Abstract
The transcriptional insulator CCCTC binding factor (CTCF)2 was shown previously to be critical for human MHC class II gene expression. Whether the mechanisms used by CTCF in humans was similar to that of the mouse and whether the three-dimensional chromatin architecture created was specific to B cells was not defined. Genome-wide CTCF occupancy was defined for murine B cells and LPS-derived plasmablasts by ChIP-seq. Fifteen CTCF sites within the murine MHC-II locus were associated with high CTCF binding in B cells. Only one third of these sites displayed significant CTCF occupancy in plasmablasts. CTCF was required for maximal MHC-II gene expression in mouse B cells. In B cells, a subset of the CTCF regions interacted with each other, creating a three-dimensional architecture for the locus. Additional interactions occurred between MHC-II promoters and the CTCF sites. In contrast, a novel configuration occurred in plasma cells, which do not express MHC-II genes. Ectopic CIITA expression in plasma cells to induce MHC-II expression resulted in high levels of MHC-II proteins, but did not alter the plasma cell architecture completely. These data suggest that reorganizing the three-dimensional chromatin architecture is an epigenetic mechanism that accompanies the silencing of MHC class II genes as part of the cell fate commitment of plasma cells.
INTRODUCTION
The murine MHC-II region spans approximately 250 kb on chromosome 17 (1, 2). In many murine haplotypes, the I-A and I-E alpha/beta heterodimeric MHC-II products are expressed on the surfaces of antigen presenting cells and function to present antigenic peptides to CD4 T lymphocytes for initiation and/or regulation of adaptive immune responses. However, due to a promoter region and first exon deletion, some haplotypes, such as the H-2b haplotype of the C57Bl/6 mouse, do not express the I-E alpha chain gene IEa (3), resulting in the expression of one functional MHC-II molecule. In addition to the classical MHC-II genes, other genes associated with antigen processing, such as those encoding H2-DM and H2-O, as well as some genes that function in MHC-I antigen presentation (Tap1, Tap2), are located in this locus (4).
Expression of MHC-II genes is cell type specific and regulated at the level of transcription [reviewed in (5-8)]. Within a few hundred bases from the transcriptional start sites of each of the MHC-II gene are conserved regions (W, X1, X2, and Y boxes) that are essential for MHC-II transcription (9, 10). A group of transcriptional factors, regulatory factor X (RFX) (11), cyclic AMP response element binding protein (CREB) (12), and nuclear factor Y (NFY) (13) bind constitutively and cooperatively to the X and Y boxes (14), respectively. Alone, these DNA binding factors are not sufficient to activate MHC-II gene expression. The transcriptional coactivator termed the MHC class II transactivator (CIITA) is also required (15). CIITA expression itself is tightly regulated and is the limiting factor governing whether these genes can be expressed or not. When expressed, CIITA is recruited to the RFX-CREB-NF-Y complex, and mediates further interactions between chromatin remodeling complexes and various components of the general transcription machinery (7, 16-19).
The mammalian insulator factor, CCCTC-binding factor (CTCF) is involved in formation of higher order chromatin structure in eukaryotes (20). When bound to DNA, CTCF functions to block the activity of enhancer elements from acting downstream and can insulate genes from heterochromatic silencing mechanisms (21). The ability to block enhancer activity from one region to another is thought to occur through CTCF's ability to form long-range chromatin loops with other sites bound by CTCF (22). The formation of such CTCF-mediated chromatin loops was particularly striking in the human MHC-II locus, where CTCF binding sites occurred at the boundaries between subregions encoding MHC-II α/β gene pairs (e.g., HLA-DR, HLA-DQ), and chromatin loops/interactions occurred between adjacent regions (23, 24). These CTCF binding site interactions were independent of the known MHC-II specific regulatory factors, including CIITA. RNAi mediated knockdown of CTCF in the human Raji B lymphoblastoid cell line resulted in decreased MHC-II gene expression and loss of the interactions between the CTCF sites, suggesting that these interactions were important for maximal MHC-II expression (Majumder et al., 2008). Thus, these sites were termed MHC-II insulators. MHC-II insulators also interacted with MHC-II promoter regions that were within ~200kb. This set of interactions was dependent on CIITA expression, suggesting that two chromatin architectural arrangements are set in the MHC-II locus of human B cells: one that is independent of expression (insulator-insulator) and a second that correlates with expression (insulator-promoter/CIITA).
To further establish a role and mechanisms of action for MHC-II insulators, the relationship between MHC-II gene expression in the mouse and the existence of MHC-II insulators was investigated. Advantage was taken of the ability to isolate primary cells and in vivo derived short-lived plasmablasts to determine if there were differences in how potential MHC-II insulators may function during this fundamental developmental stage. Plasma cell differentiation leads to the loss of both CIITA and MHC-II gene expression (25-27). ChIP-seq for CTCF demonstrated both similarities and significant differences in CTCF site occupancy occurred between the B cells and plasma cells. Notably, several CTCF binding sites identified in the I-A and I-E subregions in B cells exhibited reduced or absent CTCF binding in plasmablasts. Using chromatin conformation capture (3C) assays (28), interaction maps were generated for each of the murine CTCF-binding sites in B cells. A set of extensive interactions that defined the B cell and MHC-II expressing phenotype were observed, and included those between insulator elements, as well as interactions between the CTCF-binding sites and MHC-II gene promoters. 3C interaction profiles for a plasma cell line and in vivo derived plasmablasts displayed a distinct set of interactions when compared to B cells. Stable, ectopic expression of CIITA in a plasma cell line was used to activate MHC-II expression and potentially reprogram the architecture of the locus. Despite, the ability to generate high levels of MHC-II expression and induce interactions between MHC-II promoters and some CTCF sites, the three-dimensional architecture of the locus remained mostly in the plasma cell configuration. These results suggest that commitment to the plasma cell lineage is associated with architectural and epigenetic changes in the MHC-II locus. This novel architecture accompanies the silencing of MHC-II gene expression in this terminally differentiated cell type. Its potential contribution to this silencing is discussed.
MATERIALS AND METHODS
Mice, Primary B cell and Plasmablast Purification
C57BL/6 mice were purchased from The Jackson Laboratory. Mice were housed in the Emory University School of Medicine Facilities. All animal experiments were approved by the Emory University Institutional Animal Care and Use Committee. To obtain primary mouse B cells, spleens were isolated from 6 week-old C57BL/6 mice. Following homogenization, the CD43− B cell population was purified using a magnetic separation procedure according to the manufacturers recommendations (Miltenyi Biotec, Inc.). Purity of these preparations was verified by flow cytometry for B cell phenotypic markers using anti-B220-APC and anti-CD43-FITC. Mouse plasmablasts were obtained by injecting 50 μg LPS retro-oribitally into 6-week-old C57BL/6 mice. Three days post-injection, spleens were harvested and total splenocytes stained with anit-CD138-PE and anti-B220-APC. CD138+B220int plasmablasts and CD138+B220− plasma cells were sorted to high purity by FACS. Antibodies for FACS staining were purchased from BD Biosciences.
Cells, and cell culture
The murine P3X63Ag8 (CRL-1580; termed P3X herein) plasma cell line and A20 (TIB-208) B cell line were purchased from the American Type Tissue Collection. All murine cells were grown in RPMI 1640 medium (Mediatech, Inc.) supplemented with 10% heat-inactivated fetal bovine serum (SIGMA, Inc.), 10 mM HEPES (HyClone Laboratories), 1 mM sodium pyruvate (HyClone Laboratories), 1X non-essential amino acids (HyClone Laboratories), and 0.05 mM 2-ME (Sigma-Aldrich).
CIITA Plasmid construction
Double stranded oligonucleotides encoding a Kozak sequence, an N-terminal biotin ligase recognition site, a tobacco etch virus (TEV) site, and His 6 tag were digested with NheI and cloned into the pREP4 vector (Life technologies, Carlsbad CA) to form a base expression vector. For other purposes, an internal ribosome entry site (IRES) and the E. coli biotin ligase, BirA, were cloned into the XhoI and BamHI sites on the 3’ side of the expression cassette of the vector. The full-length cDNA sequence of CIITA isoform III was amplified using the Phusion™ High-Fidelity DNA Polymerase (New England BioLabs Inc., Ipswich, MA), and this CIITA PCR product was inserted into the NotI site of the vector using the In-Fusion™ Dry-Down PCR Cloning Kit (Clontech Laboratories Inc.) according to the manufacturers recommendations. Following the creation of this multi-tagged CIITA expression vector, the 747 bp sequence of the XL9 insulator (24) was cloned into the XbaI site present downstream of the polyadenylation signal of the CIITA-BirA coding sequence. Restriction enzyme digestion and DNA sequencing were used to verify the integrity of the construct.
Transfection and stable cells
The Amaxa® Nucleofector® kit V (Lonza, Switzerland) was used for transfection of P3X cells, and for each transfection, 4 × 106 P3X cells, and 4-5 μg of CsCl prepared DNA was used. For selection of successfully transfected cells, hygromycin (Millipore, Billerica MA) was added to the culture to a final concentration of 400 μg/ml 24 hours after transfection. Cells were grown in media supplemented with 400 μg/ml of hygromycin for 10 days until the selection was complete and then transferred to 100 μg/ml hygromycin to prevent loss of the episomal plasmid during prolonged maintenance of the stable cell lines.
Flow cytometry
For flow cytometry, the experimental and control cells were collected and washed with cold PBS and resuspended in FACS sorting buffer (1 × PBS, 1 mM EDTA, 1 % BSA, 0.2 μm filter sterilized). To measure the level of cell surface MHC-II expression, cells were labeled on ice with 1:300 dilution of PE conjugated anti-IA-E antibody (BD Pharmingen) and isotype control (BD Pharmingen) (0.5 ng/ml) was added to all cells. Prior to antibody staining, cells were incubated with anti-CD16/32 (FC block clone 2.4G2) on ice for 5 min. Stained cells were washed twice with FACS sorting buffer. A BD FACSCalibur™ flow cytometer was used for data collection and the FlowJo computer software (Tree Star, Inc., Ashland, OR) was used for analyses.
SDS-PAGE and immunoblot
SDS-PAGE and western blotting was performed following standard laboratory procedures to determine the levels of CTCF in B cells and plasma cells as indicated (29, 30). Anti-CTCF (Millipore) and anti-beta-actin (Chemicon) were diluted (1:1,000) in 5 % milk and incubated with the membrane at 4 °C overnight. Following washing, the membrane was incubated with HRP conjugated secondary goat anti-rabbit (Rockland Immunochemicals), rabbit anti-goat (Sigma-Alderich, St Louis MO) at 1:5,000 dilution for 1 hour at room temperature prior to autoradiography.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as described previously (16, 23, 24, 30). Cells were crosslinked with 1% formaldehyde for 10 m. For immunoprecipitations, 5-10 μg of anti-CTCF (Upstate, Cat# 06-917), or rabbit IgG (non-specific control) antibodies were used. Protein A beads (60 μl) were used to isolate the chromatin-antibody complexes. The DNA was purified and used as a template in real-time PCR reactions or for preparation of sequencing libraries. The immunoprecipitated DNA was quantitated by real-time PCR using a 5-point genomic DNA standard curve and an I-cycler (Biorad Laboratories, Inc.). Sequences for all primers used in the ChIP real-time PCR assays are listed in Supplemental Table 2. All ChIP experiments were performed at least three times from independent preparations of chromatin. The data were averaged and plotted with respect to the input chromatin.
ChIP-seq
Sequencing libraries were prepared from CTCF ChIP and input DNA from the indicated samples according to the manufacturers instructions (Illumina, Inc). Briefly, DNA was end-repaired, sequencing adaptors ligated, and PCR amplified for 4 cycles to ensure libraries were double stranded. 200-400 bp DNA was agarose gel purified and amplified for a further 8 cycles to PCR enrich adaptor ligated DNA into a sequencing library. Library quality was determined on an Agilent BioAnalyzer and quantitated by qPCR prior to sequencing. The B cell CTCF ChIP and input libraries were sequenced on an Illumina GAII and the plasmablast CTCF ChIP and input libraries were sequenced on a HiSeq 2000 platform.
Raw sequencing reads were mapped to the mm9 version of the mouse genome using Bowtie (31). Only uniquely mapped and non-redundant reads were used for subsequent analyses. Significantly enriched CTCF peaks over input control were determined for each sample using HOMER software (32). All sequence manipulation, annotation, and analysis was performed using HOMER software, bedTools (33), and custom R and perl scripts. All sequencing data can be found under accession number GSE44637 in the GEO database (http://www.ncbi.nlm.nih.gov/geo/).
Quantitative 3C assay
A modified 3C assay protocol was performed in this experiment as described previously (23, 34). For B cells and P3X cells, 107 cells were used. For plasmablasts and plasma cells, 0.5 ×106 cells were used. Formaldehyde was added to cells to a final concentration of 1% and incubated for 10 minutes at room temperature. Glycine (final concentration 125 mM) was added to stop the cross linking reaction. Nuclei were collected from the cross-linked cells and digested overnight with either EcoRI or BglII as indicated. Restriction enzymes were heat inactivated, diluted into ligation buffer at a ratio of ~40:1, and then ligated with T4 DNA ligase. Quantitation of the 3C products was performed by real-time PCR against a five point standard curve as described previously (23). Standard curve templates for the 3C products were generated in vitro by restriction enzyme cleaving and religating a BAC containing the region being studied. The following BACs were used to cover the murine MHC-II locus: RP24-169M8, RP24-100G18 and RP24-286E13 (purchased from Children's Hospital Oakland Research Institute). All primer combinations (Supplemental Table 2) were tested prior to use in the 3C assay to determine whether they could efficiently amplify a single product on the cleaved/religated BAC DNA. This includes the “anchor” primers, which were used to assess interactions between one restriction fragment and the others tested in the assay. All primers had a greater than 90% PCR efficiency and displayed only one product. Negative control restriction fragments were chosen with the following bias. Control fragments had to be more than two restriction sites away from the test sequence and contained within a fragment that was <10 kb in length, as we have empirically found that very large fragments score poorly in these assays. Data are presented as crosslinked frequency and represent an average derived from three independent biological replicates. Standard error is provided for the dataset. The student's t-test was used to determine if observed differences were significant.
RNAi, quantitative RT-PCR
RNAi using siRNA was performed to knockdown CTCF mRNA levels from primary B cells, using siRNA oligonucleotide SMART pools from Dharmacon Inc. (Lafayette, CO). These siRNAs were transfected into 4 × 106 primary B cells using a Nucleofection apparatus and transfecting reagents (Kit V) from Amaxa Biosystems as previously described (30, 35). Following transfection, cells were cultured for 72 h. Total RNA was isolated and used for the subsequent quantitative PCR assays. 18s primers were used in each of the experiments to normalize the data. Primers used in this experiment are listed in the Supplemental Table 2. The data presented represent the average with standard deviation of three independent experiments. Statistical significance was calculated using a student's t-test and specifically compared the values obtained using the GFP siRNAs compared to the CTCF siRNAs.
RESULTS
CTCF binding profile dynamics during B cell to plasmablast differentiation
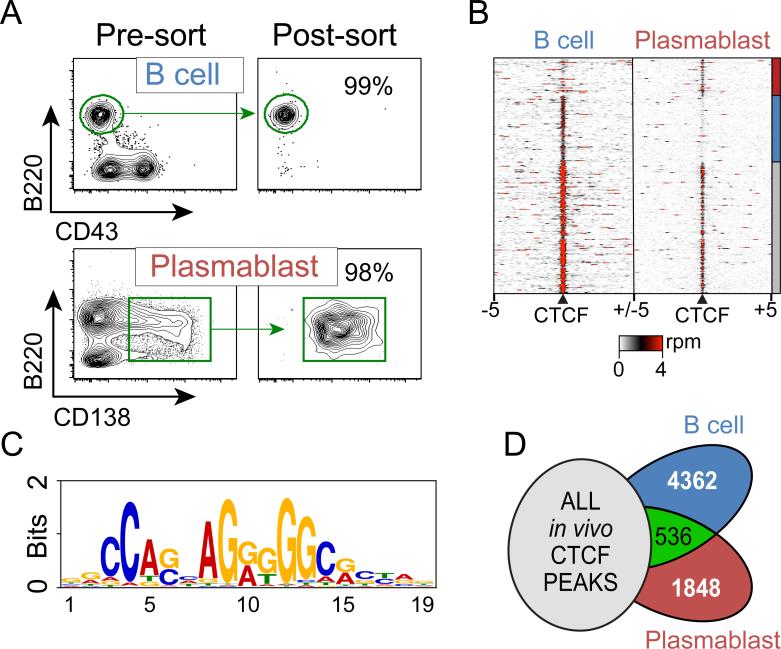
The differentiation of B cells to plasmablasts involves the repression of the B cell transcriptome and activation of genes necessary for antibody secretion (36). To determine the role and function of CTCF in this differentiation process, a high-resolution CTCF-occupancy map was generated for primary B cells and plasmablasts from C57Bl/6 mice. Splenic B cells (CD43− B220+) were isolated to high purity by magnetic bead selection, as determined by flow cytometry (Figure 1A). T-independent plasmablasts were induced by i.v. injection of LPS (27). Three days post-injection, plasmablasts (CD138+B220int) were isolated from the spleen and purified by FACS (Figure 1A). Chromatin was prepared from these cells and subjected to chromatin immunoprecipitation (ChIP) for CTCF. The isolated DNAs were analyzed by deep sequencing to generate a global CTCF binding profile for each cell type. CTCF was significantly enriched at 36,381 and 31,064 genomic loci in B cells and plasmablasts, respectively (Figure 1B). The overlap of CTCF sites between B cells and plasmablasts was computed and while the majority of sites were unchanged, a unique repertoire of CTCF sites existed for both B cells and plasmablasts (Figure 1B). CTCF bound a 19 bp motif that was identical in both cell types and matched previously reported binding sequence preferences (Figure 1C) (37-40).
Figure 1. Global CTCF binding profiles in B cells and plasmablasts.
A) A CD43− B cell population was isolated using a magnetic separation procedure and stained with anti-B220-APC and anti-CD43-FITC. 99% of the cells stained positive for the B cell marker B220. Mouse plasmablasts were induced following injection of 50 μg LPS retro-orbitally into 6-week-old C57Bl/6 mice. Splenocytes were isolated from the LPS-induced mice and stained with anit-CD138-PE and anti-B220-APC. CD138+B220int plasmablasts were isolated to high purity by FACS. B) Heat map representing CTCF ChIP-seq read density at all CTCF sites identified in B cells and plasmablasts. Each row represents 5 kb surrounding a CTCF site. Rows are clustered and annotated with a bar on the right to depict shared and cell-type specific CTCF sites. rpm – reads per million. C) Consensus sequence of core homology elements derived from the sequences of CTCF binding sites from B cells and plasmablasts as identified by the MEME-ChIP software (51). D) Venn diagram showing the overlap of in vivo derived CTCF sites identified by the ENCODE Consortium (Supplemental Table 1)(41) and B cells and plasmablasts. 4,362 and 1,848 CTCF sites were specific to B cells and plasmablasts respectively.
The ENCODE Consortium (41) has recently mapped CTCF binding in a number of primary murine tissues. To identify CTCF sites that were unique to B cells and plasmablasts, the overlap of the CTCF sites derived here and all in vivo generated CTCF maps (Supplemental Table 1) was computed. CTCF bound 4,362 and 1,848 sites that were specific to B cells and plasmablasts, respectively (Figure 1B and D). 536 peaks were shared between B cells and plasmablasts but unique from the other CTCF peaks identified. Each unique site was mapped to the nearest gene and Kegg ontology analysis was performed to identify biological processes present in the cell-type specific CTCF sites. Genes surrounding B cell unique CTCF sites were enriched in processes related to B cell receptor signaling, leukocyte migration, and survival signaling (Table 1). In contrast, plasmablast specific CTCF sites were enriched for MAP kinase signaling, cell death, and IgA antibody production processes (Table 1). These results suggest cell-type specific roles for CTCF in regulating B cell and plasmablast function as has been observed in human cell lines (38).
Table 1.
Kegg ontology processes enriched in unique B cell and plasmablast CTCF sites
| B cell | Plasmablast | ||
|---|---|---|---|
| Kegg Process | -Log10 (p-vlaue) | Kegg Process | -Log10 (p-vlaue) |
| mmu04662: B cell receptor signaling pathway | 6.29 | mmu04010: MAPK signaling pathway | 3.60 |
| mmu04660: T cell receptor signaling pathway | 5.39 | mmu04115: p53 signaling pathway | 3.19 |
| mmu04670: Leukocyte transendothelial migration | 4.87 | mmu04660: T cell receptor signaling pathway | 2.80 |
| mmu04070: Phosphatidylinositol signaling system | 4.81 | mmu04210: Apoptosis | 2.20 |
| mmu04062: Chemokine signaling pathway | 4.15 | mmu04662: B cell receptor signaling pathway | 2.11 |
| mmu05200: Pathways in cancer | 3.98 | mmu04360: Axon guidance | 1.95 |
| mmu05221: Acute myeloid leukemia | 3.92 | mmu04640: Hematopoietic cell lineage | 1.94 |
| mmu04666: Fc gamma R-mediated phagocytosis | 3.85 | mmu05014: Amyotrophic lateral sclerosis (ALS) | 1.54 |
| mmu04520: Adherens junction | 3.75 | mmu04142: Lysosome | 1.40 |
| mmu04722: Neurotrophin signaling pathway | 3.61 | mmu03420: Nucleotide excision repair | 1.35 |
| mmu04810: Regulation of actin cytoskeleton | 3.35 | mmu00534: Heparan sulfate biosynthesis | 1.28 |
| mmu04510: Focal adhesion | 2.96 | mmu04672: Intestinal imm. network for IgA production | 1.27 |
Distribution of CTCF bindings in mouse MHC-II locus
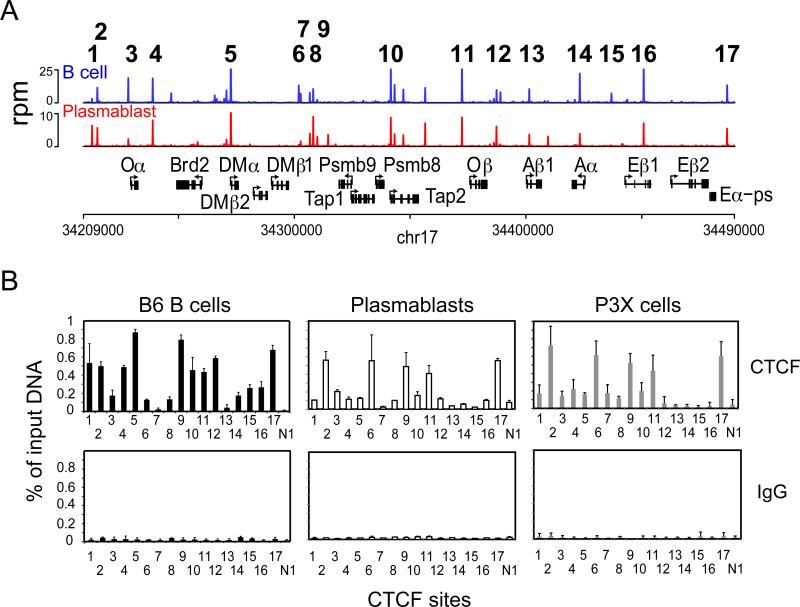
From the ChIP-seq data, 24 potential CTCF sites were observed in B cells across the 281 kb mouse MHC-II locus. Based on their proximity to an MHC-II or related gene, level of CTCF binding, or whether they were differentially bound between B cells and plasmablasts, seventeen of these potential binding sites/clusters (termed C1-C17) were chosen for further analyses (Figure 2A). Here, the MHC-II locus is defined as the region of DNA encompassing H2-Oa and H2-Ea (Figure 2A). In the C57Bl/6 strain, H2-Ea is a pseudogene due to a deletion (3). CTCF sites (C1/C2 and C17) flank the locus. Sequence alignment of the 17 sites showed a high level of homology and was consistent with the sequence logo defined above (Supplemental Figure 1 and Figure 1C). For the most part, pairs of CTCF sites surrounded many of the genes, although some of the sites were located within introns. Several clusters of sites appear to separate the H2-DM genes from the Tap genes on one side of the locus (C6-C9) from a series of sites, collectively termed C10, on the other side between Tap2 and H2-O. LPS-induced plasmablasts displayed a relatively similar pattern of putative sites with the major exception of C15, which was absent in the plasmablast data set. C15 is located between the H2-Aa and H2-Eb1 genes and is in an analogous position to the XL9 CTCF site located between the human HLA-DQA1 and HLA-DRB1 genes (24). XL9 was the first MHC-II insulator discovered in humans (24).
Figure 2. In vivo CTCF binding profile of the mouse MHC-II locus in B and plasma cells.
A) A CTCF occupancy map derived from ChIP-seq data over the MHC-II locus of C57Bl/6 mice with chromosome location is displayed for B cells and plasmablasts. CTCF sites or clusters, which were analyzed further are numbered from 1-17. Rpm, reads per million. B) Site specific quantitative ChIP assays were conducted using anti-CTCF and IgG control antisera with chromatin prepared from C57Bl/6 B cells, plasmablasts, and P3X cells. These results were averaged from three independent chromatin preparations and presented with respect to input chromatin. Standard deviation is shown.
Although ChIP-seq experiments provide exquisite and detailed datasets, they can suffer biases, such as those associated with GC content (42, 43). Thus, key regions need to be verified. ChIP coupled with qPCR was performed on chromatin prepared from purified splenic B cells, LPS-induced and FACS purified plasmablasts, and the fully differentiated P3X63Ag8 plasma cell line (referred to as P3X herein)(Figure 2B). ChIP primers (Supplemental Table 2) were centered on the CTCF-binding motif identified from the ChIP-seq data. The results showed that ~88% of the putative sites exhibited a high level of CTCF occupancy in MHC-II expressing mouse primary B cells. Low or nearly background levels of CTCF were found at C7 and C13, suggesting that these may not be true CTCF-binding sites. In both LPS-induced plasmablasts and P3X cells, the level of CTCF occupancy at sites C1, C4, C5, C10, C12, C14, and C16 was reduced when compared to B cells (Figure 2B). Unlike in B cells, C15 displayed only background binding in plasmablasts and P3X cells. As with B cells, C7 and C13 showed only background levels of CTCF binding. Importantly, this pattern was the same for primary induced plasmablasts and the P3X model plasma cell line. Thus, unique CTCF binding patterns exist for B cells and plasma cells in the MHCII locus with C1, C4, C5, C10, C12, C14, C15, and C16 displaying differential CTCF binding.
CTCF is essential for MHC-II gene expression in murine B cells
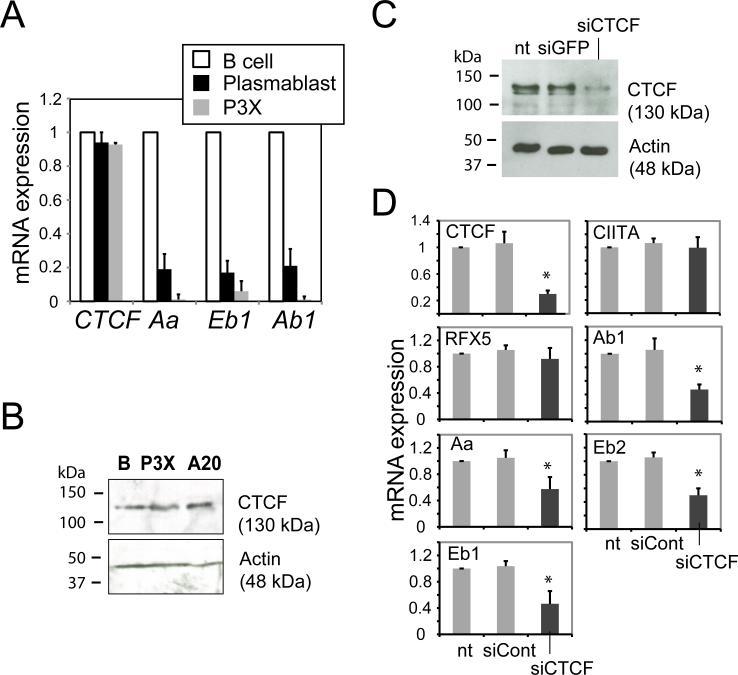
Differences in the above CTCF patterns could reflect different levels of expression of CTCF in the different cell types. However, mRNA levels of CTCF as assessed by qRT-PCR and compared between splenic B cells, LPS-induced plasmablasts, and P3X cells were similar (Figure 3A). Western blots for CTCF expression from splenic B cells, P3X and a B cell line A20 showed that similar CTCF protein levels exist between these cells (Figure 3B). Thus, CTCF expression levels between these cells were similar.
Figure 3. Expression of mouse MHC-II genes are specifically reduced in CTCF depleted splenic B cells.
A) Total RNA was purified from splenic B cells, LPS-induced plasmablasts, and P3X cells, and the mRNA level of the genes indicated was determined. B) CTCF protein levels were assessed in whole cell lysates from B cells, P3X, and A20 cells by western blotting using anti-CTCF and anti-actin antisera. C) At 72 h post-transfection of primary murine splenic B cells with CTCF or GFP (control) siRNAs, a western blot was performed to assess the degree of CTCF expression/knockdown. D) From the siRNA transfections in (C), mRNA levels of the indicated genes were determined by real-time RT-PCR using RNA prepared from untreated cells (nt) and cells transfected with CTCF or control siRNAs. Data from three biological replicates are presented as relative mRNA expression with respect to the no treatment lane (nt). Standard error is presented along with student's t-test comparisons between siRNAs to GFP and CTCF. An asterisk (*) indicates P values <0.05 between samples treated with siRNAs against GFP and CTCF.
Depletion of CTCF from human Raji B cells resulted in reduced expression of all MHC-II genes and did not affect the expression of CIITA or RFX5, two key modulators of MHC-II expression (23). To determine if the same was true for the murine system and in primary B cells, knockdown experiments in mouse primary B cells were conducted using control and mouse CTCF-specific siRNA oligonucleotide SMART pools as previously described (23). Protein and RNA were isolated from untreated (nt), siRNA-CTCF, and siRNA-Control-treated primary B cells, and the levels were analyzed by Western blot and real-time RT-PCR, respectively. The efficiency of knockdown varied between 50-75% as indicated by immunoblots using antibody against CTCF (Figure 3C). Actin levels confirmed that similar amounts of lysates were used. This degree of efficiency resulted in ~70% reduction in CTCF mRNA levels with no affect on CIITA or RFX5 mRNA levels (Figure 3D). Importantly, CTCF knockdown resulted in a significant reduction in H2-Aa, H2-Ab1, H2-Eb1, and H2-Eb2 gene expression (Figure 3D). Together, these data suggest that CTCF is required for maximal expression of the murine MHC-II genes.
MHC-II CTCF-binding sites participate in long-range interactions
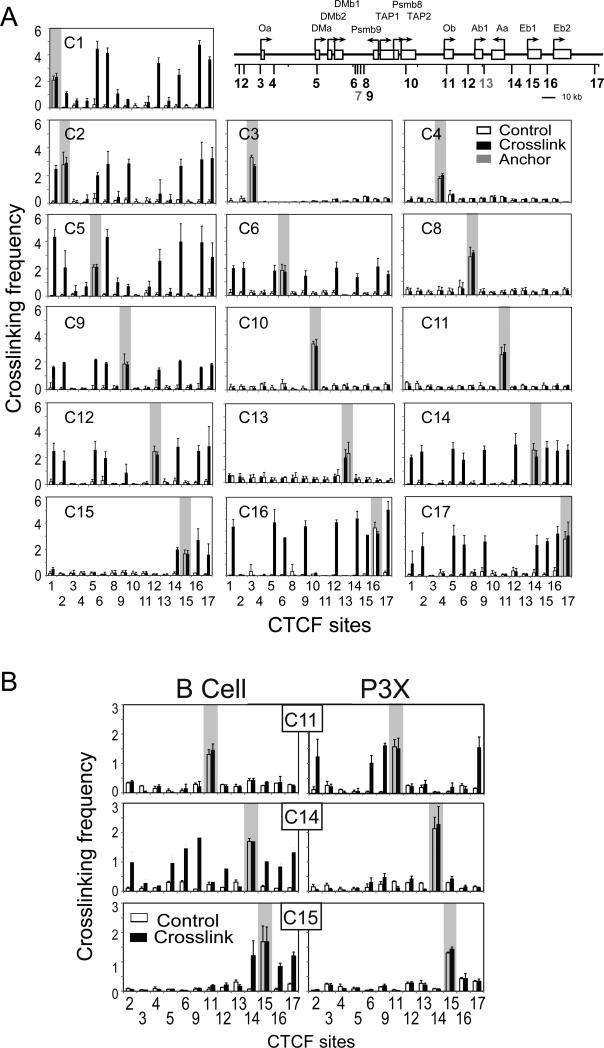
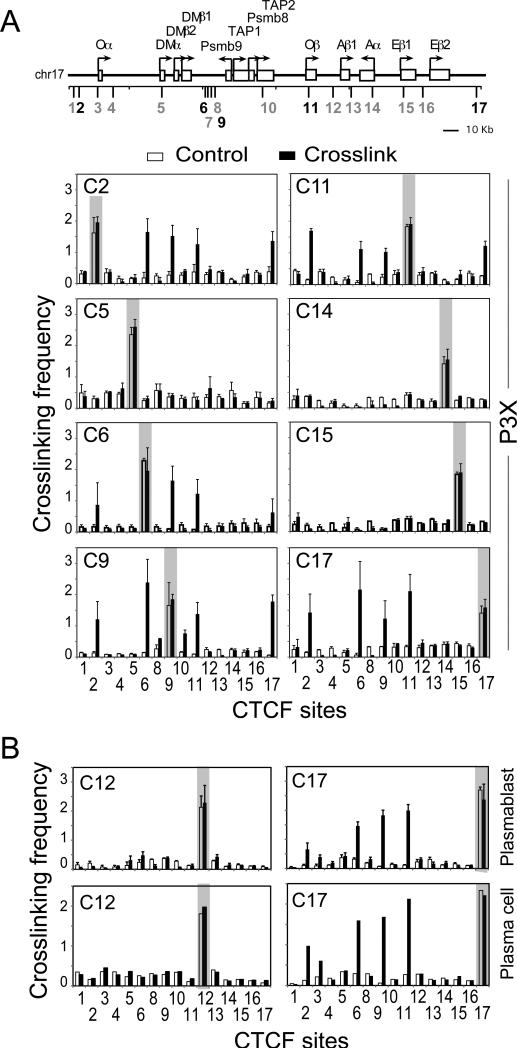
CTCF-bound DNA elements can form interacting loops both in vivo and in vitro, thereby connecting two distantly separated DNA fragments (44, 45). In the human MHC-II locus, the CTCF-bound elements interacted with one another in a distance-dependent manner with the longest interactions occurring over ~250 kb (30, 35). Interactions between the sites would be predicted to restrict gene expression/regulation, thus raising questions of which sites interacted and how the sites would demarcate the murine MHC-II locus. As the murine locus is about 2.5-fold smaller than the human locus (660 vs. 281 kb), the possibility existed that all sites within the locus could interact. To address these issues, quantitative 3C assays (46, 47), which assess the spatial relationship and proximity between DNA sequences within the nucleus of a cell, were performed in primary B cells isolated from C57BL/6 mouse spleens (Figure 4A). In this assay, formaldehyde treatment of live cells was used to fix and crosslink DNA elements. The cross-linked chromatin was isolated, digested with EcoRI, diluted, and ligated, such that only DNA ends that are in close proximity could serve as efficient ligation substrates. The novel DNA junctions produced by DNA ligation were measured by qPCR. A detailed schematic of the murine MHC-II locus with 3C primer positions and EcoRI restriction sites is shown in Supplemental Figure 2. Primer sequences used to assess the cross-linking frequencies are listed in Supplemental Table 2. Restriction fragments encompassing 16 of the CTCF sites were used as position “anchors” to interrogate interactions with the other CTCF sites (Figure 4, shaded regions). The results showed that nine of the sixteen CTCF sites (C1, C2, C5, C6, C9, C12, C14, C16, and C17) were able to interact with each other at variable frequencies in MHC-II expressing B cells (Figure 4A). In contrast to these interacting CTCF sites, the C15 site was only found to interact locally with its neighboring CTCF sites (C14, C16, and C17) – sites within the classical MHC-II gene region of the locus. As expected due to its low levels of CTCF binding, C13 demonstrated no interactions and served as a negative control. However, unlike the human MHC-II region, in which all of the characterized CTCF-bound sites interacted with their neighbors, five murine CTCF sites did not interact with any of the CTCF sites in the region (C3, C4, C8, C10, and C11). In these experiments, self-ligated anchor fragments from both formaldehyde cross-linked and untreated controls displayed equal ligation frequencies (Figure 4B, open vs. filled bars), demonstrating that similar levels of chromatin were used in the experiments and that access to the restriction sites in the locus was comparable. Additionally, no ligase control samples were unable to generate a 3C PCR product (data not shown). Because some of the EcoRI sites produced restriction fragments that contained MHC-II promoters (C11, C14, and C15), a set of 3C assays using BglII to separate the promoters and CTCF sites was performed (Figure 4B). For these three CTCF sites, similar interaction maps were observed with C11 forming no interactions, C14 interacting across the locus at numerous sites, and C15 interacting with the local CTCF sites. Thus, a specific set of interactions between a subset of CTCF sites defines the CTCF-architecture of the MHC-II locus in murine B cells.
Figure 4. Multiple chromatin loops form throughout the mouse MHC-II locus in B cells.
A) Schematic representation of the 17 CTCF site locations is provided as a guide for the location of CTCF binding regions. Gray labeled CTCF sites showed background CTCF binding as defined in Figure 2. 3C assays were conducted on C57Bl/6 B cells isolated from the spleen by negative magnetic selection. Detailed schematic maps showing the locations of the EcoRI sites are presented in Supplemental Figure 2. 3C assays were carried out as described in the Methods section. The crosslinked frequency was derived from three independent experiments. B) 3C assays conducted to assess interactions between C11, C14, and C15 and all the other CTCF sites were performed on chromatin prepared from B cells and P3X cells using BglII as the restriction enzyme as described above. In both A and B, the shaded areas represent self-ligating restriction fragments that contained the anchor primer (the single primer used to assess interactions with the other restriction fragments). These self-ligations serve as an internal control for restriction enzyme efficiency/accessibility in digesting the crosslinked chromatin. The crosslinked frequency with standard error represents the relative amount of 3C product for each set of interactions divided by a non-specific control fragment within each set of reactions. These data were derived from three independent chromatin preparations.
Plasma cells display a unique pattern of CTCF-mediated interactions
When B cells differentiate to plasma cells MHC-II gene expression is silenced (Figure 3A). This is thought to occur through the direct transcriptional repression of the MHC-II transactivator CIITA by Blimp-1 and other factors, such as ZBTB32 (26, 27). Because plasma cells represent a terminal differentiation stage that does not express MHC-II genes, and both plasmablasts and P3X cells showed CTCF-binding patterns that were distinct from the B cell pattern, it was of interest to determine if the CTCF architecture within the MHC-II locus of P3X cells shared similarities with B cells. Thus, 3C assays were performed on the P3X cells. Quite remarkably, the CTCF interaction patterns in the P3X cells were distinct from the B cell pattern (Figure 5). Although fewer anchors were used, only 5 of the CTCF sites interacted with the other CTCF sites (C2, C6, C9, C11, and C17). These interactions reflect the occupancy levels of CTCF in both P3X and plasmablasts (Figure 2A). Notable losses in interactions were observed for C5, located near the H2-M genes, C12, which separates the classical class II region from the rest of the MHC-II, and C15, which lies between H2-Aa and H2-Ea1 genes. A more intriguing difference was observed at C11. C11, which did not interact in the B cell 3C assays, interacted with CTCF sites C2, C6, C9, and C17 in the P3X cell line (Figure 5). These new interactions form a distinct structural arrangement of the CTCF-mediated three-dimensional architecture for the MHC-II locus in P3X plasma cells.
Figure 5. CTCF sites within the murine MHC-II locus form distinct interactions in plasma cells.
A) A schematic of the mouse MHC-II locus with the robust CTCF-binding sites in P3X cells shown in black and weak/background sites shown in gray is presented. 3C assays were performed in P3X cells with anchors at all the robust CTCF binding sites, as well as select differentially binding CTCF sites (C5, C14, C15) as controls. B) LPS-induced (3 days) plasmablasts (B220intCD138+) and plasma cells (B220loCD138+) from C57Bl/6 mice were induced and isolated by FACS as described above. 3C assays were conducted on C12 and C17 as described above to determine if these cells shared the same set of interactions as P3X cells. Shaded regions indicate the anchor fragment position. Crosslinking frequencies were established from three independent chromatin preparations and cell isolations as described above and in Figure 4. These data were compiled from three independent chromatin preparations.
Although P3X cells display similar CTCF occupancy patterns as LPS-induced plasmablasts, there was the concern that these cells may be exhibiting a pattern that is associated with their transformation rather than one that is associated with the plasma cell lineage. To verify that the P3X CTCF interaction pattern represents the lineage, a limited set of 3C assays (due to cell numbers) were conducted on FACS sorted, LPS-induced plasmablasts (B220intCD138+) and plasma cells (B220loCD138+) cells. In these assays, two anchors were used: C12 and C17. C17 was chosen because it showed robust interactions at multiple sites that would be diagnostic for both the B cell and P3X patterns. C12 was chosen because it displayed differential interactions between the two cell populations. The 3C data on both plasmablasts and plasma cells were very similar, with both recapitulating the data obtained in P3X cells (Figure 5B).
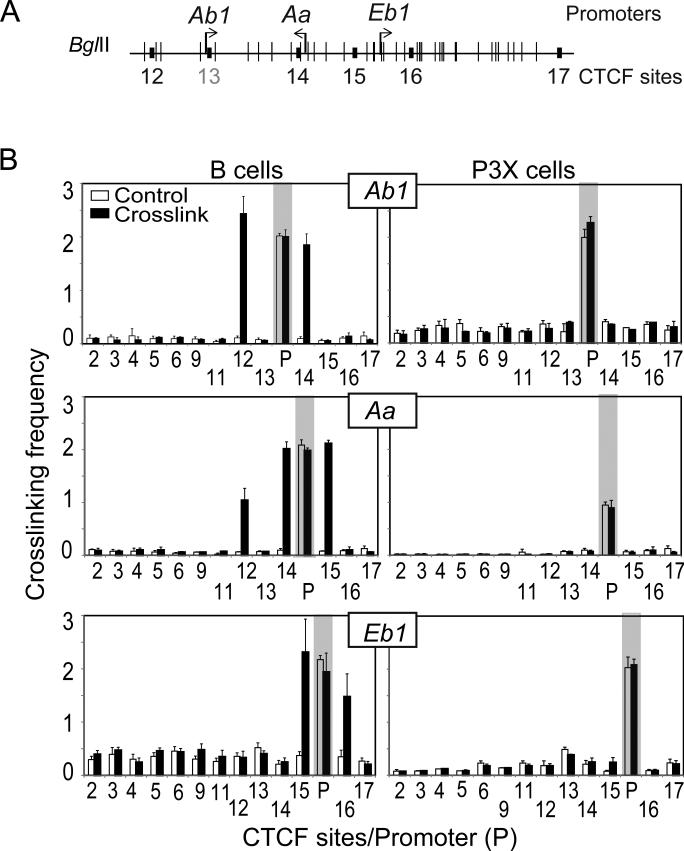
Mouse MHC-II promoters communicate with neighboring CTCF sites
Part of the mechanism involving CTCF regulation of human MHC-II genes was previously shown to involve CTCF interacting with CIITA both in cellular lysates and when bound to chromatin (35). When CIITA was bound to promoter regions, the CTCF-bound sites would interact with them forming a second set of interactions (23, 30). To determine if a similar set of events was occurring in the murine locus, 3C assays were conducted in B cells and P3X cells using the promoter regions of H2-Ab1, H2-Aa, and H2-Eb1, which contain the WXY box target sites of RFX and CIITA as 3C anchors. In these experiments, the restriction enzyme BglII was used to separate the mouse MHC-II promoters (Ab1, Aa, and Eb1 genes) from promoter proximal CTCF sites (Figure 6). Primer sequences used in this 3C assay are listed in the Supplemental Table 2. In B cells, specific interactions were observed between the H2-Ab1 promoter and the surrounding CTCF sites, C12 and C14. In a similar manner, H2-Aa interacted with C14 and C15 and to a lesser extent with C12; and H2-Eb1 interacted with C15 and C16. No significant interaction was detected in the non-MHC-II expressing P3X cells for any of the MHC-II promoters tested here (Figure 6B). Thus, like the human system, murine MHC-II promoters interact with neighboring CTCF sites in an expression dependent manner.
Figure 6. CTCF sites interact with MHC-II gene promoter regions in a manner that is coincident with MHC-II gene activity.
A) A schematic of the H2-Ab1, Aa and Eb1 classical MHC-II gene region is shown with all BglII sites indicated as vertical marks. The positions of key CTCF sites surrounding the region are indicated by small black boxes. C13 had no detectable CTCF binding (Figure 2) and is colored gray here. B) BglII-based 3C assays using the promoter regions (containing the XY box elements) as anchors were performed to determine the relative interaction frequencies between the indicated CTCF sites and their neighboring MHC-II gene promoter region restriction fragments (as indicated) in C57Bl/6 splenic B cells (MHC-II and CIITA positive) and P3X (MHC-II and CIITA negative) cells. 3C assays were performed three times from independent chromatin and cell preparations. All peaks in the B cell 3C assays were statistically significant.
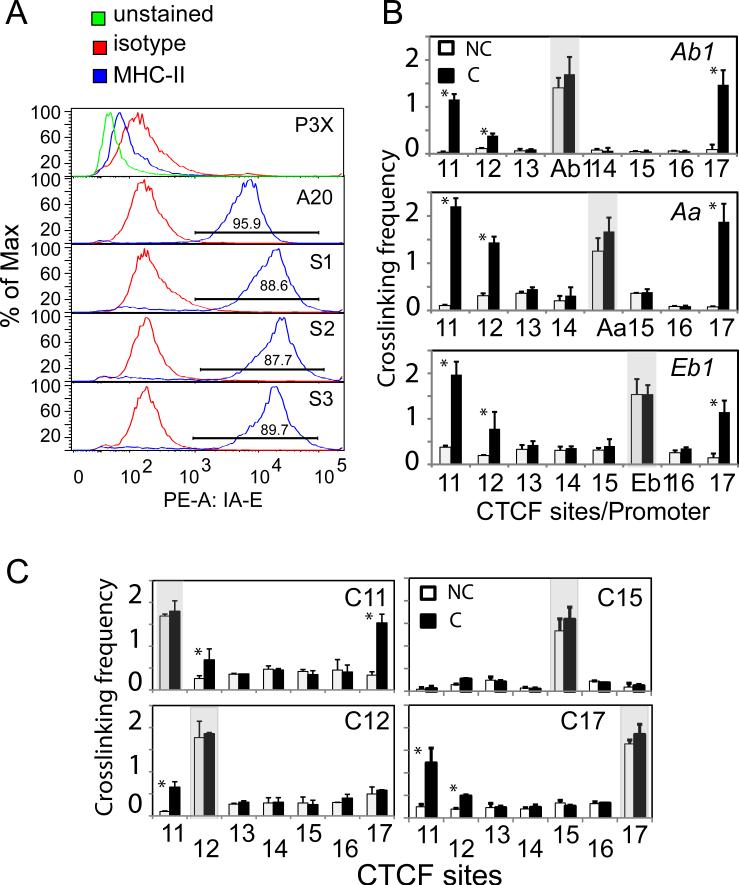
CIITA can induce interactions with CTCF sites in plasma cells but does not restore the B cell architecture
Because the levels of CTCF in B cells and the plasma cell line P3X were similar, the above data showing differences in CTCF architecture could be due to a cell fate terminal differentiation process, the lack of a CIITA, which is absent in P3X cells, or a combination of the two. To address directly whether CIITA was responsible for the observed architectural differences, a CIITA expression construct was transfected into P3X cells (Figure 7A). This construct allows long-term expression of CIITA in cells due to the inclusion of the human MHC-II XL9 insulator (24) downstream of the CIITA expression cassette (data not shown). Three independently generated CIITA-complement lines were functionally examined 21 days post transfection for surface MHC-II gene expression by flow cytometry (Figure 7A, S1, S2, and S3). In each of these lines, the expression of MHC-II surface proteins was roughly half a log higher than the MHC-II/CIITA positive A20 B cell line, demonstrating that these lines are fully complemented.
Figure 7. Ectopic expression of CIITA in P3X plasma cells restores MHC-II expression, but does not fully induce the CTCF architecture observed in B cells.
A) Flow-cytometry panels depicting the surface expression of MHC-II gene expression from three independent CIITA-stably transfected P3X cell lines (S1, S2, and S3). A20 and P3X cells were used as positive and negative controls, respectively. B) BglII-based 3C assays designed to observe CTCF-promoter interactions were performed as in Figure 6 on the CIITA-complemented P3X cells using 3C anchors at the promoters of I-Ab, I-Aa, and I-Eb1. Interactions were assessed only at CTCF sites C11-C17. C) EcoRI-based 3C assays were conducted in CIITA-complemented P3X cells as above using C11, C12, C15, and C17 as anchors. Filled and open bars indicate Crosslinked (C) and non-crosslinked (NC) samples, respectively. All 3C assays were performed three times from independent chromatin preparations and cultures. Significant 3C values are indicated (*, p<0.05).
To evaluate whether the complemented lines regained/reestablished chromatin interactions between promoter regions or CTCF sites, 3C assays were performed. The introduction of CIITA into the P3X cells resulted in interactions between MHC-II promoters at H2-Ab1, H2-Aa, and H2-Eb1 with CTCF sites (C11, C12, and C17 (Figure 7B)). However, these were not completely identical to the interactions observed in B cells, and most likely represent the closest possible CTCF-bound site. Intriguingly, although at lower frequencies, C12 (which exhibited reduced levels of binding of CTCF in P3X cells) interacted with each of the three MHC-II promoters as in B cells (Figure 7B). Examination of the local CTCF-CTCF interactions showed that C11 and C17 interactions were still the major plasma cell interacting CTCF sites within the region (Figure 7C). However, C12 displayed some interactions with C11, but not with C17. C15 did not interact with any of the other CTCF sites tested. Thus, while CIITA can restore expression of MHC-II, in part by forming interactions with available CTCF sites, it cannot fully reprogram the CTCF architecture within the locus to reflect the B cell state.
DISCUSSION
In this study, high-resolution maps of CTCF-binding sites in murine B cells and LPS-induced plasmablasts were generated and used to define the CTCF-centric three-dimensional base chromatin structure of the MHC-II locus in these cell types. Although the majority of the CTCF-binding sites were the same between the two cell types, a set of B-cell specific and plasmablast specific binding sites were identified. Within the MHC-II locus, differences between the binding patterns were revealed to a higher degree upon quantitative traditional ChIP analyses, which showed that the plasmablasts had a lower level of CTCF occupancy within the region and in particular in the classical MHC-II gene subregion. This was further documented by analysis of the P3X plasma cell line, which showed even greater differences at each of the sites.
The analysis of the murine locus revealed three common features between the human and murine MHC-II loci that may reflect cross-species three-dimensional gene organization features that are coupled with regulatory function. First, despite the fact that the MHC-II genes themselves are expressed coordinately, both species have numerous CTCF sites separating the genes. In the human system, the CTCF sites previously characterized (23, 24, 30, 35, 48) are intergenic and occur between isotype specific α/β MHC-II genes; whereas in the mouse, there were more sites, and the sites surround/insulate the promoter regions of each of the MHC-II genes. The exception to this is in the H2-DMa, -DMb2, and -DMb1 genes, which are flanked by a single set of CTCF sites, C5-C6, in the mouse vs. each gene being surrounded in the human. Second, in both humans and mice, CTCF is required for maximal MHC-II expression in B cells as shown by the CTCF knockdown experiments. This finding was also observed in macrophages (49). Third, CTCF interacts with CIITA in a manner that forms sub-locus chromatin loops that are associated with maximal gene expression. The reintroduction of CIITA into the plasma cell line demonstrated that this CIITA-CTCF interaction is an integral part of the expression mechanism and is directed by the presence of CIITA in the system.
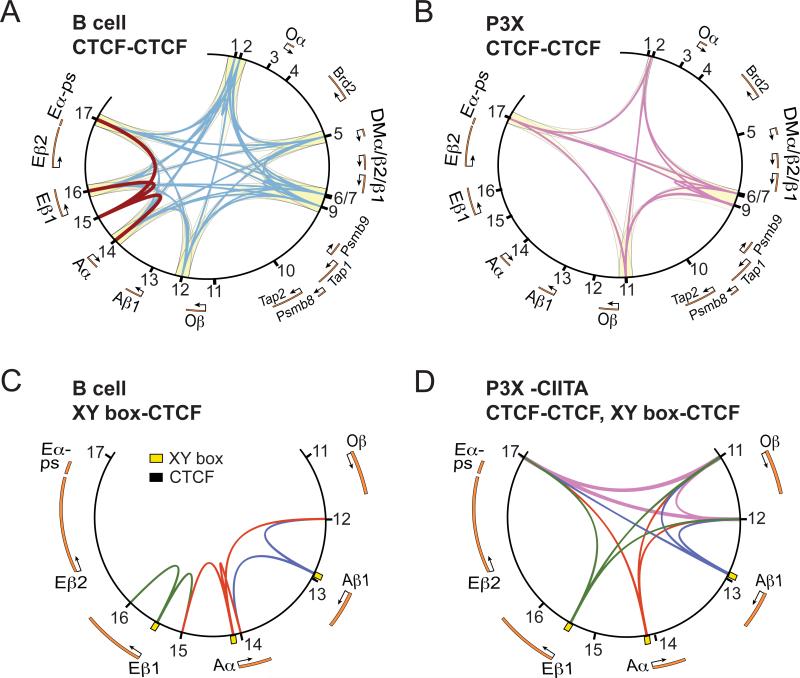
The 3C data provided information regarding interactions between the CTCF-binding sites themselves and how the promoter regions interacted with the established CTCF-CTCF architecture. By plotting the data from each of the four 3C experiments onto a set of Circos plots (50), a higher order overview of the interactions takes form (Figure 8). In viewing the B cell CTCF-CTCF interaction plot (Figure 8A), one immediately observes four major compartments that are defined by the nine CTCF sites that displayed CTCF-CTCF interactions with each other across the entire locus: 1) H2-Oa and Brd2; 2) H2-DMa and -DMb; 3) H2-Ob, Tap1, Tap2, PSMB8, and PSMB9; and 4) the classical MHC-II gene region. The classical MHC-II subregion is further divided by several CTCF sites, including C15, which interacts only with its three neighboring CTCF sites (C14, C16, and C17). From this view, these sites architecturally define the entire MHC-II locus. The longest interactions observed spanned the entire locus (C1/C2 – C17), a distance of ~281 kb. This distance was essentially the maximum distance observed for any interaction in the human MHC-II locus.
Figure 8. Circos plot of 3C interactions among CTCF sites and MHC-II promoters in both MHC-II expressing and non-expressing cells.
Results obtained from 3C interactions were summarized into four Circos plots (50) with a schematic of the relevant region of the MHC-II locus shown. CTCF-binding sites are indicated by black lines and are numbered accordingly. A) CTCF–CTCF interaction data from B cells are presented. Blue line connections show the CTCF-CTCF interactions across the locus, whereas the maroon lines indicate the interactions of C15. B) CTCF–CTCF site interaction data from P3X cells with lilac lines representing the interactions. Yellow background shading in panels A and B highlight the higher-order domains identified in each cell type. C) CTCF–MHC-II promoter interactions in B cells are shown for the classical MHC-II genes. Yellow boxes represent the position of the WXY box regions. D) CTCF–CTCF and CTCF–MHC-II promoter interactions for the classical MHC-II genes are shown in P3X cells that have been complemented with a CIITA expression vector. The lines/interactions in panel D use the same coloring scheme as B and C.
In sharp contrast to the B cell CTCF architecture was that of the plasmablast / plasma cells (Figure 8B). In plasma cells, MHC-II locus border interactions favored C2 and C17, with no interactions with C1. Other interactions throughout the locus were missing or unique to this cell type as well. Instead of four major compartments, there were only three. On one end of the locus, the H2-Oa, Brd2, and H2-DM genes were grouped into a single compartment as the interaction with C5, which segregates the B cell CTCF compartments 1 and 2 was missing. The Tap and PSMB genes were still located in a single compartment, but unlike in the B cells, this plasma cell specific compartment no longer included H2-Ob. This was due to new interactions using C11, which resides between Tap2 and H2-Ob, with the other interacting CTCF sites (C2, C6, C9, and C17) and a loss of interactions with C12. The third plasma cell compartment clustered H2-Ob and all of the classical MHC-II genes together in a single compartment with no additional CTCF interactions subdividing the subregion. The mechanism for the changes in compartments correlates directly with the overall occupancy levels of CTCF. In the plasma cells, CTCF binding at C1, C5, and C12 is greatly reduced compared to B cells. It should be noted that CTCF occupancy in P3X cells and LPS-induced plasmablasts was similar to each other, and importantly, overall CTCF protein and RNA levels were similar between the B cells and the plasma cells. This suggests that the loss of CTCF binding reflects a cell fate decision and locus specific effect and not a general loss of CTCF in the cell. Aside from DNA sequence, the differential in CTCF occupancy is likely due to changes in accessibility of the site to CTCF. Accessibility at these regions could be controlled/regulated by epigenetic mechanisms such as DNA methylation or repressive histone modifications.
Of the CTCF sites, C15 was distinct from the others in that it did not interact across the entire locus, but instead formed local interactions with the CTCF sites within the MHC-II subregion. One reason for this could be that C15 bound modest levels of CTCF compared to the other sites. CTCF binding to C15 was also absent in plasma cells. C15 was considered an important site because its location (between H2-Eb1 and H2-Aa) was similar to that of XL9, the first MHC-II insulator discovered (24). XL9 resides between HLA-DRB1 and HLA-DQA1 and is important for their activity. Like XL9, C15 appears to function in an analogous manner by interacting with neighboring CTCF sites and with the promoters of its flanking genes.
For the three expressed MHC-II gene promoters (H2-Eb1, -Aa and -Ab1), interactions with local CTCF sites were observed in B cells. Like the human system, the mouse MHC-II gene promoters could interact with more than one CTCF site. For H2-Ab1 and H2-Eb1, the 3C data suggest that there is no preference for which site is chosen. H2-Aa, which resides in the middle of the MHC-II subregion showed an equal preference for the adjacent CTCF sites but could also interact with C12 to some extent. Thus, we propose that the choice of which CTCF site to interact with may be availability of a “free” CTCF protein rather than proximity to one site or the other. This conclusion is supported by the experiments in which CIITA was ectopically expressed in the P3X cell line. In addition to inducing the MHC-II genes to a high level, the presence of CIITA resulted in establishment of MHC-II promoter–CTCF site interactions. With the exception of a modest level of MHC-II promoter–C12 interactions, the strongest interactions were with C11 and C17; sites that displayed the highest level of CTCF occupancy in P3X cells. We speculate that C12 may be more accessible than the other regions and thus, during the reintroduction of CIITA, can be occupied in some of the cells and contribute to the interactions and architecture of the locus that were observed. The presence of CIITA-CTCF interactions may stabilize or contribute to the occupancy of C12 in this experimental system. Intriguingly, MHC-II promoters did not interact with C11 or C17 in B cells, even though CTCF was present at high levels. Together these data indicate that the MHC-II promoters can interact with CTCF sites that are further away, but supports the proposed availability model.
The 3C data suggest that interactions across the locus may form a number of architectures, including a complex rosette-like structure with all of the CTCF interacting sites forming a central hub. For such a structure to form, multiple CTCF-CTCF interactions must take place per site. A CTCF hub/rosette coupled with the additional CIITA-mediated promoter loops is appealing in that it could allow specific transcription machinery for the antigen processing pathway genes to be highly concentrated, which would presumably allow for high levels of transcription initiation. Alternatively, it is possible that each CTCF site interacts with a single additional site. Such architecture would introduce considerable heterogeneity among chromosomes within each cell and within the population of cells. If the CTCF-CTCF interactions serve to limit gene expression to genes within a “single regulatory-loop”, dysregulation of some genes could occur through some CTCF-CTCF interactions. For example, if the constitutive Tap2 gene promoter regulatory mechanism was in the same loop as the I-A genes, expression of the I-A genes might occur in all cells containing that loop. Considering this scenario, perhaps it is for this reason that both the B cell and the plasma cell architecture cordoned off the Tap and PSMB genes in their own compartment. Thus, it is likely that CTCF-CTCF interactions and compartments serve several purposes. The first is to prevent dysregulation of neighboring genes in non-antigen presenting cells. The second is to provide ready and available access of CTCF to MHC-II promoters when CIITA is bound. Together, these interactions could lead to a high concentration of coordinately regulated gene promoters in defined nuclear spaces that could increase the efficiency of transcription factor binding and transcription initiation in antigen presenting cells.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to acknowledge Joshua Lee and Royce Butler for excellent technical assistance and to thank members of the laboratory for their critical comments on the manuscript. We also thank Benjamin Barwick for his genomePlots R scripts.
Abbreviations
- 3C
chromatin conformation capture
- ChIP
chromatin immune precipitation
- CTCF
CCCTC binding factor
- IRES
internal ribosome entry site
- NF-Y
Nuclear factor-Y
- RFX
Regulatory factor X
- P3X
P3X63Ag8 plasma cells
Footnotes
This work was supported by NIH grants AI43000 and GM47310.
Conflict of Interest Statement:
The authors have no financial conflict of interest.
REFERENCES
- 1.Steinmetz M, Minard K, Horvath S, McNicholas J, Srelinger J, Wake C, Long E, Mach B, Hood L. A molecular map of the immune response region from the major histocompatibility complex of the mouse. Nature. 1982;300:35–42. doi: 10.1038/300035a0. [DOI] [PubMed] [Google Scholar]
- 2.Hood L, Steinmetz M, Goodenow R. Genes of the major histocompatibility complex. Cell. 1982;28:685–687. doi: 10.1016/0092-8674(82)90046-0. [DOI] [PubMed] [Google Scholar]
- 3.Dembic Z, Ayane M, Klein J, Steinmetz M, Benoist CO, Mathis DJ. Inbred and wild mice carry identical deletions in their E alpha MHC genes. The EMBO journal. 1985;4:127–131. doi: 10.1002/j.1460-2075.1985.tb02326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Bouteiller P. HLA class I chromosomal region, genes, and products: facts and questions. Critical reviews in immunology. 1994;14:89–129. doi: 10.1615/critrevimmunol.v14.i2.10. [DOI] [PubMed] [Google Scholar]
- 5.Boss JM, Jensen PE. Transcriptional regulation of the MHC class II antigen presentation pathway. Current opinion in immunology. 2003;15:105–111. doi: 10.1016/s0952-7915(02)00015-8. [DOI] [PubMed] [Google Scholar]
- 6.Choi NM, Majumder P, Boss JM. Regulation of major histocompatibility complex class II genes. Current opinion in immunology. 2010;23(1):81–87. doi: 10.1016/j.coi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reith W, Mach B. The bare lymphocyte syndrome and the regulation of MHC expression. Annu Rev Immunol. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 8.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109(Suppl):S21–33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 9.Boss JM, Strominger JL. Regulation of a transfected human class II major histocompatibility complex gene in human fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:9139–9143. doi: 10.1073/pnas.83.23.9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorn A, Durand B, Marfing C, Le Meur M, Benoist C, Mathis D. Conserved major histocompatibility complex class II boxes--X and Y--are transcriptional control elements and specifically bind nuclear proteins. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:6249–6253. doi: 10.1073/pnas.84.17.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reith W, Satola S, Herreo-Sanchez C, Amaldi I, Lisowska-Grospierre B, Griscelli C, Hadam MR, Mach B. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF X. Cell. 1988;53:897–906. doi: 10.1016/s0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 12.Moreno CS, Beresford GW, Louis-Plence P, Morris AC, Boss JM. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 13.Dorn A, Bollekens J, Staub A, Benoist C, Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987;50:863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- 14.Louis-Plence P, Moreno CS, Boss JM. Formation of a regulatory factor X/X2 box-binding protein/nuclear factor-Y multiprotein complex on the conserved regulatory regions of HLA class II genes. J Immunol. 1997;159:3899–3909. [PubMed] [Google Scholar]
- 15.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 16.Beresford GW, Boss JM. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat Immunol. 2001;2:652–657. doi: 10.1038/89810. [DOI] [PubMed] [Google Scholar]
- 17.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 18.Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat Immunol. 2003;4:132–137. doi: 10.1038/ni883. [DOI] [PubMed] [Google Scholar]
- 19.Choi NM, Boss JM. Multiple histone methyl and acetyltransferase complex components bind the HLA-DRA gene. PloS one. 2012;7:e37554. doi: 10.1371/journal.pone.0037554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol Cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Labrador M, Corces VG. Setting the boundaries of chromatin domains and nuclear organization. Cell. 2002;111:151–154. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- 22.Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majumder P, Gomez JA, Boss JM. The human major histocompatibility complex class II HLA-DRB1 and HLA-DQA1 genes are separated by a CTCF-binding enhancer-blocking element. J Biol Chem. 2006;281:18435–18443. doi: 10.1074/jbc.M601298200. [DOI] [PubMed] [Google Scholar]
- 25.Chang C-H, Fodor WL, Flavell RA. Reactivation of a major histocompatibility complex class II gene in mouse plasmacytoma cells and mouse T cells. J.Exp.Med. 1992;176:1465–1469. doi: 10.1084/jem.176.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piskurich JF, Lin KI, Lin Y, Wang Y, Ting JP, Calame K. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- 27.Yoon HS, Scharer CD, Majumder P, Davis CW, Butler R, Zinzow-Kramer W, Skountzou I, Koutsonanos DG, Ahmed R, Boss JM. ZBTB32 is an early repressor of the CIITA and MHC class II gene expression during B cell differentiation to plasma cells. J Immunol. 2012;189:2393–2403. doi: 10.4049/jimmunol.1103371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 29.Majumder P, Boss JM. DNA methylation dysregulates and silences the HLA-DQ locus by altering chromatin architecture. Genes and immunity. 2011;12:291–299. doi: 10.1038/gene.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majumder P, Boss JM. CTCF controls the expression and the chromatin architecture of the human major histocompatibility complex class II locus. Mol Cell Biol. 2010;30(17):4211–4223. doi: 10.1128/MCB.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 35.Majumder P, Boss JM. Cohesin Regulates MHC Class II Genes through Interactions with MHC Class II Insulators. J Immunol. 2011;187:4236–4244. doi: 10.4049/jimmunol.1100688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Seminars in immunology. 2011;23:341–349. doi: 10.1016/j.smim.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Chen H, Tian Y, Shu W, Bo X, Wang S. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PloS one. 2012;7:e41374. doi: 10.1371/journal.pone.0041374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, Thurman RE, Kaul R, Myers RM, Stamatoyannopoulos JA. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome research. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Consortium EP. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheung MS, Down TA, Latorre I, Ahringer J. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucleic acids research. 2011;39:e103. doi: 10.1093/nar/gkr425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic acids research. 2012;40:e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartkuhn M, Renkawitz R. Long range chromatin interactions involved in gene regulation. Biochim Biophys Acta. 2008;1783:2161–2166. doi: 10.1016/j.bbamcr.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Pant V, Kurukuti S, Pugacheva E, Shamsuddin S, Mariano P, Renkawitz R, Klenova E, Lobanenkov V, Ohlsson R. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol Cell Biol. 2004;24:3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 47.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 48.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Nikolic T, Movita D, Lambers ME, de Almeida CR, Biesta P, Kreefft K, de Bruijn MJ, Bergen I, Galjart N, Boonstra A, Hendriks R. The DNA-binding factor Ctcf critically controls gene expression in macrophages. Cellular & molecular immunology. 2013 doi: 10.1038/cmi.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.