, Cancer Research Center of Hawaii
-
○Shoji Ikeda, BA, University of Hawaii, Cancer Research Center of Hawaii
-
○Michael Green, CTR, University of Hawaii, Cancer Research Center of Hawaii
-
○Catherine Grafel-Anderson, BS, University of Hawaii, Cancer Research Center of Hawaii
-
○Rayna Weise, MPH, University of Hawaii, Cancer Research Center of Hawaii
-
○
- Iowa:
-
○Freda Selk, AAS, University of Iowa
-
○Dan Olson, MS, University of Iowa
-
○
- Kentucky:
-
○Thomas Tucker, PhD, MPH, University of Kentucky
-
○Claudia Hopenhayn, PhD, MPH, University of Kentucky
-
○Amy Christian, MSPH, University of Kentucky
-
○
- Los Angeles County, California:
-
○Joe House, University of Southern California
-
○Myles G. Cockburn, PhD, University of Southern California
-
○Andre Kim, MPH, University of Southern California
-
○
- Louisiana:
-
○Edward Peters, DMD, SM, ScD, Louisiana State University
-
○Lauren Cole, PhD, MPH Candidate, Louisiana State University
-
○Tara Ruhlen, MPH, Carolinas Rehabilitation, Charlotte, NC
-
○
- Michigan:
-
○Glenn Copeland, MS, Michigan Department of Community Health
-
○Lana Ashley, Michigan Department of Community Health
-
○Jetty Alverson, Michigan Department of Community Health
-
○Michelle Hulbert, Michigan Department of Community Health
-
○Won Silva, MA, Michigan Department of Community Health
-
○Samuel Hirsch, MD, St Joseph Mercy Hospital, Ann Arbor, Michigan
-
○
- Battelle:
-
○Christopher Lyu, MPA, Battelle, Durham, NC
-
○Bruce Ellis, MS, Battelle, Arlington, VA
-
○Natalie Madero, BS, Battelle, Baltimore, MD
-
○Emily Reid, BA, Battelle, Durham, NC
-
○Donna Little, BS, Battelle, Baltimore, MD
-
○April Greek, PhD, Battelle, Seattle, WA
-
○Dale Rhoda, PhD, Battelle, Columbus, OH
-
○Susan Brossoie, CTR/RHIT, UNC Hospitals Cancer Registry, Chapel Hill, NC
-
○Katherine Gideon, Battelle, Toxicology Northwest, Richland, WA
-
○Linda Delma Gieseke, Battelle, Columbus, OH
-
○Stephanie Ashcraft, Battelle, Columbus, OH
-
○
Financial Disclosures: The support for collection of original specimens from non-repositories (Kentucky, Florida, Michigan, Louisiana), coordination of genotyping data from both SEER and NPCR registries, and genotyping was largely supported by CDC intramural funds and Vaccine For Children Funds. This project has been supported in part with Federal funds by the Centers for Disease Control and Prevention (CDC) under grant number NO. 5U58DP000810-5 (Kentucky), 5U58DP000844-5 (Florida), 5U58DP000812-5 (Michigan), and 5U58DP000769-5 (Louisiana) and with Federal funds for Residual Tissue Repositories from the National Cancer Institute SEER Population-based Registry Program, National Institutes of Health, Department of Health and Human Services, under Contract No. N01-PC-35139 (Los Angeles), N01-PC-35143 (Iowa) and N01-PC-35137 (Hawaii).
The collection of data from California used in this publication was largely supported by the California Department of Health Services as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; by the National Cancer Institute, National Institutes of Health, Department of Health and Human Services under Contract No. N01-PC-2010-00035; and grant number 1U58DP000807-3 from the Centers for Disease Control and Prevention.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
There are no financial disclosures from any of the authors.
Reference List
- 1.National Program of Cancer Registries. Cancer Incidence 1999 – 2009 (States only) 2012 Available at http://wondercdcgov/cancerhtml.
- 2.Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Waldron W, et al. National Cancer Institute; Bethesda, MD: 2011. SEER Cancer Statistics Review, 1975–2008. http://seer.cancer.gov/csr/1975_2008/, based on November 2010 SEER data submission, posted to the SEER web site. [Google Scholar]
- 3.De Vuyst H, Clifford GM, Nascimento MC, Madeline MM, Franceschi S. Prevalence and type distribuion of human paoillomavirus in carcinoma and intraepithelial neoplasia of the vulva, vagina and anus: A meta-analysis. Int J Cancer. 2009;124:1626–1636. doi: 10.1002/ijc.24116. [DOI] [PubMed] [Google Scholar]
- 4.Hoots BE, Palefsky JM, Jeanne M, Pimenta JM, Smith JS. Human papillomavirus type distribution in anal cancer and anal intraepithelial lesions. Int J Cancer. 2010;124:2375–2383. doi: 10.1002/ijc.24215. [DOI] [PubMed] [Google Scholar]
- 5.Kreimer AR, Gonzalez P, Katki HP, Porras C, Schiffman M, Rodriguez AC, et al. Efficacy of a bivalent HPV16/18 vaccine against anal HPV 16/18 infection among young women: a nested analysis within the Costa Rica Vaccine Trail. Lancet Oncol. 2011;12(9):862–870. doi: 10.1016/S1470-2045(11)70213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palefsky JM, Giuliano AR, Goldstone S, Moreira ED, Aranda C, Jessen H, et al. HPV vaccine against anal HPV infection andanal intraepithelial neoplasia. N Engl J Med. 2011;365:1576–1585. doi: 10.1056/NEJMoa1010971. [DOI] [PubMed] [Google Scholar]
- 7.Joseph DA, Miller JW, Wu X, Chen VW, Morris CR, Goodman MT, et al. Understanding the burden of human papillomavirus-associated anal cancers in the US. Cancer. 2008;113(10 Suppl):2892–2900. doi: 10.1002/cncr.23744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frisch M, Fenger C, van den Brule AJ, Sorensen P, Meijer CL, Walboomers JM, et al. Variants of squamous cell carcinoma of the anal canal and perianal skin and their relation to human papillomaviruses. Cancer Res. 1999;59(3):753–757. [PubMed] [Google Scholar]
- 9.Abramowitz L, Jacguard AC, Jaroud F, Haesebaert J, Siproudhis L, Pradat P, et al. Human papillomavirus genoype distribution in anal cancer in France: The EDiTH V study. Int J Cancer. 2011;129(2):433–499. doi: 10.1002/ijc.25671. [DOI] [PubMed] [Google Scholar]
- 10.Da Costa MM, Hogeboom CJ, Holly EA, Palefsky JM. Increased risk of high-grade anal neoplasia associated with a Human Papillomaivirus type 16 E6 sequence variant. J Infect Dis. 2002;185:1229–1237. doi: 10.1086/340125. [DOI] [PubMed] [Google Scholar]
- 11.Xi LF, Critchlow CW, Wheeler CM, Koutsky LA, Galloway DA, Kuypers J, et al. Risk of anal carcinoma in situ in relation to human papillomavirus type 16 variants. Cancer Res. 1998;58(17):3839–3844. [PubMed] [Google Scholar]
- 12.Steinau M, Patel SS, Unger ER. Efficient DNA extraction for HPV genotyping in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2011;13(4):377–381. doi: 10.1016/j.jmoldx.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swan DC, Limor JR, Duncan KL, Rajeevan MS, Unger ER. Human papillomavirus type 16 variant assignment by pyrosequencing. J Virol Methods. 2006;136:166–170. doi: 10.1016/j.jviromet.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Swan DC, Rajeevan MS, Tortolero-Luna G, Follen M, Tucker RA, Unger ER. Human papillomavirus type 16 E2 and E6/E7 variants. Gynecol Oncol. 2005;96:695–700. doi: 10.1016/j.ygyno.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 15.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Sha KV, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 16.Tachezy R, Jirasek T, Salakova M, Lufvidova V, Kubecova M, Horak L, et al. Human papillomavirus infection and tumours of the anal canal: correlation of histology, PCR detection in paraffin sections and serology. APMIS. 2007;115(3):195–203. doi: 10.1111/j.1600-0463.2007.apm_526.x. [DOI] [PubMed] [Google Scholar]
- 17.Vol. 90, Human Papillomaviruses. Lyon: IARC; 2007. IARC Monographs on the Evaluation of carcinogenic risks to Humans. [PMC free article] [PubMed] [Google Scholar]
- 18.Zuna RE, Moore WE, Shanesmith RP, Dunn ST, Wang SS, Schiffman M, et al. Association of HPV16 E6 variants with diagnostic severity in cervical cytology samples of 354 women in a US population. Int J Cancer. 2009;125(11):2609–2613. doi: 10.1002/ijc.24706. [DOI] [PMC free article] [PubMed] [Google Scholar]