Abstract
Cytosolic nucleotidase II (cN-II) from Legionella pneumophila (Lp) catalyzes the hydrolysis of GMP and dGMP displaying sigmoidal curves while catalysis of IMP hydrolysis displayed a biphasic curve in the initial rate versus substrate concentration plots. Allosteric modulators of mammalian cN-II did not activate LpcN-II while GTP, GDP and the substrate GMP were specific activators. Crystal structures of the tetrameric LpcN-II revealed an activator binding site at the dimer interface. A double mutation in this allosteric binding site abolished activation, confirming the structural observations. The substrate GMP acting as an activator and partitioning between the allosteric and active site is the basis for the sigmoidicity of the initial velocity versus GMP concentration plot. The LpcN-II tetramer showed differences in subunit organization upon activator binding that is absent in the activator-bound human cN-II structure. This is the first observation of a structural change induced by activator binding in cN-II that may be the molecular mechanism for enzyme activation.
Keywords: 5’-nucleotidase, Allostery, Heterotropic activation, Substrate activation, GMP-complexed LpcN-II structure
Introduction
5’-nucleotidases (EC 3.1.3.5) are a large family of enzymes that catalyze the dephosphorylation of ribo- and deoxyribo nucleoside monophosphates to their corresponding nucleosides [1]. Based on their cellular localization, 5’-nucleotidases are broadly classified as membrane bound and soluble forms with the latter form further subdivided into categories based on their substrate specificity. Cytosolic nucleotidase-I (cN-I) shows specificity for AMP and pyrimidine nucleoside monophosphates, cytosolic nucleotidase-II (cN-II) for IMP and GMP, cytosolic nucleotidase III (cN-III) for pyrimidine nucleoside monophosphates and ISN1, a new class of 5’-nucleotidase, for IMP [2, 3]. All soluble 5’-nucleotidases are members of the haloacid dehalogenase (HAD) superfamily of enzymes [4, 5].
The cN-II class of nucleotidases play critical roles in maintaining the flux through purine nucleotide cycle and oxypurine cycle, pathways which generate critical metabolites like fumarate, ammonia, phosphoribosyl pyrophosphate and ribose-1-phosphate [6]. Given the critical role of cN-IIs, this enzyme from mammalian sources shows multiple levels of regulation. Mammalian cN-IIs display sigmoidal velocity versus substrate concentration plots for the hydrolysis of their substrate IMP, inhibition by inorganic phosphate [7] and heterotropic activation by ATP [8], 2,3-BPG [9], polyphosphates [10] and diadenosine polyphosphates [11]. Though reports establishing unambiguously the mechanism of activation of mammalian cN-II are lacking, it has been speculated that the binding of the activator facilitates formation of higher order enzyme oligomers [12]. Structures of the apo and various complexed forms of the human cN-II identify the effector-molecule binding site, though no conformational change is seen upon activator binding [13, 14]. It was proposed that the effector-induced disorder-to-order transition, generating rearrangements within the catalytic site and the subsequent coordination of the catalytically essential magnesium, is the probable mechanism of activation. However, it should be noted that the human cN-II used for crystallographic studies corresponds to 1-536 out of 561 residues [13, 14]. It has been shown earlier that truncation of 13 residues from the carboxy terminus in human cN-II leads to a 33-fold reduction in Vmax and 2.5-fold increase in Km value for the substrate IMP along with a 55-fold reduction in the activation of IMP hydrolysis by ATP [12].
Here we report the detailed biochemical, kinetic and structural characterizations of recombinant L. pneumophila cN-II (LpcN-II), the first prokaryotic cN-II to have been studied. LpcN-II prefers GMP over IMP as its substrate, and has an obligate requirement for the divalent cation Mg2+. GTP was found to be a novel activator of the enzyme, modulating the Km without significantly affecting the Vmax for the substrate GMP. The enzyme displayed a unique feature of substrate activation by GMP. To understand the mechanism of activation, we report the PO4- and GMP-complexed crystal structures of the enzyme. Though crystallization was in the presence of GMP-PNP, electron density was evident only for GMP moiety. GMP binds at the dimer interface and this binding site has been validated by mutation of the contacting residues R86 and Y421. A tilt of one subunit with respect to another was observed when the PO4-complexed and GMP-complexed structures were superposed, a feature unlike the human cN-II structures that do not exhibit altered organization upon ligand binding. The possible peptide connectivities between the active and effector sites that may serve as structural conduits are discussed.
L. pneumophila is the causative agent of Legionnaires' disease. Understanding the metabolism of the pathogen is a prerequisite for devising effective intervention strategies. cN-II activity is indispensable for the survival of astrocytoma cells [15]. Being a pivotal enzyme in nucleotide metabolism, differences in the structure and regulation of human and L. pneumophila cN-II could be exploited to devise appropriate intervention strategies for Legionnaires' disease.
Results
Biochemical characterization, substrate specificity and kinetic parameters of LpcN-II
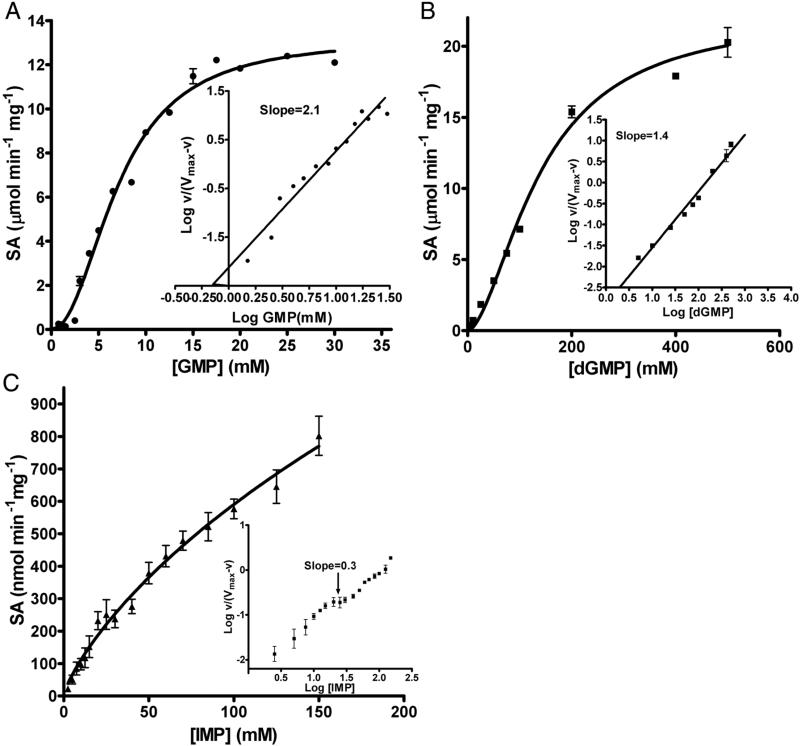
Purified recombinant hexa-histidine tagged full-length LpcN-II (Figure S1A-C), upon examination by size-exclusion chromatography, was a tetramer at 20 μM protein concentration (Figure S1D). LpcN-II readily catalyzed the breakdown of selected nucleoside monophosphates (Table S1) and the preferred cofactor for the reaction was Mg2+ followed by Mn2+. GMP was found to be the most preferred substrate followed by dGMP and IMP while activity on AMP, XMP, CMP and UMP was poor. The initial velocity versus substrate concentration plots for GMP and dGMP were sigmoidal while IMP curves were biphasic (Figure 1A-C). Summarized in Table 1 are the kinetic parameters of LpcN-II for various substrates and cofactors. The Km values for all the substrates are in the mM range similar to other cN-Is and cN-IIs that have high Km values (1-15 mM) for AMP/IMP [1]. The enzyme displayed highest catalytic efficiency for GMP with a kcat/Km value of 1.7 mM−1 sec−1. The sigmoidicity of the initial velocity plot is evident in the high values of Hill coefficient (nH). When compared to mammalian cN-IIs, LpcN-II exhibits lower affinity for IMP (Table S2). LpcN-II also hydrolyzed p-nitrophenyl phosphate (pNPP), at much lower catalytic efficiency, with hyperbolic velocity versus substrate concentration plots. It should be noted that pNPP is not a physiologically relevant substrate. However, this activity of LpcN-II was amenable to continuous monitoring of product formation and hence was exploited to understand the kinetic behavior of LpcN-II activation.
Figure 1.
Substrate titration plots of LpcN-II for (A) GMP, (B) dGMP and (C) IMP. Plots in A and B show sigmoidal kinetics while C shows biphasic kinetics. Insets in panels A and B show Hill plot of log (v/ (Vmax-v)) versus log [substrate] with the slope value of the line indicated. Inset in panel C shows Hill plot of log (v/ (Vmax-v)) versus log [IMP] indicative of negative homotropic cooperativity. The slope indicated is for the mid-region (highlighted by arrow) of the plot. Data were fitted to the Hill equation (1, Methods, supplemental information) using GraphPad prism Software, version 4.0 (GraphPad Software, Inc., San Diego, CA). ‘v’ refers to the initial velocity. SA stands for specific activity.
Table 1.
Summary of kinetic parameters of LpcN-II for various substrates and cofactors
| S.No. | Substrate/pseudo substrate | S0.5 or Km (mM)c | Vmax (μmol min−1 mg−1) | kcat (sec−1) | kcat/Km or kcat/S0.5 (mM−1 sec−1)c | nHd | Type of kinetic profile |
|---|---|---|---|---|---|---|---|
| 1. | GMP1 | 7.2 ± 0.3 | 13.2 ± 0.4 | 12.4 | 1.7 | 2.1 | Sigmoidal |
| 2. | dGMP1 | 142 ±17 | 23 ± 2 | 20.7 | 0.15 | 1.7 | Sigmoidal |
| 3. | IMP1 | 18 ± 9a 118 ± 10a |
0.4 ± 0.1b 1.3 ±0.2b |
0.3 1.2 |
0.02 0.01 |
---- | Biphasic |
| 4. | pNPP1 | 5.1 ± 0.9 | 1.0 ± 0.1* | 1.0 × 10−3 | 0.1 × 10−3 | 1 | Hyperbolic |
| 5. | MgCl22 | 1.1 ± 0.1 | 0.9 ± 0.0* | ----- | -------- | ------ | Hyperbolic |
| 6. | MnCl22 | 0.5 ± 0.2 | 0.3 ± 0.0* | ------- | --------- | ------ | Hyperbolic |
Footnotes.
nmol min−1 mg−1
Metal ion used was 30 mM MgCl2
substrate was 15 mM pNPP
lower and higher Km values for IMP from ν versus [S] biphasic curves
lower and higher Vmax values for IMP from ν versus [S] biphasic curves
Either S0.5 or Km is used depending on the kinetic profile
nH is Hill coefficient. The parameters were derived from extrapolation of intercepts from Eadie-Hofstee plots.
Heterotropic allosteric activation of LpcN-II by GTP, dGTP and GDP
The list of compounds screened for their effect on LpcN-II's pNPP hydrolyzing activity is provided in Table S3. While activators of the mammalian enzyme had no effect on the prokaryotic enzyme, GTP, dGTP, and GDP showed activation of LpcN-II activity. The dissociation constant (KD) of GTP was 4.2 ± 0.4 mM under catalytic conditions and 4.1 ± 0.7 mM under equilibrium conditions (Figure S2A and B). Absence of activation by ATP and ITP with potent activation by GTP, dGTP and GDP indicates that C2 amino group on the purine ring is indispensible for activation of LpcN-II. It should also be noted that GTP is not hydrolyzed by the enzyme as evident from Chen's assay and ion-pair reverse phase HPLC (IP-RP-HPLC).
a) K-type activation of GMP hydrolysis by GTP
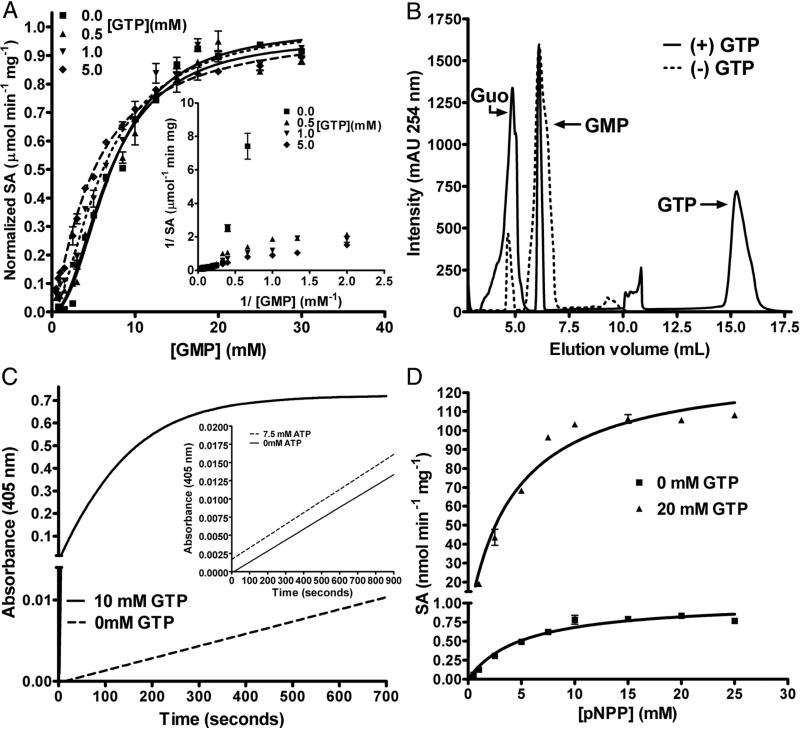
Activation of GMP hydrolysis by GTP was checked by estimating for the liberated phosphate. With increasing concentration of GTP, the sigmoidicity of velocity versus [GMP] plot reduced (the Hill coefficient reduced from 2.1 in the absence of GTP to 1.2 at 5mM GTP) (Figure 2A, Table S4). This is also evident in the double reciprocal plot of 1/v versus 1/ [GMP] where, with increasing concentration of GTP, the nonlinearity was abolished (Fig. 2A inset). Further, in the v versus [S] plot for GMP hydrolysis, the maximum difference of ~8 fold in product formation between the unactivated and activated LpcN-II was seen at 1.5 mM substrate and 1 mM GTP concentration. This observation was confirmed by IP-RP-HPLC, wherein under similar conditions a 9-fold increase in guanosine formation was seen (Figure 2B). ATP, under similar assay conditions, did not show increase in rate of GMP hydrolysis.
Figure 2.
Activation of GMP and pNPP hydrolysis by GTP. (A) Initial velocity versus [GMP] plots at different fixed GTP concentrations. Inset: The double reciprocal plot shows the disappearance of the non-linear nature of the curves with increasing GTP concentrations. The velocity on the y-axis is normalized to 1. The curves were fitted to the Hill equation. SA stands for the specific activity of the enzyme. (B) Activation of GMP hydrolysis by GTP examined by IP-RP-HPLC. The reactions were carried out in Tris HCl, pH 7.4, 30 mM MgCl2, and 1.5 mM GMP. 1 mM of GTP was used in the reaction where the effect of the activator was to be assessed. Dotted line represents hydrolysis of GMP in the absence of GTP and solid line represents hydrolysis of GMP in the presence of 1 mM GTP. For the unactivated reaction, the area under the peak for GMP and guanosine was 1053 ± 62 and 129 ± 14, respectively. For the activated reaction, the area under the peak for GMP and guanosine was 342 ± 48 and 813 ± 220, respectively. The values are an average of 3 measurements. The peak areas were integrated using the ÄKTA™ Basic HPLC software UNICORN, version 5.11. (C) Activation of pNPP hydrolysis by GTP. Time scans of pNPP hydrolysis in the absence and presence of GTP is represented by dotted and solid lines, respectively. The y-axis is broken for ease of visualization. The inset shows pNPP titration in the absence (solid line) and presence (dotted line) of ATP. (D) pNPP titration in the absence and presence of GTP. The y-axis is broken for ease of visualization. The data points were fit using the linear and non-linear curve fitting algorithms of GraphPad Prism software, version 4.0 (GraphPad Software, Inc., San Diego, CA).
Allosteric systems that demonstrate altered substrate affinity upon effector binding are referred to as ‘K-type’ systems [16]. Activation of GMP hydrolysis by GTP is ‘K-type’ since the activator reduces the Km without altering the Vmax. The Km for GMP in the absence of added GTP was 7.2 ± 0.3 mM while it reduced to 5.1 ± 0.3 mM in the presence of 5 mM GTP. Qax, expressed as Kia/Kia/x, (where Kia is the KD of the substrate from ES complex and Kia/x is the KD of the substrate from ES complex in the presence of the activator) is 1.41, which shows that GTP causes an increase in the enzyme's affinity for the substrate.
b) V-type activation of pNPP hydrolysis by GTP
Time course of pNPP hydrolysis approached saturation within 200 sec. of the initiation of reaction in the presence of GTP while ATP did not activate the enzyme (Figure 2C and inset). The Vmax for pNPP hydrolysis increased from 1.0 ± 0.1 nmol min−1 mg−1, in the absence of GTP, to 134 ± 6 nmol min−1 mg−1, in the presence of 20 mM GTP, showing a ~130 fold enhancement (Figure 2D) while the Km values were not appreciably different (Table S4). Systems that demonstrate altered catalytic rates (Vmax or kcat) are described as ‘V-type’ systems [16]. Activation of pNPP hydrolysis by GTP is ‘V-type’ since the activator increases the Vmax without altering the Km. The intersection patterns in the plots of 1/v versus 1/ [pNPP] at different fixed [GTP] and 1/v versus 1/ [GTP] at different fixed [pNPP] (Figure S2C and D) are indicative of random binding of GTP and pNPP to the enzyme and also confirm that the activation of pNPP hydrolysis brought about by GTP is non-essential ‘V’-type, without significant variation in Km values.
Effect of GMP, GDP, IMP and ITP on pNPP hydrolysis
a) GMP activation of pNPP hydrolysis
GMP activates pNPP hydrolysis by approximately 15 fold and the initial velocity plot becomes sigmoidal in the presence of this nucleotide (Fig 3A and inset). The Km for pNPP in the absence of GMP is 4.5 ± 0.7 mM and in the presence of 15 mM GMP, the S0.5 becomes 7.0 ± 0.4 mM. The marginal increase in the Km/ S0.5 for pNPP might be because of competition at the active site.
Figure 3.
Activation of pNPP hydrolysis by GMP. (A) pNPP titration was carried out at 0 mM and 15 mM GMP concentration and monitored for pNP formation at 405 nm. The inset shows the double reciprocal plot for pNPP titration in the presence and absence of GMP. Note the non-linearity of the curve in the presence of GMP, characteristic of enzymes exhibiting sigmoidal behavior. pNPP titration, in the absence of GMP, was stopped at 20 mM because pNPP exhibits substrate inhibition. (B) Reduction in GTP activation of pNPP hydrolysis by GMP. The liberation of pNP was monitored at 405 nm. Each line corresponds to the titration of [GTP-Mg2+] at various fixed concentration of GMP. (C) Reduction in GTP activation of pNPP hydrolysis by IMP. The liberation of pNP was monitored at 405 nm. Each line corresponds to the titration of [GTP-Mg2+] at various fixed [IMP]. Data were fitted by linear regression and non-linear curve fitting algorithms of GraphPad prism Software, version 4.0 (GraphPad Software, Inc., San Diego, CA).
b) GMP and GDP bind to the GTP binding site
GMP was found to bring about a 2-fold reduction in the activation of pNPP hydrolysis by GTP as evident from comparison of time course measurements of substrate hydrolysis in the presence of GTP alone, GMP alone and combined equimolar quantities of GMP and GTP (Figure S3A). To understand the mechanism of reduction in GTP activation and the site to which GMP binds to bring about activation, a competition assay was carried out wherein [GTP] was varied at several fixed concentrations of GMP and saturating [pNPP], whose hydrolysis was monitored at 405 nm. The plot showed lines intersecting on the 1/v axis (Figure 3B) indicative of competitive displacement of GTP by GMP at the activator-binding site. The reverse plot, wherein GMP is titrated at several fixed concentrations of GTP with the hydrolysis of pNPP monitored at 405 nm, shows a concentration-dependent decrease in pNPP hydrolysis with increasing GMP (Figure S3B) attesting to the observation that GMP competitively reduces the activation brought about by GTP. GDP was also found to compete for binding to the GTP binding site and bring about a concentration dependent reduction in activation of pNPP hydrolysis by GTP (Figure S3C). Guanosine at a concentration of 5 mM was also found to compete weakly for GTP binding site (Figure S3D) while guanine alone due to poor solubility could not be tested. These results suggest that at least one phosphate is necessary for effective activation, with the potency of activation being highest for GTP, followed by GDP and GMP.
c) IMP and ITP bind to the GTP binding site
Though IMP and ITP do not activate pNPP hydrolysis, they reduce the activation of pNPP hydrolysis brought about by GTP. The competitive intersection pattern of the lines in Figure 3C and Figure S3E indicate that IMP and ITP bind to the GTP-binding site. Taken together, these observations indicate that the oxo-group at C6 is essential for binding to the activator site while the amino group at C2 is essential for bringing about activation. On the contrary, ATP does not affect the activation of pNPP hydrolysis brought about by GTP (Figure S3F).
Crystallographic studies
The structure of L. pneumophila was solved in complex with PO4-(PDB ID 4G63) and GMP (PDB ID 4OHF) at 2.7 Å and 2.5 Å, respectively. The PO4-complexed structure at 2.7 Å resolution contains two phosphate ions with both present at sites on the protein that do not have known functional significance. The GMP-complexed structure at 2.5 Å resolution has one PO4 at a location similar to one of the phosphates in the PO4-complexed structure while the second PO4 is present in the active site. The structure of LpcN-II in complex with GMP contains four molecules in the asymmetric unit (ASU) of space group C2 and the PO4-complexed structure contains one molecule in the ASU of space group I4122, with the tetramer being generated from crystallographic symmetry.
Tertiary and Quaternary structures of LpcN-II
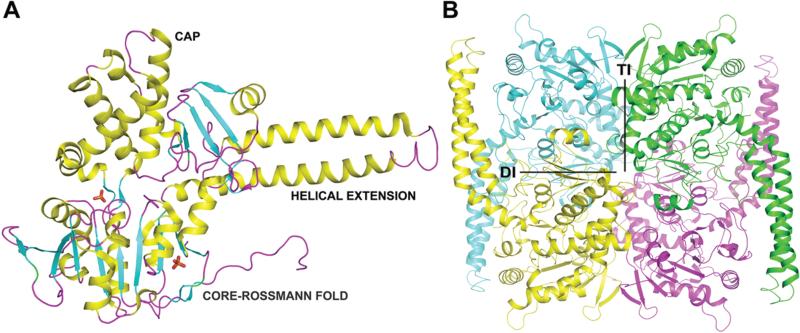
LpcN-II exhibits a two domain organization with a core domain and a cap domain. The core (D2 - R30, K183 - E325, D424 - I459) is an α/β-domain containing an eight-stranded parallel β-sheet surrounded by seven α-helixes, characteristic of the α/β Rossmann fold (Figure 4A). The cap domain (Y31- K182, N403-V423) has a mixed α/β organization with eight α-helices and seven β-strands clustering separately (PDBsum). There is a helical extension of approximately 76 residues (from E326 to Y402) characteristic of cN-II nucleotidases. There are two distinct interfaces constituting the LpcN-II tetramer, the dimeric interface (DI) and the tetrameric interface (TI) (Figure 4B). The subunits are related by 180° rotations about DI and TI. The interface across non-adjacent subunits makes 19 contacts at 4 Å cutoff involving 5 residues (K77, R110, N120, Q154 and Q158).
Figure 4.
PO4-complexed monomeric (A) and tetrameric (B) LpcN-II structures. In panel A, the respective domains have been marked as CAP, CORE-ROSSMANN FOLD and HELICAL EXTENSION. In panel B, DI represents dimer interface and TI, the-tetramer interface. All structure figures were generated with PyMOL (DeLano Scientific LLC).
Active site of LpcN-II
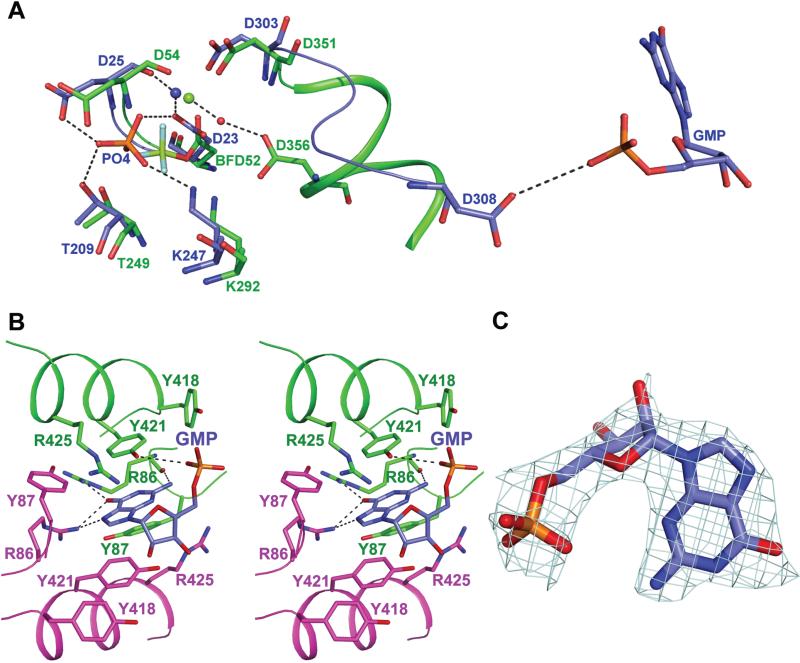
The GMP-complexed LpcN-II structure has a phosphate ion coordinated to Mg2+ present in the active site, located between the core and the cap domains (Figure 5A). Electron density for Mg2+ is absent in A subunit while electron density for phosphate is seen in all 4 subunits even at 6σ. The four catalytic motifs, motif I (DXDX [T/V]), motif II-([T/S), motif III ([K/R]) and motif IV (DX0-4D), characteristic of HAD superfamily enzymes [17], are conserved in LpcN-II. The contacts of phosphate with active site residues Asp23 and Asp25 (motif I), Thr209 (motif II), Lys247 (motif III) and Asp303 (motif IV) are largely similar across subunits.
Figure 5.
Active site and effector binding site in LpcN-II. (A) Active site of GMP-complexed LpcN-II structure (blue) superposed with human BeF3-complexed structure (green, PDB ID 2JCM). Shown also is Motif IV in ribbon format and GMP (shown in CPK color) in the effector site of LpcN-II. BFD52 represents BeF3 covalently linked to D52 in the human structure. Dashed lines that indicate hydrogen bonds are shown only for LpcN-II except for the water-mediated hydrogen bond from Asp356 to Mg2+(green sphere) from the human cN-II structure. Water molecule is shown as red sphere. Mg2+ from the LpcN-II structure is shown as blue sphere. (B) Residues in the activator-binding site that make contact with GMP. GMP is colored in blue for carbon atoms, and residues from the two monomers are colored in green and magenta. Dashed lines indicate hydrogen bonds. (C) The electron density for GMP from omit 2Fo-Fc map seen at 1 σ.
Effector binding site
The effector binding site, located at the dimer interfaces (DI), could be seen in the GMP-bound structure (Figure 5B), in which there is sufficient electron density for only GMP moiety (Figure 5C) of GMP-PNP despite soaking the crystals in fresh GMP-PNP and MgCl2 prior to data collection. Moreover, while the electron density for the α-phosphate and the ribose of GMP-PNP were poor, the guanine group could be clearly visualized. Despite the presence of Mg2+ in the crystallization buffer, absence of Mg at the effector binding site could have contributed to disorder of beta and gamma phosphates of GMP-PNP. In the structure of SRP GTPase complexed to GMP-PNP, it has been suggested that an increase in the temperature factor values for β/γ phosphates is due to the absence of Mg2+ ions [18]. Further, in the ATP/IMP-complexed human cN-II structure, the β/γ phosphates that are well ordered are coordinated to Mg2+ [14].
The effector site consists of residues Arg86, Tyr87 and Tyr421 from both subunits and Asp308 and Arg425 from only one subunit of the dimer (Figure 5B). Tyr421 and Tyr87 stack above and below the purine ring of GMP. Arg86 from both subunits contacts O6 of GMP. The corresponding residue in human enzyme is Ala114. Interactions of O6 with Arg86 may be the key determinant of the binding specificity of ITP and GTP over ATP. The N2 amino group of GMP is involved in forming a hydrogen bond with backbone O of Arg86. This interaction would be absent in ITP as it lacks the C2 amino group. These interactions may possibly be the reason why guanine nucleotides both bind and activate LpcN-II, while ATP, the human cN-II activator, does not. Most importantly, the site of GMP binding is ~25 Å away from the catalytic aspartate of the active site reinforcing the observation from kinetics that the substrate also binds to a site distal from the active site to bring about activation.
Validation of effector binding site by mutagenesis
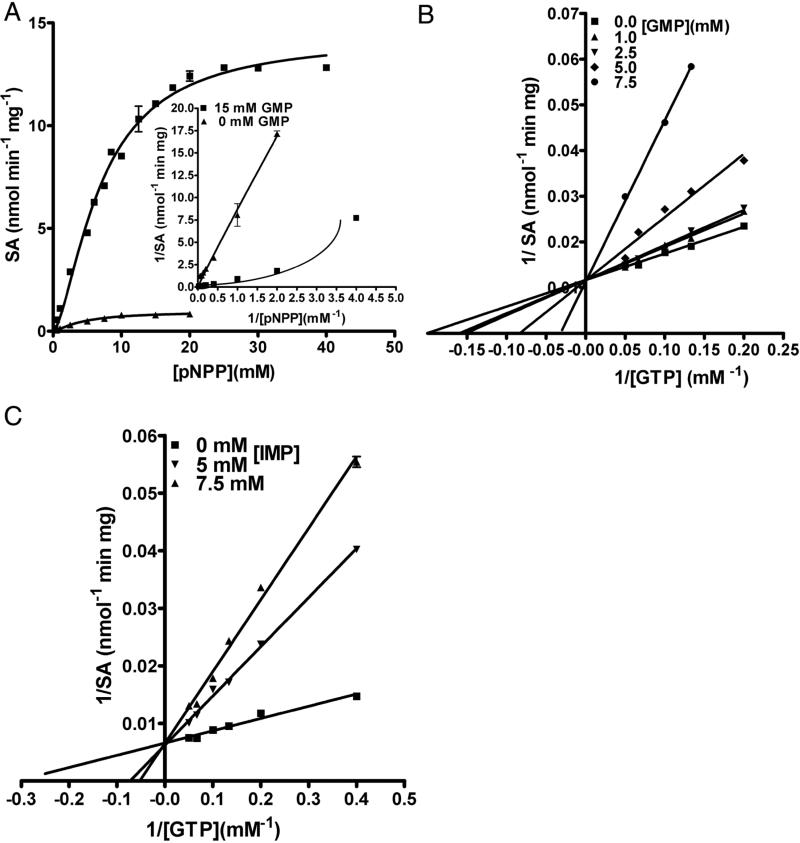
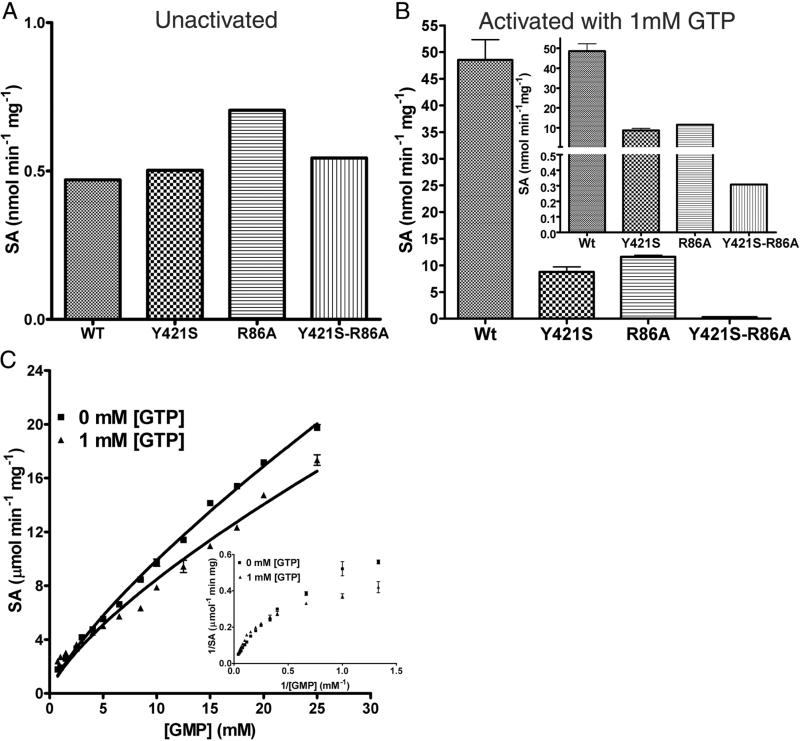
Given the poor electron density for the ligand molecule in the GMP-complexed LpcN-II structure, and given that the mode of activator binding in the human and Legionella enzymes are different, it was important to validate the ligand binding site by site-directed mutagenesis. Based on contact analysis, two residues viz., Y421 and R86, were selected and mutated to their respective human counterparts, Ser and Ala, respectively. A double mutant Y421S/R86A was also generated. The mutants were purified to homogeneity and found to elute as tetramers on analytical size-exclusion chromatography indicating that introduction of mutation/s at the interface had not perturbed their oligomeric status (Figure S4A and B). Though the single and double mutants displayed the same level of basal activity as the wild-type for the substrate pNPP, activation of pNPP hydrolysis by GTP was compromised in the case of the single mutants (Y421S and R86A) and abolished in the case of the double mutant (Y421S/R86A) (Figure 6A and B). ATP was still unable to activate the mutant enzyme. Further, the double mutant displayed biphasic kinetics and non-linear double reciprocal plots (indicative of negative cooperativity) for GMP hydrolysis both in the presence and absence of the heterotropic activator GTP (Figure 6C and inset). These kinetic features, taken together, confirmed the activator-binding site and also supported the substrate activation model, wherein substrate binds to an alternative site on the enzyme and allosterically modulates its affinity for the active site. It should be noted that the initial velocity curves for IMP were also biphasic (Figure 1C) given that it's binding to the allosteric site (reduces the activation of pNPP hydrolysis by GTP, Figure 3C) does not activate the enzyme.
Figure 6.
Activity of activator-site mutants of LpcN-II. (A) Activity of wild type (wt), Y421S, R86A and double mutant Y421S/R86A on pNPP. Note that the unactivated wild type and various mutants display comparable activity for the substrate pNPP. (B) Activity of the activated wild-type and mutants for pNPP hydrolysis at 1 mM GTP. The inset shows split y-axis to see the activity of double mutant. Note that as compared to the wild-type enzyme, GTP-activation of the single mutants is compromised and abolished for the double mutant. (C) Biphasic initial velocity versus [GMP] plots for the double mutant Y421S/R86A in the absence and presence of 1mM GTP. Inset shows the non-linear double reciprocal plot characteristic of negative cooperativity.
b) Structure alteration upon activator binding and probable mode of activation
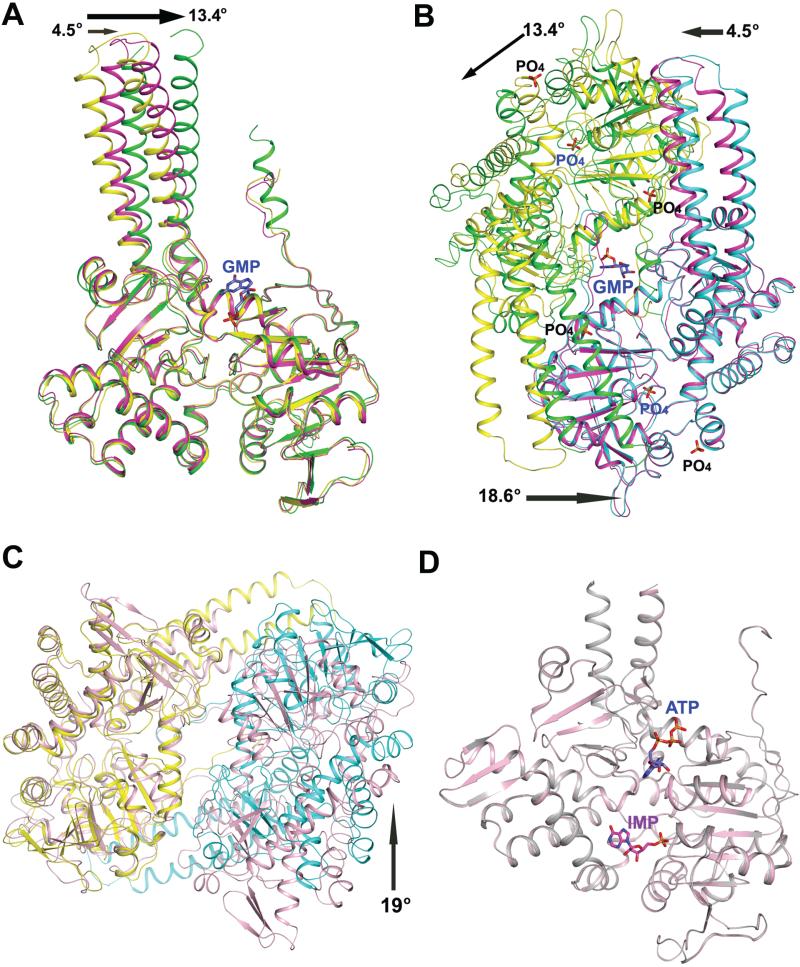
When the Cα trace (excluding the residues 339-397 and 450-462, constituting the helical extension and the carboxy terminus of the protein, respectively) of the monomers of the PO4-complexed and GMP-complexed structures from LpcN-II were superposed, an RMSD of 0.84 Å was obtained. Though the core and the cap domains of the two structures superposed well, the helical extension showed a rotation in the GMP-complexed structure vis-a-vis the PO4-complexed structure. Moreover, this rotation of the helical extension, respectively, was 13.4° and 4.5° for subunit A (green in Figure 7A) and B (magenta in Figure 7A) of the GMP-complexed dimer with respect to that of PO4-complexed structure (yellow in Figure 7A). This asymmetry in rotation is a reflection of the asymmetry of the ligand bound at the symmetric dimeric interface. Further structural alterations became obvious on comparison of the dimer organization of the PO4-complexed and GMP-complexed LpcN-IIs (Figure 7B), wherein significant differences in terms of buried surface area and number of salt bridges at subunit interfaces were observed. The PO4-complexed structure buries 4,696 Å2 at the dimer interface and contains 6 salt bridges, whereas the equivalent buried surface area at the dimer interface of the GMP-complexed structure is 5,985 Å2 and contains 12 salt bridges [19]. This tightly associated reorganization of the LpcN-II oligomer brought about by GMP binding to the effector site could serve as a structural basis for allosteric activation.
Figure 7.
Structural changes upon activator binding in human cN-II and LpcN-II. (A) Superposition of the PO4-complexed subunit A (yellow) and subunit A (green) and subunit B (magenta) of the GMP-complexed LpcN-II structures. The rotation (in °) of the helical extension in each subunit of the GMP-complexed structure relative to that of the PO4-complexed structure is shown by an arrow. (B) Superposition of the PO4-complexed subunit B (cyan) and the GMP-complexed subunit B (magenta) of LpcN-II, showing the activator binding site and the rearrangement of subunit A (green) in the GMP-complexed structure relative to the PO4-complexed structure (yellow). The activator binds in a groove at the dimer interface. The rotation (ino) of the helical extension in each subunit and the core of the GMP-complexed structure relative to the position of each subunit of the PO4-complexed structure is shown by an arrow. (C) Superposition of the human apo dimer structure (pink, PDB ID 2XCX) and LpcN-II PO4-complexed (yellow/cyan) dimer structure (PDB ID 4G63). Only subunit A from each structure (yellow in PO4-complexedstructure ) was included in the structural alignment. The arrow indicates a rotation (in o) of the subunit B of the PO4-complexed LpcN-II structure with respect to human apo structure. (D) Structural alignment of human cN-II apo form (in pink, PDB ID 2J2C) and in complex with IMP and ATP (in gray, PDB ID 2XCW).
Examination of the structure shows the presence of connectivity between the effector and active sites through the segment Arg70 to Tyr87 that forms part of a β-hairpin structure with the guanidinium group of Arg70 making contact with the catalytic Asp303 and His304 of motif IV. This entire segment is part of the cap domain and bridges the active site and the activator site (Figure S5A). This 17 residue peptide segment, when examined across cN-IIs from different species shows significant degree of conservation (Figure S5B), suggesting its possible role in modulating allostery in other cN-IIs also. Further, residues 303 to 308 that are part of motif IV form connectivity between the effector binding site and active site which is established by an Asp308-GMP (PO4 group) contact at one end of motif IV in the effector site and Asp303-Mg2+ contact at the active site. This connectivity is seen only in one subunit of the dimer since a single effector molecule is present at the dimeric interface.
Comparision of human cN-II structure with LpcN-II
Phylogenetic analysis of cN-II sequences from different kingdoms of life shows that LpcN-II is distantly related to mammalian cN-IIs (Figure S6A). Prokaryotic cN-IIs lack the carboxy terminal acidic stretch (made of polyaspartates or polyglutamates) that is found in cN-II sequences from the animal kingdom (Figure S6B). This acidic stretch in mammalian cN-IIs has been implicated in mediating enzyme aggregation and activation [12]. The human and L. pneumophila cN-IIs share 32% sequence identity and superposition of the PO4-complexed LpcN-II and human cN-II monomeric structures shows an RMSD of 1.3 Å. However, superposition of the two dimers leads to an increased RMSD of 1.9 Å indicating significant differences in the dimeric organization between human and LpcN-II structures (Figure 7C). Two key differences between the structures are the presence of approximately 30 additional residues at the amino terminus of the human protein that do not have a counterpart in the Legionella enzyme and the absence of the stretch from N174-F191 (human numbering) in the Legionella enzyme (Legionella numbering T145-M150). This segment, only in the human structure, makes extensive contacts at the tetramer interface. In addition, the LpcN-II structure has contacts across non-adjacent sub-units that are absent in the human structure. Further, a substantial part (approximately 18 residues) of the helical extension is not modeled in the human cN-II structure.
The active site residues seen in LpcN-II are conserved in human cN-II and other HADs. In the BeF3 complexed human cN-II structure, the side chain of Asp52 is covalently bonded to BeF3, a phosphate mimic. The side-chain of the corresponding residue, Asp 23 in LpcN-II has a different orientation in the active site of the GMP-complexed structure (Figure 5A). A notable exception is the position of Asp308 in LpcN-II, which is significantly away from the active site, while in the various nucleotide and BeF3-complexed human cN-II structures, the corresponding residue, Asp356, contacts Mg2+ at the active site through a water mediated hydrogen bond.
Comparison of the effector conformation and the binding site shows that LpcN-II has novel features that are distinct from human cN-II. LpcN-II has one ligand at each dimer interface leading to a stoichiometry of 2 protein subunits to 1 nucleotide. This is in stark contrast to ATP-complexed human cN-II where 2 ligand molecules bridged by a single Mg2+ are seen at the dimer interface. The conformation of GMP is syn in LpcN-II while the ATP is anti in human cN-II. Syn conformation of the guanine nucleotide is seen in the structure of GDP-fucose complexed to MUR1 GDP-Mannose 4,6-dehydratase from Arabidopsis thaliana [20].
The residues contacting adenosine [13], ATP, AP4A and 2,3-BPG in the various complexed human cN-II structures [14] are not conserved in LpcN-II. Comparison of the residues contacting the ligand in the GMP-complexed LpcN-II structure with their human counterpart shows that only Tyr87 of LpcN-II is conserved as Tyr115 in the human enzyme while Tyr421 and Arg86 of LpcN-II correspond to Ser 452 and Ala114, respectively, in the human enzyme.
Though human cN-II does not show a change in subunit organization upon effector binding (Figure 7D), the segment corresponding to motif IV referred to as helix A undergoes a disorder to order transition upon ligand binding. The ordering of this helix results in repositioning of Asp356 such that it contacts the active site Mg2+ through a water bridge. Though this ordering has been suggested to be induced only upon activator binding, BeF3-complexed structure (PDB ID 2JCM) lacking the activator also has the helix ordered and Asp356 repositioned. A distinct difference from the human activator bound structure is the absence of Asp308 movement to the active site upon effector binding in LpcN-II. The conformation of motif 4 in LpcN-II remains undisturbed upon effector binding. With the observation of these differences between human and LpcN-II, the role of Asp308 in activation needs examination by mutagenesis. Finally, it should be noted that while LpcN-II is full length and active, the human cN-II clone studied has a truncation at the C-terminus resulting in almost complete loss of activity [12].
Discussion
Nucleotides are components of several vital cellular processes and enzymes involved in nucleotide metabolism are tightly regulated to ensure homeostasis. The cN-II class of 5'-nucleotidases from mammalian sources play several roles essential in oxypurine cycling, nucleotide monophosphate degradation, maintanance of NMP pools through their phosphotransferase reaction [2] and, hence, are tightly regulated allosterically. The current study throws light on multiple regulatory mechanisms of LpcN-II, viz., substrate activation for substrates GMP and dGMP, negative homotropic cooperativity for substrate IMP and heterotropic allosteric activation by GTP and GDP.
LpcN-II is allosterically activated by guanine nucleotides with activation by GTP being ‘K’ type for GMP hydrolysis and ‘V’ type for pNPP hydrolysis. Though at this stage, the molecular basis for the difference is not clear, it should be noted that the two substrates, GMP and pNPP differ in their molecular size and structure. Variation in mechanisms of activation with different substrates has also been reported in the case of rat brain IMP-GMP specific 5’ nucleotidase that is allosterically activated by Ap4A. This activator displays a mixed-type mechanism with IMP as substrate with increase in Vmax and decrease in Km and an exclusive K-type mechanism on the substrates GMP, AMP and XMP [7-11, 21, 22]. Unexpectedly, the ‘K’ type activation in LpcN-II has not resulted in a large drop in Km value for GMP, the reason for which is due to GMP partitioning to both the activator and active site. Hence, a true reduction in Km value in the presence of GTP could not be estimated and the experimentally derived Km is an ‘apparent’ value.
Activator binding at the dimeric interface brings about structural alterations as is evident from our crystallographic studies. These alterations were also evident in solution studies of LpcN-II, wherein partial trypsin proteolysis and CD spectroscopy were used as probes on the complexed and apo enzymes (data not shown). On the other hand, ligand binding did not confer any thermodynamic stability to the protein as seen in urea and temperature denaturation studies (data not shown). Feedback inhibition by a downstream metabolite of an upstream anabolic enzyme to control the flux through a pathway is a phenomenon well documented in literature[23] while feedback activation, wherein a downstream metabolite activates an upstream catabolic enzyme, is sparsely mentioned. In the case of LpcN-II, we present a case of feedback activation by GTP to deplete a precursor (GMP) and keep its level in control.
The promiscuous nature of the activator binding site, whereby it can accommodate all guanine nucleotides, raises further interesting aspects. By far, the most interesting of these interactions is that by the substrate GMP activating its own hydrolysis. Based upon observations that GMP activates LpcN-II's pNPP hydrolysis and competitively reduces activation of pNPP hydrolysis by GTP, we surmise that substrate GMP binds to the allosteric GTP binding site. Further, we propose that the sigmoidal nature of initial velocity versus [GMP] plots arises from GMP partitioning to both active and allosteric sites. Further, the crystallographic data which shows GMP density at the interfacial site provides unequivocal structural evidence for GMP mediated activation. The high S0.5 values obtained for substrate GMP and dGMP might be a reflection on the fact that substrate is partitioning between the allosteric activator binding site and the active site. Substrate activation is a very rare phenomenon with only a few examples present of enzymes getting activated by substrates binding to a site distal from the active site [24, 25]. The double mutant Y421S/R86A, wherein activation of pNPP hydrolysis by GTP is completely abolished, also shows abolishment of activation of pNPP hydrolysis by GMP (data not shown) validating that both GMP and GTP bind to the same allosteric activator site. Substrate activation by GMP of its own hydrolysis in LpcN-II, a catabolic enzyme, can be understood as a mechanism akin to inhibition of an anabolic enzyme by the product of its reaction.
The IMP titration curves for the wild-type LpcN-II displays biphasic kinetics indicative of negative cooperativity whereby substrate binding to one active site reduces the affinity of the substrate for the adjacent active site. This shows that substrate binding to the activator binding site and subsequent activation is mandatory for the initial velocity versus [S] plots to show saturation. It should be recalled that though IMP binds to the activator-binding site (Fig. 3C), it does not activate the pNPP hydrolysis activity of LpcN-II. Further, upon abolishing the activator binding site in the double-mutant Y421S/R86A, the initial velocity versus [GMP] curve also showed biphasic kinetics. All the above results lead us to speculate that the biphasic behavior seen in the initial velocity plots for IMP is switched to a sigmoidal response curve in GMP plots as the latter substrate is also capable of bringing about allosteric activation. This behavior is interesting from the perspective that the catabolic role of the enzyme might be directed towards depleting GMP/IMP pools depending on the levels of guanine nucleotides.
In conclusion, we have shown that LpcN-II is regulated at multiple levels. Biochemical and structural evidence suggests that the prokaryotic enzyme is modulated differently from its mammalian counterpart. Moreover, we postulate that the structural demonstration of subunit reorganization upon activator binding is the probable mechanism for homotropic and heterotropic activation by substrate and the activator, respectively.
Experimental procedures
Reagents
All reagents and chemicals, unless mentioned otherwise, were of high quality and were procured from Sigma-Aldrich Co., USA or Merck & Co., USA. The purity of GTP, GDP and GMP were confirmed by HPLC. Gel filtration matrices were from Amersham Biosciences, UK. Media components were from Himedia, Mumbai, India. Macrosep centrifugal devices were from Pall Co., USA. Crystallization cocktail buffers were from Hampton Research, USA.
Bioinformatics analysis
The sequence of LpcN-II was obtained from NCBI. The non-redundant database at NCBI was used to search for homologs of LpcN-II using the algorithm BLASTP. Distant homology searches were carried out using PSI-BLAST [26]. T-Coffee[27] was used for generating multiple sequence alignment profiles. Phylogenetic and molecular evolutionary analyses were done using MEGA, version 3.1[28]. Molecular visualization and structure analysis were done using various tools like SPDBV[29], PyMOL (pymol.sourceforge.net) and CCP4 suite of programs [30].
Cloning, expression and purification of wild-type LpcN-II
The gene expressing the protein was cloned in a pET21 derivative, expressed by transforming in E. coli BL21 (DE3) pMgK cells and purified by immobilized nickel affinity chromatography. The detailed protocol is given in supplemental methods.
The oligomeric status of the protein was probed by size exclusion chromatography on a pre-calibrated analytical superdex-200 column (See supplemental methods for detailed protocol).
Construction of Y421S, R86A and Y421S/R86A LpcN-II mutants
Site directed mutants of the wild type LpcN-II were generated by the quick change PCR method using a single mutagenic oligonucleotide primer [31]. Primers 5' AAT ATT TTA AAA CTG AGC GCA TAT GGT GCC ATT AGA TTG 3' and 5' GAG AGC TAC TTC GCT AGC CAA GTT GAT AGA TTC 3' were used for the generation of R86A and Y421S mutants, respectively, using wild-type gene as template. The double mutant Y421S/R86A was generated by using Y421S mutant LpcN-II as template. PCR product was digested with DpnI and transformed into XL-1 Blue strain of E. coli. The site of mutation was confirmed by restriction analysis for the introduced restriction site and by sequencing. The plasmid was isolated and transformed into E. coli XL1Blue strain for maintenance and in E. coli Rosetta (DE3) pLysS for expression studies with ampicillin and chloramphenicol as selection markers. Conditions for expression and purification of all three mutant enzymes were kept same as that for the wild-type enzyme.
Activity Assays
Phosphate ester hydrolysis of various nucleotide monophosphates by LpcN-II was monitored using Chen's assay [32] with slight modifications. pNPP hydrolysis assays were done in 50 mM Tris HCl, pH 8.0 in a final reaction volume of 250 μl using a double beam Hitachi UV 2010 (Hitachi High Technologies America, Inc., San Jose, CA, USA) spectrophotometer. The details of the assay are provided in supplemental methods.
Screening for activators of LpcN-II activity
Various activators were screened to assess their effect on pNPP hydrolyzing activity of LpcN-II. The detailed protocol and the metabolites tested are summarized in supplemental methods and Table S2.
Determination of the dissociation constant (KD) of [Activator-Mg2+] complexes of LpcN-II
The KD of GTP-Mg2+-enzyme complex, dGTP-Mg2+-enzyme complex and GDP-Mg2+-enzyme complex was determined kinetically (under catalytic conditions) while that of GTP-Mg2+-enzyme complex alone was also determined under conditions of equilibrium with radiolabeled GTP. The details of the protocol are specified in supplemental methods.
Kinetics of GTP activation
(a) Activation of GMP hydrolysis by GTP- Activation of GMP hydrolysis by GTP was monitored by IP-RP-HPLC [33] and Chen's assay. The details of the protocol are specified in supplemental methods. (b) Activation of pNPP hydrolysis by GTP- Activation of pNPP hydrolysis by GTP was monitored by performing activation kinetics. The details of the protocol are specified in supplemental methods.
Crystallization and data collection
Summary of crystallization and data collection strategies for the PO4- complexed and GMP-PNP-complexed LpcN-II are provided in supplemental methods and Table S5. Data collection and model building statistics are as summarized in Table 2.
Table 2.
Crystallization, data collection and refinement parameters for the PO4-complexed and GMP-complexed crystal forms of LpcN-II.
| PO4-complexed LpcN-II | GMP-complexed LpcN-II | |
|---|---|---|
| Resolution | 20-2.70 | 44.70-2.53 |
| No. of reflections | 29199 | 71361 |
| No. of reflections for Rfree | 2920 | 6753 |
| Rvalue | 0.222 | 0.231 |
| Rfree | 0.258 | 0.272 |
| RMS deviation-Bond length | 0.007 | 0.007 |
| RMS deviation-Bond angle | 1.4 | 1.4 |
| Mean B-value (overall) | 61.7 | 54.3 |
| B-value from Wilson plot | 60.1 | 60.2 |
| % residues (most favourable) | 90.6 | 89.9 |
| % residues (additionally allowed) | 8.0 | 9.2 |
| % residues (generously allowed) | 1.4 | 0.9 |
| % residues (disallowed) | 0.0 | 0.0 |
| PDB ID | 4G63 | 4OHF |
Phasing, Model building and refinement
The diffraction images of both SO4- and PO4- complexed LpcN-II were processed with the HKL package [34] and the selenium sites of SO4- complexed LpcN-II were located with the program SnB [35]. SOLVE/RESOLVE [36] was used for phasing the reflections and automated model building, which correctly placed 15 % of the residues with side chains. The majority of the model was built manually with the program XtalView [37]. Structure refinement was performed with CNS [38]. The model of the SO4- complexed structure (PDB entry: 2BDE) was subsequently used to determine the structure of PO4-complex using the molecular replacement method with the program COMO [39]. The data processing and refinement statistics are summarized in Table 2.
Structure of SO4-complexed LpcN-II was also used as the search model for structure solution of GMP-PNP-complexed LpcN-II by molecular replacement with the program MOLREP [40]. Reflections in the resolution range 44.7-2.53 Å of this crystal form were used for the rotation and translation searches. The highest peak of the translation search had a correlation coefficient of 45.311 % and an R-factor of 51.035 %. The structure of the GMP-PNP-complexed cN-II was solved with four polypeptides per asymmetric unit. Refinement was initiated with the program CNS 1.3. The 2Fo–Fc map calculated after initial positional refinement showed reasonable fit for the majority of the main chain atoms except for those that belonged to the region comprising residues 360-400. Subsequent rounds of model building and refinement were carried out using the programs XtalView [37] and CNS in which specification of NCS restraints helped reduce both the R factor and Rfree of the model considerably. Inspection of difference (Fo−Fc) map showed positive electron density for the GMP moiety of GMP-PNP. Potential sites for solvent molecules were identified both manually and by automatic water-picking algorithm of CNS 1.3. The structure was refined to a final R/Rfree 0.23/0.27 to resolution 2.53 Å. Refinement statistics are as specified in Table 2.
Supplementary Material
Acknowledgements
We would like to acknowledge the X-ray crystallographic facility at the Molecular biophysics unit, Indian institute of science, Bangalore for providing machine time to facilitate the collection of data. We would also like to thank Dr. Kiran Kulkarni for assistance with structure solution. We thank Angela Lauricella and George DeTitta for setting up initial crystal screenings; Randy Abramowitz and John Schwanof for setting up the X4A beamline. Research at the NESG was supported by the Protein Structure Initiative of the National Institutes of Health (U54 GM074958 and U54 GM094597).
The work was supported by funding from Department of Biotechnology and Department of Science and Technology, New Delhi, India. BS was supported by CSIR (Council for Scientific and Industrial Research) senior research fellowship, CS was supported by DBT-Postdoctoral fellowship, AS was supported by CSIR junior research fellowship. BS and FF have contributed equally to the work.
Abbreviations
- LpcN-II
Legionella pneumophila cytosolic nucleotidase II
- ISN1
IMP-specific 5’-nucleotidase
- IP-RP-HPLC
ion pair reverse phase HPLC
- pNPP
p-Nitrophenyl phosphate
- 2,3-BPG
2,3-Bisphosphoglyceric acid
Footnotes
Accession numbers
The PDB accession numbers for the coordinates and structure factors reported in this paper are 2BDE and 4G63. The accession number of GMP complexed LpcN-II is 4OHF.
References
- 1.Zimmermann H. 5′-Nucleotidase: molecular structure and functional aspects. The Biochemical journal. 1992;285(Pt 2):345–65. doi: 10.1042/bj2850345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunsucker SA, Mitchell BS, Spychala J. The 5′-nucleotidases as regulators of nucleotide and drug metabolism. Pharmacology & therapeutics. 2005;107:1–30. doi: 10.1016/j.pharmthera.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Itoh R, Saint-Marc C, Chaignepain S, Katahira R, Schmitter JM, Daignan-Fornier B. The yeast ISN1 (YOR155c) gene encodes a new type of IMP-specific 5′-nucleotidase. BMC biochemistry. 2003;4:4. doi: 10.1186/1471-2091-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srinivasan B, Balaram H. ISN1 nucleotidases and HAD superfamily protein fold: in silico sequence and structure analysis. In silico biology. 2007;7:187–93. [PubMed] [Google Scholar]
- 5.Bianchi V, Spychala J. Mammalian 5′-nucleotidases. The Journal of biological chemistry. 2003;278:46195–8. doi: 10.1074/jbc.R300032200. [DOI] [PubMed] [Google Scholar]
- 6.Barsotti C, Pesi R, Felice F, Ipata PL. The purine nucleoside cycle in cell-free extracts of rat brain: evidence for the occurrence of an inosine and a guanosine cycle with distinct metabolic roles. Cellular and molecular life sciences : CMLS. 2003;60:786–93. doi: 10.1007/s00018-003-2371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spychala J, Madrid-Marina V, Fox IH. High Km soluble 5′-nucleotidase from human placenta. Properties and allosteric regulation by IMP and ATP. The Journal of biological chemistry. 1988;263:18759–65. [PubMed] [Google Scholar]
- 8.Bontemps F, Van den Berghe G, Hers HG. 5′-Nucleotidase activities in human erythrocytes. Identification of a purine 5′-nucleotidase stimulated by ATP and glycerate 2,3-bisphosphate. The Biochemical journal. 1988;250:687–96. doi: 10.1042/bj2500687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bontemps F, Vincent MF, Van den Bergh F, van Waeg G, Van den Berghe G. Stimulation by glycerate 2,3-bisphosphate: a common property of cytosolic IMP-GMP 5′-nucleotidase in rat and human tissues. Biochimica et biophysica acta. 1989;997:131–4. doi: 10.1016/0167-4838(89)90144-1. [DOI] [PubMed] [Google Scholar]
- 10.Marques AF, Teixeira NA, Gambaretto C, Sillero A, Sillero MA. IMP-GMP 5′-nucleotidase from rat brain: activation by polyphosphates. Journal of neurochemistry. 1998;71:1241–50. doi: 10.1046/j.1471-4159.1998.71031241.x. [DOI] [PubMed] [Google Scholar]
- 11.Pinto RM, Canales J, Gunther Sillero MA, Sillero A. Diadenosine tetraphosphate activates cytosol 5′-nucleotidase. Biochemical and biophysical research communications. 1986;138:261–7. doi: 10.1016/0006-291x(86)90274-3. [DOI] [PubMed] [Google Scholar]
- 12.Spychala J, Chen V, Oka J, Mitchell BS. ATP and phosphate reciprocally affect subunit association of human recombinant High Km 5′-nucleotidase. Role for the C-terminal polyglutamic acid tract in subunit association and catalytic activity. European journal of biochemistry / FEBS. 1999;259:851–8. doi: 10.1046/j.1432-1327.1999.00099.x. [DOI] [PubMed] [Google Scholar]
- 13.Wallden K, Stenmark P, Nyman T, Flodin S, Graslund S, Loppnau P, Bianchi V, Nordlund P. Crystal structure of human cytosolic 5′-nucleotidase II: insights into allosteric regulation and substrate recognition. The Journal of biological chemistry. 2007;282:17828–36. doi: 10.1074/jbc.M700917200. [DOI] [PubMed] [Google Scholar]
- 14.Wallden K, Nordlund P. Structural basis for the allosteric regulation and substrate recognition of human cytosolic 5′-nucleotidase II. Journal of molecular biology. 2011;408:684–96. doi: 10.1016/j.jmb.2011.02.059. [DOI] [PubMed] [Google Scholar]
- 15.Careddu MG, Allegrini S, Pesi R, Camici M, Garcia-Gil M, Tozzi MG. Knockdown of cytosolic 5′-nucleotidase II (cN-II) reveals that its activity is essential for survival in astrocytoma cells. Biochimica et biophysica acta. 2008;1783:1529–35. doi: 10.1016/j.bbamcr.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. Journal of molecular biology. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 17.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. Journal of molecular biology. 2006;361:1003–34. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 18.Padmanabhan S, Freymann DM. The conformation of bound GMPPNP suggests a mechanism for gating the active site of the SRP GTPase. Structure. 2001;9:859–67. doi: 10.1016/s0969-2126(01)00641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costantini S, Colonna G, Facchiano AM. ESBRI: a web server for evaluating salt bridges in proteins. Bioinformation. 2008;3:137–8. doi: 10.6026/97320630003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulichak AM, Bonin CP, Reiter WD, Garavito RM. Structure of the MUR1 GDP-mannose 4,6-dehydratase from Arabidopsis thaliana: implications for ligand binding and specificity. Biochemistry. 2002;41:15578–89. doi: 10.1021/bi0266683. [DOI] [PubMed] [Google Scholar]
- 21.Le Hir M. A soluble 5′-nucleotidase in rat kidney. Stimulation by decavanadate. The Biochemical journal. 1991;273(Pt 3):795–8. doi: 10.1042/bj2730795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bretonnet AS, Jordheim LP, Dumontet C, Lancelin JM. Regulation and activity of cytosolic 5′-nucleotidase II. A bifunctional allosteric enzyme of the Haloacid Dehalogenase superfamily involved in cellular metabolism. FEBS letters. 2005;579:3363–8. doi: 10.1016/j.febslet.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Gerhart JC, Pardee AB. The enzymology of control by feedback inhibition. The Journal of biological chemistry. 1962;237:891–6. [PubMed] [Google Scholar]
- 24.Konig S, Spinka M, Kutter S. Allosteric activation of pyruvate decarboxylase. A never-ending story? Journal of Molecular catalysis B enzymology. 2009;61:100–110. [Google Scholar]
- 25.Lu G, Dobritzsch D, Baumann S, Schneider G, Konig S. The structural basis of substrate activation in yeast pyruvate decarboxylase. A crystallographic and kinetic study. European journal of biochemistry / FEBS. 2000;267:861–8. doi: 10.1046/j.1432-1327.2000.01070.x. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of molecular biology. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in bioinformatics. 2004;5:150–63. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 29.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography, Acta crystallographica Section D. Biological crystallography. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Shenoy AR, Visweswariah SS. Site-directed mutagenesis using a single mutagenic oligonucleotide and DpnI digestion of template DNA. Analytical biochemistry. 2003;319:335–6. doi: 10.1016/s0003-2697(03)00286-0. [DOI] [PubMed] [Google Scholar]
- 32.Chen PS, Jr., Toribara TY, Huber W. Microdetermination of Phosphorous. Analytical chemistry. 1956;28:1756–1758. [Google Scholar]
- 33.Werner A, Schneider W, Siems W, Grune T, Schreiter C. Ion-pair reversed phase HPLC determination of nucleotides, nucleosides and nucleobases-application to nucleotide metabolism in hepatocytes. Chromatographia. 1989;27:639–643. [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Weeks CM, Miller R. The design and implementation of SnB v2.0. J Appl Cryst. 1999;32:120–124. [Google Scholar]
- 36.Terwilliger TC. SOLVE and RESOLVE: Automated structure solution and density modification. Meth Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 37.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. Journal of structural biology. 1999;125:156–65. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 38.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.Jogl G, Tao X, Xu Y, Tong L. COMO: a program for combined molecular replacement, Acta crystallographica Section D. Biological crystallography. 2001;57:1127–34. doi: 10.1107/s0907444901006783. [DOI] [PubMed] [Google Scholar]
- 40.Vagin AA, Teplyakov A. MOLREP: an automated program for molecular replacement. journal of applied crystallography. 1997;30:1022–1025. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.