Abstract
Dysferlin is a calcium-binding transmembrane protein involved in membrane fusion and membrane repair. In humans, mutations in the dysferlin gene are associated with muscular dystrophy. In this study, we isolated plasma membrane-enriched fractions from full-grown immature oocytes of the sea star, and identified dysferlin by mass spectrometry analysis. The full-length dysferlin sequence is highly conserved between human and the sea star. We learned that in the sea star Patiria miniata, dysferlin RNA and protein are expressed from oogenesis to gastrulation. Interestingly, the protein is highly enriched in the plasma membrane of oocytes. Injection of a morpholino against dysferlin leads to a decrease of endocytosis in oocytes, and to a developmental arrest during gastrulation. These results suggest that dysferlin is critical for normal endocytosis during oogenesis and for embryogenesis in the sea star and that this animal may be a useful model for studying the relationship of dysferlin structure as it relates to its function.
Keywords: Dysferlin, Sea star, Plasma membrane, Oocytes, Endocytosis, Gastrulation
Introduction
Dysferlin is a transmembrane protein associated with rare forms of muscular dystrophies, such as limb-girdle muscular dystrophy type 2B (Bashir et al., 1998), Miyoshi myopathy (Liu et al., 1998), and distal myopathy (Illa et al., 2001). Loss of dysferlin from the plasma membrane of muscle fibers leads to abnormalities in vesicular trafficking and membrane repair (Kobayashi et al., 2012). Moreover, freshly isolated monocytes of dysferlinopathy patients show deregulated expression of fibronectin and fibronectin-binding integrins which is recapitulated by transient knockdown of dysferlin in the human monocytic cell line, THP1. Dysferlin regulates cellular interactions by forming a complex with integrins at the cell membrane (de Morree et al., 2013). In humans, the dysferlin transcript is highly expressed in skeletal muscle, heart, placenta, and more weakly in liver, lung, kidney and pancreas (Bashir et al., 1998). More recently, it was also found in cultured vascular endothelial cells (Leung et al., 2011; Sharma et al., 2010). Dysferlin is a member of the ferlin family of proteins, based on the similarity to the C. elegans protein Fer-1 (Fertilization defective-1). In C. elegans, the full-length dysferlin homolog Fer-1 (230 kDa) is present early in spermatogenesis and becomes less abundant in spermatids, perhaps because it is proteolytically processed into smaller isoforms (195 and 180 kDa) (Washington and Ward, 2006). Fer-1 is required for the fusion of specialized vesicles, called membranous organelles, with the plasma membrane during spermatogenesis (Achanzar and Ward, 1997; Washington and Ward, 2006).
Two other ferlin proteins have been identified in humans, myoferlin and otoferlin. Myoferlin was first considered as a muscle specific protein and its deletion results in impaired mouse myoblast fusion into mature skeletal myotubes (Doherty et al., 2005). However, myoferlin has been shown to be critical for endocytosis in endothelial cells (Bernatchez et al., 2009). Myoferlin is also abundantly expressed in invasive breast tumor cells and remarkably, its depletion stalls the invasion of these cells (Li et al., 2012). Otoferlin is essential for hearing and pathogenic mutations are associated with nonsynchronic autosomal recessive deafness (Yasunaga et al., 1999). Otoferlin is required in the priming and fusion of synaptic vesicles during sound encoding, which occurs at synapses between cochlear inner hair cells and the auditory nerve (Pangrsic et al., 2012).
Ferlin proteins function in various developmental and reproductive processes in several organisms. While dysferlin and myoferlin are co-expressed in the human placenta, only dysferlin expression is positively correlated to cell fusion in trophoblastic cells (Robinson et al., 2009). In Drosophila melanogaster, only one ferlin gene, called misfire (mfr), has been found. The transcript is expressed in the testis and ovaries of adult flies. In males, mfr expression is required for efficient breakdown of the sperm plasma membrane and completion of sperm activation during fertilization. In females, it is required for egg patterning by limiting the spread of proteins such as Gurken. Mutations in misfire delay embryogenesis (Ohsako et al., 2003; Smith and Wakimoto, 2007). In zebrafish, the dysferlin transcript is first detected at 11h post-fertilization, remains detectable until 96h, and is expressed in different tissues including muscle, brain and eye (Kawahara et al., 2011). Reduction of dysferlin expression in this animal by morpholino injection causes abnormal formation of muscle structures (Kawahara et al., 2011). The dysferlin gene is also present in the sea urchin genome (Sodergren et al., 2006) and this animal was used to test the role of dysferlin in membrane healing (Covian-Nares et al., 2010). Laser wounding of one cell in a 4-cell stage embryo triggered calcium spikes in neighboring cells through voltage-gated calcium channels whereas embryos injected with a dysferlin morpholino lost their calcium spike activity in neighboring cells after wounding. The authors conclude that the dysferlin-depleted embryos have lost an ability to receive a signal, possibly a nucleotide, released by the wounded cell.
Here, we found that dysferlin is enriched in the plasma membranes of oocytes and embryos from the sea star, an animal with oocytes and embryos tractable for studying cellular dynamics (Fresques et al., 2013; Hinman et al., 2003a; Wessel et al., 2010). Sea stars are invertebrate deuterostomes, a sister group to chordates. Oocytes, eggs and embryos of the species used here, Patiria miniata, are optically clear, enabling excellent observations of development. Developing oocytes are plentiful, manipulatable, are arrested in prophase of first meiotic division and meiotic reinitiation is induced by the hormone 1-methyladenine (Kanatani et al., 1969). After fertilization, embryos develop quickly and reach the blastula stage in one day, and gastrulate in two days. In this study, we show that in the starfish Patiria miniata (Pm), knockdown of the dysferlin protein results in reduced endocytosis during oogenesis and significant developmental defects in embryos.
Materials and methods
Animals
P. miniata and A. forbesi were housed in aquaria with artificial seawater (ASW) at 16°C (Coral Life Scientific Grade Marine Salt; Carson, CA). Gametes were acquired by opening up the animals. Oocytes were collected in filtered seawater and sperm was collected dry. To obtain mature oocytes, the full-grown immature oocytes were incubated for an hour in filtered sea water containing 2 µM 1-methyladenine. After addition of sperm, fertilized eggs were cultured in filtered seawater at 16°C (Foltz et al., 2004; Wessel et al., 2010).
Isolation of plasma membrane-enriched fractions
Full-grown immature or mature oocytes were isolated in calcium-free sea water (CFSW), resuspended in buffer A (Sucrose 0.36M, 50 mM Tris pH8.5mM EDTA) and homogenized on ice by five up and down strokes of a loose-fitting glass pestle in a 40mL glass dounce tissue grinder. Following a 15-minute incubation on ice, the sample was homogenized with an additional five strokes and the homogenate was then incubated at 4°C overnight to settle the plasma membrane-enriched fraction (PMEF) by gravity. This fraction was washed three times with buffer A and the membranes were collected for analysis, or pelleted in a microfuge at 14 000g for 5 minutes. For SDS-PAGE, the pellet was resuspended in Laemmli Sample Buffer (LSB) containing 5mM DTT, and the samples were boiled for 5 min at 100°C.
Rhodamine-B-Isothiocynate (RITC) labeling of oocyte surface proteins
Full-grown immature oocytes were isolated in CFSW and all labeling steps were performed in CFSW. Oocytes were pelleted by gentle centrifugation and rotated at 16°C for about 90 min in a 15 ml conical centrifuge tube containing 25µl of RITC/1ml CFSW. Oocytes were then rinsed with filtered SW from the Marine Biological Laboratories (MBL; Woods Hole, MA) three times. An aliquot (200 µl) of RITC-labeled whole oocytes was pelleted and suspended in 200 µl of 2× LSB containing 5 mM DTT for SDS-PAGE analysis. PMEFs were isolated from these labeled oocytes using the protocol described above. Images were taken with a fluorescence dissecting stereoscope (Olympus SZX16) connected to an Olympus X cite 120 lamp and to a camera (Nikon D90), managed by Pro 2 software. Samples were electrophoresed through a 4–12% SDS-PAGE, stained with Coomassie Brilliant Blue for 1h and the RITC signal was detected using a Typhoon Imager (GE Healthcare, Typhoon 9410).
Mass spectrometry and protein analysis
PMEF was loaded on a SDS-PAGE for Coomassie Brilliant Blue staining. The two main high molecular weight proteins were obtained and processed for in gel digestion using the In gel tryptic digestion kit (Pierce). Proteins were digested overnight at 37°C in presence of 10 ng/µl trypsin. Samples were analyzed using a Thermo-Finnigan LTQ linear ion trap mass spectrometer. Tryptic peptides were fractionated on a reversed phase column and introduced directly onto an LTQ mass spectrometer via electrospray ionization. Pm-dysferlin was identified via database matching with the program SEQUEST (Eng, 1994), using the Pm ovary transcriptome (Adrian Reich and Gary Wessel, unpublished data).
EMBOSS Needle was used to obtain protein identities and similarities between the species (EMBL-EBI). SOSUI was used to identify the transmembrane domains (Mitaku et al., 2002). CDD (Conserved Domain Database) was used to identify the protein domains (Marchler-Bauer et al., 2011). The DysF domains of the human dysferlin protein were also defined according to a previous analysis (Lek et al., 2010). YinOYang (Gupta and Brunak, 2002), NetPhos (Blom et al., 1999), Sulfinator (Monigatti et al., 2002), and ProP (Duckert et al., 2004) were used to analyze the potential post-translational modifications of the proteins.
Antibody production
Rabbit polyclonal antibody against the peptide sequence ALPRPKFSDSTGKI of dysferlin from P. miniata was custom developed and affinity purified by GenScript (Piscataway, NJ). This peptide, specific to dysferlin, is located in the DysFN domain (amino-acids 889 to 902).
Western blot
Western blot analysis was performed following electrophoretic transfer of proteins from SDS-PAGE onto 0.22µm nitrocellulose membranes (Towbin et al., 1979). Membranes were incubated with antibodies directed against Pm-dysferlin (1/5000) in 20mM Tris-HCl (pH7.6), 3% BSA, and 0.1% Tween-20, overnight at 4°C. The antigen-antibody complex was measured by chemiluminescence using horseradish peroxidase-coupled secondary antibodies according to the manufacturer’s instruction (ECL; GE Healthcare Biosciences, Pittsburgh, PA, USA).
Real-time quantitative PCR (QPCR)
RNA was extracted from the different developmental stages using the RNeasy Micro Kit (Qiagen; Valencia, CA). cDNA was prepared using the TaqMan ® Reverse Transcription Reagents kit (Applied Biosystems; Foster City, CA). QPCR was performed on a 7300 Real-Time PCR system (Applied Biosystems; Foster City, CA) with the FastStart Universal SYBR Green Master Kit (Roche). Pm-dysferlin was amplified using the following primers: F (5’-AACGAAGCCCTGGAAGAGAT-3’) and R (5’- GCATGGAGACCGGAGTAGAG-3’). Experiments were run in triplicate, and the data were normalized to ubiquitin RNA levels (Hinman and Davidson, 2003; Hinman et al., 2003b). Pm-ubiquitin was amplified using the primers F (5’-TTCGGTGAAAGCCAAGATTC-3’) and R (5’-CCCACCTCTCATGGCTAGAA-3’).
Whole mount RNA in situ hybridization (WMISH)
The sequence used to make antisense WMISH probes for Pm-dysferlin was amplified from Pm ovary cDNA and cloned into pGEM T-Easy (Promega). Pm-dysferlin was amplified using the following primers: F (5’-ATACGCGTGTGGGTTGCCTGG-3’) and R (5’-AGCGCGCTACACGCTGATCG-3’). The pGEM T-Easy plasmids were linearized using NdeI (T7 transcription) (Promega; Madison, WI). Antisense, DIG-labeled RNA probes were constructed using a DIG RNA labeling kit (Roche; Indianapolis, IN). WMISH experiments were performed as described previously (Minokawa et al., 2004), and the alkaline phosphatase reaction was carried out for 1h. A non-specific DIG-labeled RNA probe complementary to neomycin resistance gene, obtained from the pSport 18 (Roche; Indianapolis, IN) was used as a negative control. Samples were imaged on a Zeiss Axiovert 200M microscope equipped with a Zeiss color AxioCam MRc5 camera (Carl Zeiss, Inc.; Thornwood, NY).
Immunofluorescence
Oocytes and embryos were cultured as described above, and samples were collected at the indicated stages for whole-mount antibody labeling. The fertilization envelope was manually removed in the early stages. Cells were fixed in 4% paraformaldehyde in ASW and washed with PBS before being treated with 0.1% Triton X-100 for 20 minutes. After 3 washes with PBS-Tween 20, oocytes and embryos were blocked for an hour at room temperature in blocking buffer: PBS-0.05% Tween 20–3% BSA (Sigma; St. Louis, MO). For labeling, the cells were incubated overnight at 4°C with either the anti Pm-dysferlin purified antibody diluted by 1:500 (final concentration: 2µg/ml) in blocking buffer, or this antibody pre-incubated for 2 hours at room temperature with the corresponding blocking peptide used at the final concentration of 40µg/ml. The cells were washed 3 times with PBS-Tween 20, and then incubated with an anti-rabbit Alexa Fluor 488 (green) (Life technologies, Grand Island, NY), diluted by 1:500 in blocking buffer for two hours at room temperature. Oocytes and embryos were then washed 3 times with PBS-Tween 20. Images were captured using a LSM 510 laser scanning confocal microscope (Carl Zeiss, Inc.; Thornwood, NY).
Morpholino injection
Oocytes were injected with the dysferlin morpholino 5’- TCGACACAATCACCATCAGC GACAT-3’ and incubated for 36h before in vitro maturation. An irrelevant morpholino (the sea urchin, Strongylocentrotus purpuratus, SCP2) was used as a control: 5'-GGACATACTTGTCAGGTCGTGCCAA-3'. The injection solution contained 1mM of morpholino. Each young and full-grown oocyte received approximately 1.4 and 0.5 nM of morpholino respectively. Embryos were imaged on a Zeiss Axiovert 200M microscope equipped with a Zeiss color AxioCam MRc5 camera (Carl Zeiss, Inc.; Thornwood, NY).
In vitro endocytosis assay
Morpholinos injected oocytes were incubated in FSW for 36–48 h at 16°C and then transferred to filtered sea water containing 15µg/ml of FM1–43 (Molecular Probes, Grand Island, NY) and incubated for additional 3–4h at 16°C. Oocytes were then washed in FSW and observed at the confocal microscope for monitoring the presence of endocytic vesicles.
Results
Dysferlin is enriched in the plasma membrane of oocytes
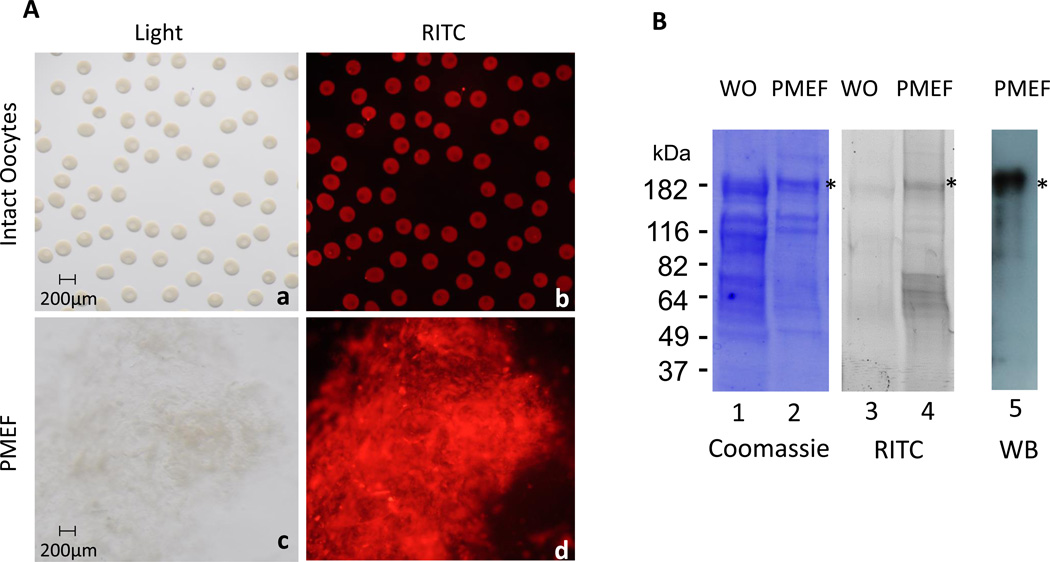
To identify the proteins expressed in the plasma membrane of the full-grown immature oocytes, the surface was labeled with RITC and the labeled membranes were isolated (Figure 1A). Proteins obtained from whole oocytes or plasma membrane-enriched fractions (PMEFs) were electrophoresed in a 4–20% SDS-PAGE and visualized by Coomassie Blue staining and by RITC fluorescence imaging (Figure 1B). Two bands of high molecular mass, flanking the 182 kDa marker (*) were reproducibly observed in the plasma membrane fraction. These were prepared for mass spectrometry analysis and the resulting peptide sequences were identified by a SEQUEST search against the P. miniata ovary transcriptome (Adrian Reich and Gary Wessel, unpublished data). Dysferlin was identified with two dysferlin peptides from both bands (Figure S1). The largest dysferlin transcript (Pm 1104) found in the Pm transcriptome contains an open reading frame of 6240 nucleotides that encodes for a 2080 aa protein, with an expected size of 233 kDa. An affinity-purified rabbit polyclonal antibody was generated against Pm-dysferlin and immunoblot analysis on the PMEF led to a specific signal surrounding the 182kD marker (Figure 1B), supporting the identity of the isolated protein. PMEFs were also isolated from a second sea star species, Asterias forbesi (Af), and a main band, also present at 182 kDa, was observed (Figure S2). An Af-dysferlin transcript was identified from the Af ovary transcriptome (Adrian Reich and Gary Wessel, unpublished data), and similar to the Pm-dysferlin, it contained an open reading frame of 6231 nucleotides, encoding a cognate protein of 2077 amino acids in length.
Figure 1. Dysferlin is expressed in the cortex of sea star oocytes.
(A) Proteins present at the surface of Pm oocytes were labeled with RITC. Images were taken on intact oocytes (b), and after purification of the plasma membranes (d). The corresponding bright field images are respectively shown in a and c. (B) RITC labeled proteins from whole oocytes (WO) or from the plasma membrane-enriched fraction (PMEF) were loaded on SDS-PAGE and visualized by either Coomassie staining (lanes 1–2) or RITC fluorescence (lanes 3–4). Two bands (*) near the 182 kDa marker were reproducibly found enriched in the plasma membrane fraction and identified as Pm-dysferlin by mass spectrometry. A signal at the similar molecular weight was obtained by western blot using the affinity-purified antibody against Pm-dysferlin on the PMEF (lane 5).
Dysferlin is highly conserved between human and sea star
The alignment of amino acid sequences indicates that Pm-dysferlin is highly conserved with the human dysferlin: Pm and Af-dysferlin are 85.6% identical and 92.4% similar. Human dysferlin shares 46.9% of identity and 63.2% of similarity with Pm, and 46.9% of identity and 63.7% of similarity with Af. Moreover, the same domains are found in the dysferlins of these three species (Figure S3). In both human and sea star, dysferlin contains six main C2 domains. The C2 domain (approximately 130 residues) was first identified in the Ca2+-dependent protein kinase C (PKC) and is involved in Ca2+-dependent membrane binding. It is also found in presynaptic proteins such as synaptotagmins (Cho and Stahelin, 2006). Dysferlin also contains 2 highly conserved DysF domains that are tandemly repeated. The presence or absence of this domain in ferlin family members is used to define two subgroups: dysferlin and myoferlin (presence) and otoferlin (absence) (Lek et al., 2012; Lek et al., 2010), although the role of this domain is not fully understood. However, the DysF domain appears more susceptible to retain mutations and is involved in dysferlinopathy (Patel et al., 2008). FerA and FerB, which lie in the long stretch between C2C and C2D, are also conserved between human and sea star and both domains are unique to ferlin proteins. FerB is conserved in all ferlins, while FerA is only found in DysF containing ferlins (Type 1), suggesting FerA and DysF may have complementary or additive function (Lek et al., 2010). The FerI domain, which lies between the C2B and C2C domains, is conserved in Hs, Pm and Af. This domain was also identified in ferlin-like proteins of protozoan Apicomplexan parasites Plasmodium (malaria), Theileria (East Coast fever), Babesia (tick fever), and Toxoplasma (toxoplasmosis), that account for significant worldwide mortality and morbidity amongst humans and livestock (Lek et al., 2010). Finally, human and sea star dysferlin contain a transmembrane domain similarly placed near the C-terminus of the protein (Bansal and Campbell, 2004).
The dysferlin transcript is expressed throughout oogenesis and embryogenesis in sea star
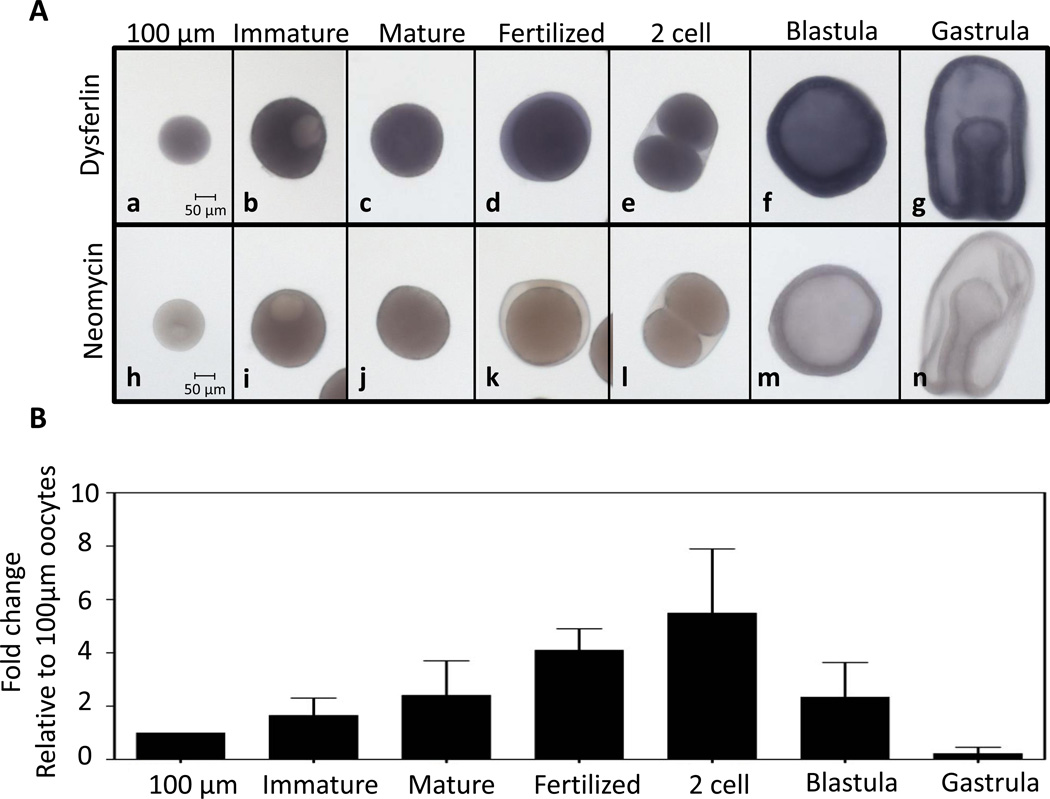
To determine the temporal expression of the dysferlin transcript in Pm, an RNA probe was synthesized for in situ hybridization and a probe against the neomycin-resistance gene was used as a negative control (Figure 2A). Pm-dysferlin mRNA accumulates uniformly in the stages tested, from young oocytes (100µm diameter) to gastrula. Quantitative PCR was used to measure the relative RNA level in young oocytes, full-grown immature oocytes (>160 µm), mature oocytes, fertilized eggs, two-cell stage, blastula and gastrula (Figure 2B). All values were normalized against Pm-ubiquitin mRNA. Although differences were seen in qPCR signals, these were not significantly different (p>0.05) between the 100µm oocytes and each of the other stages. These results indicate that the transcript level of dysferlin in Pm uniformly accumulates throughout oogenesis and early development.
Figure 2. Pm-dysferlin mRNA is uniformly expressed throughout oogenesis and embryogenesis.
(A) Whole mount in situ hybridization in the sea star P. miniata, using probes against Pm-dysferlin in 100µm oocytes (a), immature full-grown oocytes (b), mature oocytes (c), 30 minutes after fertilization (d), at the two-cell stage (e), in blastula (f), and in gastrula (g). Neomycin resistance gene is used as a negative control (h-n). (B) qPCR was used to measure the RNA levels of Pm-dysferlin at the indicated developmental stages: 100 µm oocytes, full-grown immature oocytes, mature oocytes, 30 minutes after fertilization, two-cell stage, at blastula, and gastrula. All values were normalized against the Pm-ubiquitin mRNA and represented as a fold-change relative the amount of RNA present in the 100 µm oocytes. Significance was assessed using One-Way ANOVA test. No significant differences were obtained at p<0.05.
The dysferlin protein is enriched in the plasma membranes of oocytes and embryos
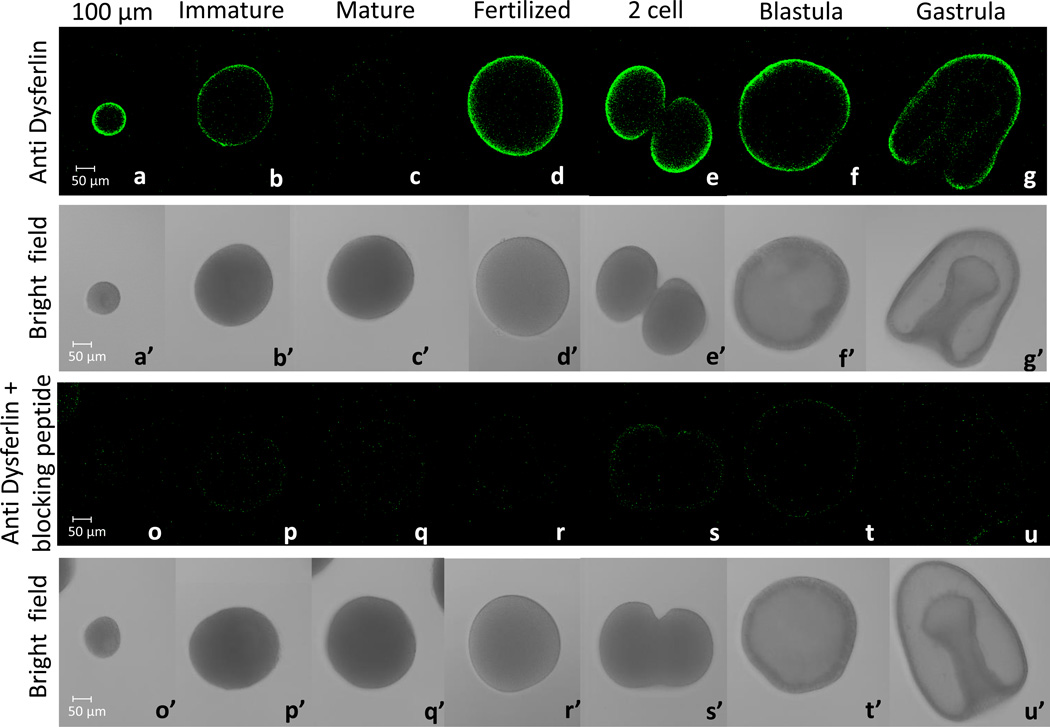
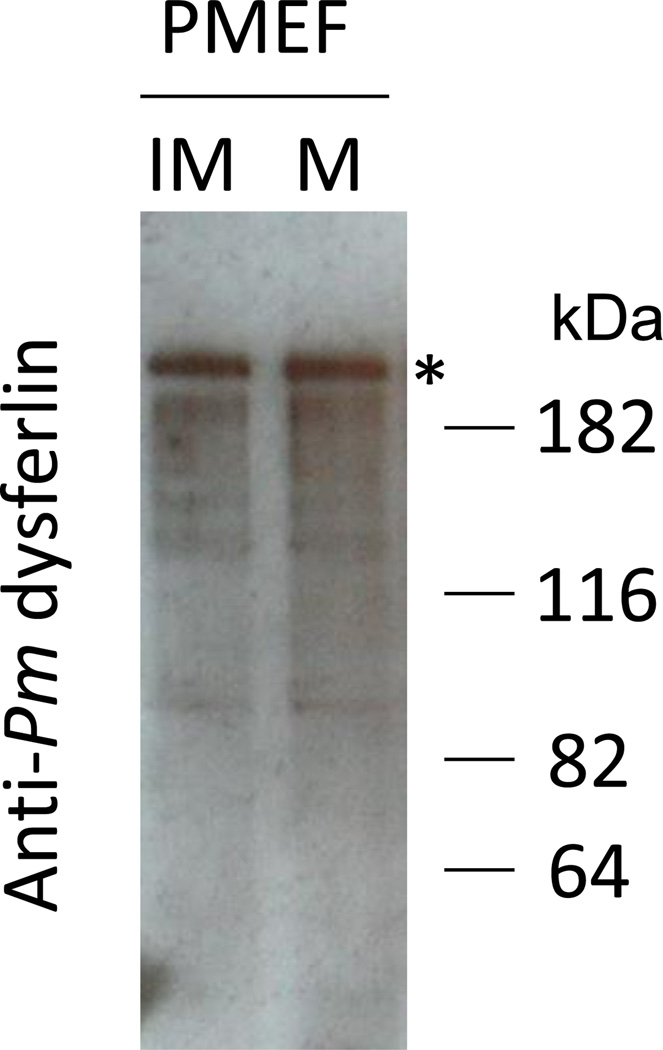
An affinity-purified rabbit polyclonal antibody was generated against Pm-dysferlin and used to determine dysferlin localization by immunofluorescence (Figure 3). Pm-dysferlin is detected at the cortex of the oocytes during oogenesis. Interestingly, dysferlin is highly expressed in young oocytes (Figure 3a) and decreases in full-grown immature oocytes (Figure 3b). Pm-dysferlin is barely detectable in mature oocytes (Figure 3c) but is readily detectable at the plasma membrane right after fertilization (Figure 3d) and at the two-cell stage (Figure 3e). The protein is also detected in blastula (Figure 3f) and gastrula stages (Figure 3g). No immunofluorescence signal was detected under the same conditions when the cells were exposed to the Pm-dysferlin antibody preincubated with the corresponding blocking peptide (Figure 3o–u). Moreover, the expression of Pm-dysferlin protein in oocytes was further tested by Western blot analysis after enrichment of the plasma membranes (Figure 4). A similar level of Pm-dysferlin was obtained in the fractions from immature and mature oocytes, suggesting that in mature oocytes, even though the protein cannot be detected by immunofluorescence, Pm-dysferlin is present and associated with the plasma membrane.
Figure 3. The Pm-dysferlin protein is expressed during oogenesis and embryogenesis.
Immunofluorescence using the antibody against Pm-dysferlin (a,b,c,d,e,f,g) on 100 µm diameter oocytes (a), full-grown immature oocytes (b), mature oocytes (c), fertilized eggs (d), two-cell stage (e), blastula (f), and gastrula (g). The corresponding bright field images are respectively shown in a’ to g’. To test the specificity of the Pm-dysferlin antibody, a control of immunofluorescence was performed by pre-incubating this antibody with its corresponding blocking peptide (o - u). The corresponding bright field images are respectively shown in o’ to u’. Images were taken using the same microscope settings (laser intensity, pinhole opening).
Figure 4. The Pm-dysferlin protein is expressed in the plasma membrane of both immature and mature oocytes.
A signal above 182kDa was obtained by western blot using the affinity-purified antibody against Pm-dysferlin on PMEFs from immature (IM) or mature (M) oocytes.
Endocytosis is reduced in Pm-dysferlin knockdown oocytes
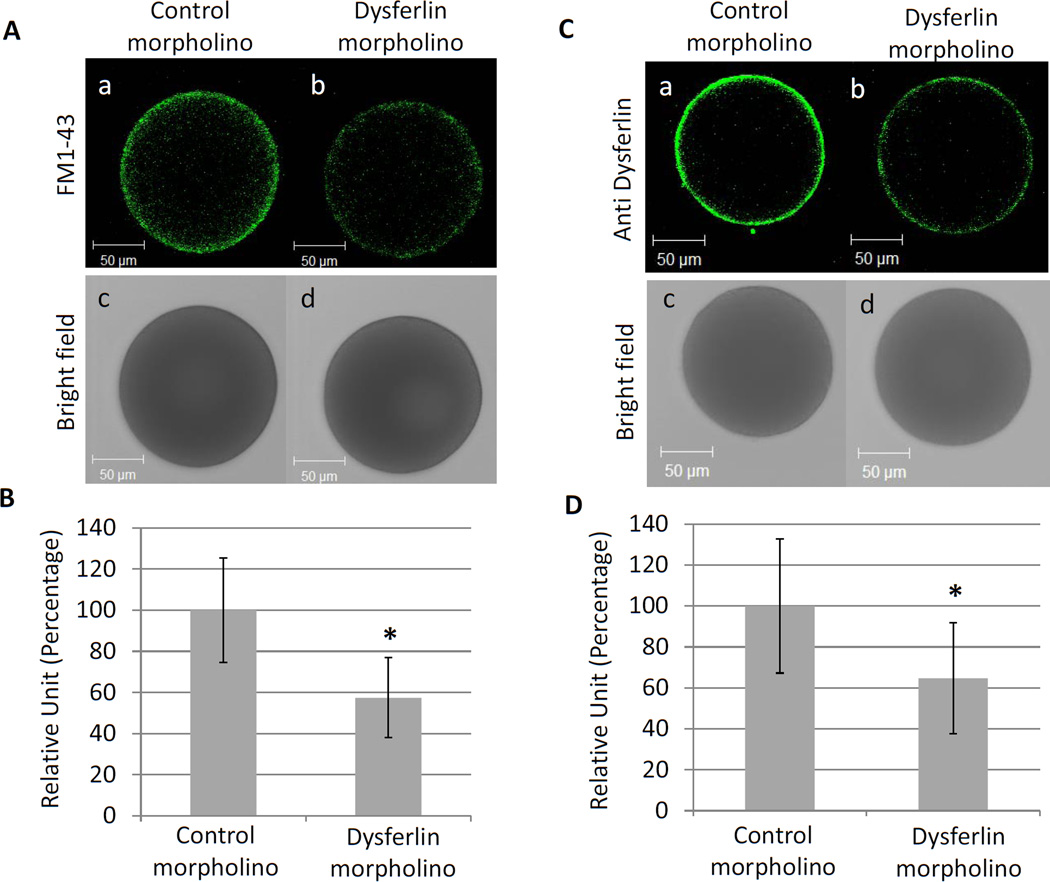
To test if dysferlin plays a role in endocytosis during oogenesis in the sea star, a morpholino was injected into young oocytes to block dysferlin translation (Figure 5). Although these dysferlin-depleted oocytes remained viable, endocytosis was decreased by more than 40% in the dysferlin injected oocytes, compared to oocytes injected with a control morpholino (Figure 5A and 5B). The efficiency of the morpholino was tested by immunofluorescence using the antibody against Pm-dysferlin, and quantification revealed that the morpholino reduced the protein expression by 35% (Figure 5C and 5D), suggesting that not only was dysferlin actively being synthesized in the oocyte, it was also being actively degraded. A strong correlation was seen between the decrease in endocytosis and the decrease in dysferlin expression.
Figure 5. Endocytosis is reduced in dysferlin knockdown oocytes.
(A) Young oocytes were injected by either a control (a) or the dysferlin morpholino (b) and incubated with FM1–43 to test endocytosis. The corresponding bright field images are respectively shown in (c) and (d). (B) The fluorescence obtained in (A) was quantified using Metamorph. Twenty four (with an average size of 137.8 ± 8.6 µm) and twenty five (with an average size of 139.1 ± 9.4 µm) injected oocytes were quantified respectively for the control and the dysferlin morpholino. Significant difference (*) was observed between the control and the dysferlin morpholino using Student’s t-test, P<0.05. (C) The expression of Pm-dysferlin decreases in oocytes after morpholino injection. Immunofluorescence on young oocytes injected with either a control (a) or the dysferlin morpholino (b). The corresponding bright field images are respectively shown in (c) and (d). (D) The fluorescence obtained in (C) was quantified using Metamorph. Twenty (with an average size of 138 ± 7.2 µm) and twenty two oocytes (with an average size of 140.7 ± 8.2 µm) injected oocytes were used respectively for the control and for dysferlin morpholino. Significant difference (*) was observed between the control morpholino and the dysferlin morpholino using Student’s t-test, P<0.05.
Pm-dysferlin knockdown induces a developmental arrest at gastrula stage
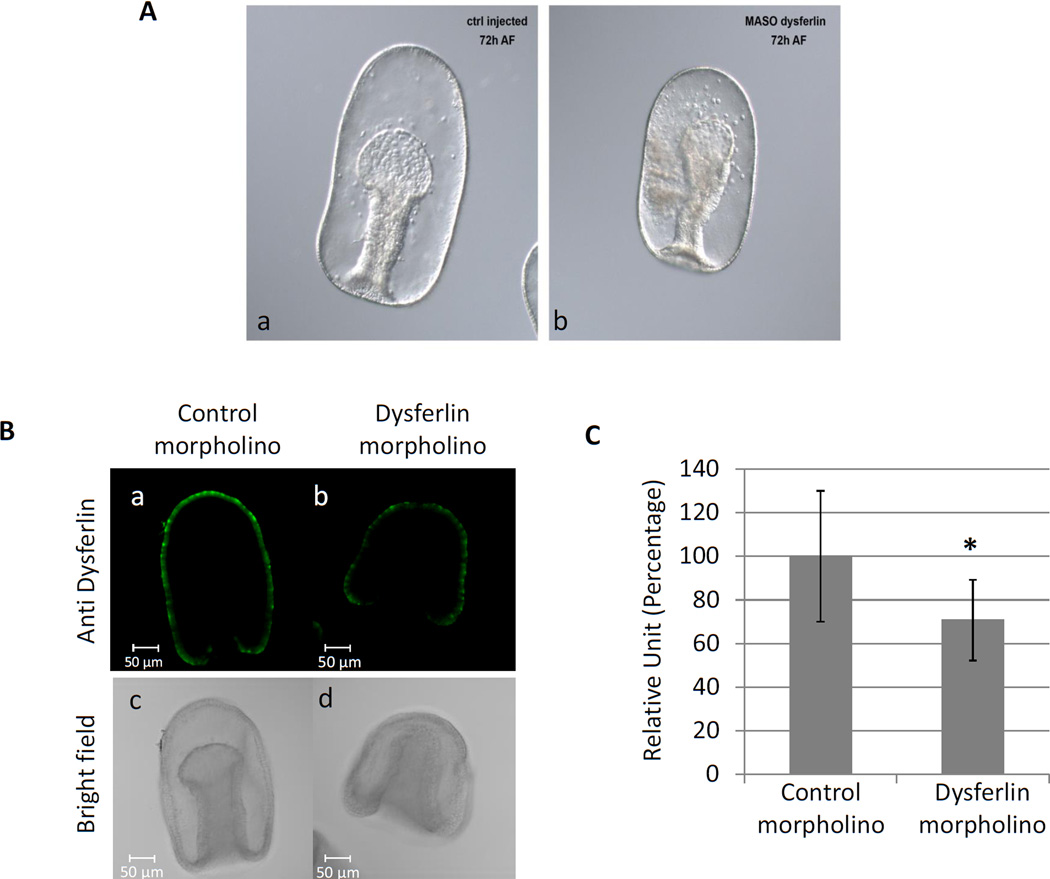
To determine the function of dysferlin during embryonic development in the sea star, the same morpholino was injected into full-grown immature oocytes. Injection of dysferlin morpholinos did not affect oocyte maturation, fertilization, or early development in Pm when compared to the control (data not shown). Interestingly, dysferlin morpholino-injected embryos were smaller, arrested at gastrula (Figure 6A), and led to a disorganization of the mesenchyme cells and the archenteron. The efficiency of the morpholino was tested by immunofluorescence using the antibody against Pm-dysferlin (Figure 6B); the morpholino decreased the expression of the protein significantly, on average by 30% during this developmental window (Figure 6C).
Figure 6. Knockdown of Pm-dysferlin leads to a developmental arrest at gastrula stage.
(A) Immature oocytes were injected by either a control (a) or the dysferlin morpholino (b). After maturation and fertilization, the embryos were cultured until the gastrula stage, 72 hours after fertilization (72 AF). (B) The expression of Pm-dysferlin decreases in embryos after morpholino injection. Immunofluorescence on embryos injected with either a control (a) or the dysferlin morpholino (b). The corresponding bright field images are respectively shown in (c) and (d). (C) The fluorescence obtained in (B) was quantified using Metamorph. Nine and eight injected embryos were used respectively for the control and for dysferlin morpholino. Significant difference (*) was obtained between the control morpholino and the dysferlin morpholino using Student’s test, P<0.05.
Discussion
Dysferlin is a transmembrane protein involved in membrane fusion and membrane repair but a paucity of information limits our understanding of its function during oogenesis and embryogenesis. Only one dysferlin transcript was found in the Pm ovary transcriptome (Pm_1104), but since the complete Pm genome has not been annotated yet, additional dysferlin isoforms could be expressed at a lower level and not detected in the transcriptome. However, three transcripts were found for Pm-otoferlin (Pm_10061, Pm_54312, Pm_21293) although we did not follow-up on their function in these cells. We found that dysferlin is highly enriched in the cortex of oocytes (Figure 3) though it may also be present below detectable thresholds in other cellular compartments. In bovine aortic endothelial cells (BAEC), a GFP-tagged dysferlin was also found in the nucleus and in the Golgi apparatus (Sharma et al., 2010). Ferlin domains also have similarity to several nucleolar protein domains (Staub et al., 2004). The nucleolus is the center of ribosome biogenesis (Tschochner and Hurt, 2003) but is also able to immobilize proteins as a post translational regulatory mechanism (Audas et al., 2012).
We learned that dysferlin could be knocked down in young sea star oocytes by morpholinos (Figure 5). This result is important since it shows that the oocyte is constantly undergoing protein turnover and synthesis of dysferlin, and by extension likely other proteins as well. Even with dysferlin being a transmembrane protein, turnover is robust in these cells, opening the opportunity to knock-down and test other oocyte-associated factors for functionality in for example, maturation, fertilization, and early development. Dysferlin could be required for the fusion of specific vesicles with the plasma membrane to add extra membrane at the fusion site during oocyte growth, as described in C. elegans during spermiogenesis (Washington and Ward, 2006) but how this mechanism might work is not well understood. We could envision a feedback system whereby if exocytosis is reduced by the dysferlin knock-down, then endocytosis would be reduced to retain total plasma membrane surface area. Endocytosis is a critical process utilized by growing oocytes to import yolk protein precursors (Brooks and Wessel, 2004; Opresko and Wiley, 1987). In C.elegans, endocytosis of cell surface proteins was also found to be critical for the regulation of meiotic maturation in oocytes (Cheng et al., 2008). Studies in both Xenopus (El-Jouni et al., 2007) and mouse (Lowther et al., 2011) reveal that endocytosis occurs in oocytes, and contributes to cAMP signaling to regulate meiotic arrest. The physiological function of endocytosis in the sea star oocyte is currently under investigations.
Interestingly, we found by Western blot that the dysferlin protein is expressed at the plasma membrane in mature oocytes, even though it is undetectable by immunofluorescence in situ at this, but not other stages. The antibody is directed against a single peptide (amino acids 889 to 902) contained in the DysFN domain (Figure S3) and one possible explanation for this paradox is that in mature oocytes, the protein might have either changed its conformation or bound a new partner, preventing the interaction with the antibody. The DysF domains have maintained high sequence conservation during evolution (Lek et al., 2010) and in humans, mutations of this domain leads to dysferlinopathy, almost certainly due to degradation of the unfolded protein (Patel et al., 2008). The solution structure of the inner DysF domain of the dysferlin paralogue myoferlin has been solved. It consists of two long β-strand (residues 929–939 and 1018–1028) oriented antiparallel with each other. The intervening 77 residues form a long loop which packs against both sides of the central β-hairpin; several regions of this loop form short stretches of β-strand. The region annotated by SMART as DysFN supplies the first strand and around two-thirds of the loop and the region annotated as DysFC provides a third of the loop and the second strand (Patel et al., 2008). In Pm mature oocytes, the protein also could be post-translationally modified near the immune peptide site, resulting in a change of its conformation. A YinOYang prediction analysis suggests that serine 896 included in the targeted peptide can be modified by phosphorylation and by addition of O-linked β-N-acetylglucosamine. An additional NetPhos analysis predicts that serines 896 and 898 can be phosphorylated. These specific modifications might have been lost during the plasma membrane enrichment, leading to the recognition of the protein by immunoblot but not in situ. Moreover, additional modifications or cleavage of the protein, outside of the peptide sequence, could also change the conformation of the entire protein. Sulfinator predicts that four tyrosines in the protein can be sulfated, and ProP predicts three arginine or lysines that could be used as cleavage sites. In mouse, dysferlin has been found to interact with the histone deacetylase 6 via its C2D domain, and with the alpha tubulin through its C2A and C2B domain during myogenesis (Azakir et al., 2010; Di Fulvio et al., 2011), Dysferlin can also form a dimer in vitro, and in living adult skeletal muscle fibers isolated from mice Calcium insensitive C2 domains mediate a high affinity self-association of dysferlin in a parallel homodimer, leaving the Ca-sensitive C2A domain free to interact with membranes (Xu et al., 2011). In Pm mature oocytes, dysferlin could be interacting with new partners and/or forming dimers, reducing its accessibility to the antibody and thereby making this reagent a functional reporter.
Dysferlin is also expressed during early development after fertilization, and through blastula and gastrula stages (Figure 3). Reduction of its expression did not lead to any developmental defects during the early developmental periods. This may be a result of low knockdown efficiency, but it could also suggest that dysferlin is not essential right after fertilization, during normal development. Taking into account the role of dysferlin in membrane wounding in sea urchin 4-cell stage embryos (Covian-Nares et al., 2010), the main function of this protein, right after fertilization, could be in membrane sealing during the rapid cellular divisions, but is expendable, and perhaps compensated by other membranous proteins. Interestingly, this reduction of dysferlin expression leads to a developmental arrest during gastrulation, as shown by the morpholino injection experiments (Figure 6). Injection of the dysferlin morpholino in sea star seems to disorganize the mesenchyme cells and the archenteron. These data suggest that dysferlin could be required for the cellular rearrangements needed for the elongation of the archenteron, and/or for cellular signaling that may be important for stimulating this process. Sea stars appear to be a good model to study the function of dysferlin and the role of each domain within a context of oogenesis and embryogenesis.
Supplementary Material
Highlights.
A new protocol is documented for isolating the cell surface from sea star oocytes.
Cell surface labeling and proteomic analysis identified the calcium – binding protein dysferlin.
Protein knock down approaches in the oocyte revealed dysferlin as an important element in the endocytic process.
Acknowledgments
We thank Jim Clifton, Proteomics Facility Manager, for his help with mass spectrometry and Adrian Reich for sharing his transcriptome sequences and expertise in their screening. TMO was a Visiting Professor sponsored by the American Society for Cell Biology Minorities Affairs Committee from 2010–2012 (funding provided from NIH grant #GM008622). We appreciate C. Ookie for his sharing with us the biotintylation protocol. We gratefully acknowledge NSF grant IOS-1120972 to GMW. This research is based in part upon work conducted using the Rhode Island NSF/EPSCoR Proteomics Share Resource Facility, which is supported in part by the National Science Foundation EPSCoR Grant No. 1004057, National Institutes of Health Grant No. 1S10RR020923, S10RR027027 (Orbitrap XL ETD Mass Spectrometer), a Rhode Island Science and Technology Advisory Council grant, and the Division of Biology and Medicine, Brown University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achanzar WE, Ward S. A nematode gene required for sperm vesicle fusion. Journal of cell science. 1997;110(Pt 9):1073–1081. doi: 10.1242/jcs.110.9.1073. [DOI] [PubMed] [Google Scholar]
- Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Molecular cell. 2012;45:147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Azakir BA, Di Fulvio S, Therrien C, Sinnreich M. Dysferlin interacts with tubulin and microtubules in mouse skeletal muscle. PloS one. 2010;5:el0122. doi: 10.1371/journal.pone.0010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal D, Campbell KP. Dysferlin and the plasma membrane repair in muscular dystrophy. Trends in cell biology. 2004;14:206–213. doi: 10.1016/j.tcb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Bashir R, Britton S, Strachan T, Keers S, Vafiadaki E, Lako M, Richard I, Marchand S, Bourg N, Argov Z, Sadeh M, Mahjneh I, Marconi G, Passos-Bueno MR, Moreira Ede S, Zatz M, Beckmann JS, Bushby K. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nature genetics. 1998;20:37–42. doi: 10.1038/1689. [DOI] [PubMed] [Google Scholar]
- Bernatchez PN, Sharma A, Kodaman P, Sessa WC. Myoferlin is critical for endocytosis in endothelial cells. Am J Physiol Cell Physiol. 2009;297:C484–C492. doi: 10.1152/ajpcell.00498.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. Journal of molecular biology. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Brooks JM, Wessel GM. The major yolk protein of sea urchins is endocytosed by a dynamin-dependent mechanism. Biology of reproduction. 2004;71:705–713. doi: 10.1095/biolreprod.104.027730. [DOI] [PubMed] [Google Scholar]
- Cheng H, Govindan JA, Greenstein D. Regulated trafficking of the MSP/Eph receptor during oocyte meiotic maturation in C. elegans. Current biology : CB. 2008;18:705–714. doi: 10.1016/j.cub.2008.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochimica et biophysica acta. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Covian-Nares JF, Koushik SV, Puhl HL, 3rd, Vogel SS. Membrane wounding triggers ATP release and dysferlin-mediated intercellular calcium signaling. J Cell Sci. 2010;123:1884–1893. doi: 10.1242/jcs.066084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Morree A, Flix B, Bagaric I, Wang J, van den Boogaard M, Grand Moursel L, Frants RR, Ilia I, Gallardo E, Toes R, van der Maarel SM. Dysferlin regulates cell adhesion in human monocytes. The Journal of biological chemistry. 2013;288:14147–14157. doi: 10.1074/jbc.M112.448589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fulvio S, Azakir BA, Therrien C, Sinnreich M. Dysferlin interacts with histone deacetylase 6 and increases alpha-tubulin acetylation. PloS one. 2011;6:e28563. doi: 10.1371/journal.pone.0028563. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Doherty KR, Cave A, Davis DB, Delmonte AJ, Posey A, Earley JU, Hadhazy M, McNally EM. Normal myoblast fusion requires myoferlin. Development. 2005;132:5565–5575. doi: 10.1242/dev.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein engineering, design & selection : PEDS. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- El-Jouni W, Haun S, Hodeify R, Hosein Walker A, Machaca K. Vesicular traffic at the cell membrane regulates oocyte meiotic arrest. Development. 2007;134:3307–3315. doi: 10.1242/dev.005454. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., Ill An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Foltz KR, Adams NL, Runft LL. Echinoderm eggs and embryos: procurement and culture. Methods Cell Biol. 2004;74:39–74. doi: 10.1016/s0091-679x(04)74003-0. [DOI] [PubMed] [Google Scholar]
- Fresques T, Zazueta-Novoa V, Reich A, Wessel GM. Selective accumulation of germ-line associated gene products in early development of the sea star and distinct differences from germ-line development in the sea urchin. Developmental dynamics : an official publication of the American Association of Anatomists. 2013 doi: 10.1002/dvdy.24038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. 2002:310–322. [PubMed] [Google Scholar]
- Hinman VF, Davidson EH. Expression of AmKrox, a starfish ortholog of a sea urchin transcription factor essential for endomesodermal specification. Gene expression patterns : GEP. 2003;3:423–426. doi: 10.1016/s1567-133x(03)00083-8. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proceedings of the National Academy of Sciences of the United States of America. 2003a;100:13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman VF, Nguyen AT, Davidson EH. Expression and function of a starfish Otx ortholog, AmOtx: a conserved role for Otx proteins in endoderm development that predates divergence of the eleutherozoa. Mechanisms of development. 2003b;120:1165–1176. doi: 10.1016/j.mod.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Illa I, Serrano-Munuera C, Gallardo E, Lasa A, Rojas-Garcia R, Palmer J, Gallano P, Baiget M, Matsuda C, Brown RH. Distal anterior compartment myopathy: a dysferlin mutation causing a new muscular dystrophy phenotype. Annals of neurology. 2001;49:130–134. [PubMed] [Google Scholar]
- Kanatani H, Shirai H, Nakanishi K, Kurokawa T. Isolation and indentification on meiosis inducing substance in starfish Asterias amurensis. Nature. 1969;221:273–274. doi: 10.1038/221273a0. [DOI] [PubMed] [Google Scholar]
- Kawahara G, Serafini PR, Myers JA, Alexander MS, Kunkel LM. Characterization of zebrafish dysferlin by morpholino knockdown. Biochemical and biophysical research communications. 2011;413:358–363. doi: 10.1016/j.bbrc.2011.08.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Izawa T, Kuwamura M, Yamate J. Dysferlin and animal models for dysferlinopathy. J Toxicol Pathol. 2012;25:135–147. doi: 10.1293/tox.25.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek A, Evesson FJ, Sutton RB, North KN, Cooper ST. Ferlins: regulators of vesicle fusion for auditory neurotransmission, receptor trafficking and membrane repair. Traffic. 2012;13:185–194. doi: 10.1111/j.1600-0854.2011.01267.x. [DOI] [PubMed] [Google Scholar]
- Lek A, Lek M, North KN, Cooper ST. Phylogenetic analysis of ferlin genes reveals ancient eukaryotic origins. BMC evolutionary biology. 2010;10:231. doi: 10.1186/1471-2148-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C, Utokaparch S, Sharma A, Yu C, Abraham T, Borchers C, Bernatchez P. Proteomic identification of dysferlin-interacting protein complexes in human vascular endothelium. Biochemical and biophysical research communications. 2011;415:263–269. doi: 10.1016/j.bbrc.2011.10.031. [DOI] [PubMed] [Google Scholar]
- Li R, Ackerman WEt, Mihai C, Volakis LI, Ghadiali S, Kniss DA. Myoferlin depletion in breast cancer cells promotes mesenchymal to epithelial shape change and Stalls invasion. PloS one. 2012;7:e39766. doi: 10.1371/journal.pone.0039766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Aoki M, Ilia I, Wu C, Fardeau M, Angelini C, Serrano C, Urtizberea JA, Hentati F, Hamida MB, Bohlega S, Culper EJ, Amato AA, Bossie K, Oeltjen J, Bejaoui K, McKenna-Yasek D, Hosier BA, Schurr E, Arahata K, de Jong PJ, Brown RH., Jr Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nature genetics. 1998;20:31–36. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- Lowther KM, Nikolaev VO, Mehlmann LM. Endocytosis in the mouse oocyte and its contribution to cAMP signaling during meiotic arrest. Reproduction. 2011;141:737–747. doi: 10.1530/REP-10-0461. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr Patterns. 2004;4:449–456. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Mitaku S, Hirokawa T, Tsuji T. Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics. 2002;18:608–616. doi: 10.1093/bioinformatics/18.4.608. [DOI] [PubMed] [Google Scholar]
- Monigatti F, Gasteiger E, Bairoch A, Jung E. The Sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics. 2002;18:769–770. doi: 10.1093/bioinformatics/18.5.769. [DOI] [PubMed] [Google Scholar]
- Ohsako T, Hirai K, Yamamoto MT. The Drosophila misfire gene has an essential role in sperm activation during fertilization. Genes & genetic systems. 2003;78:253–266. doi: 10.1266/ggs.78.253. [DOI] [PubMed] [Google Scholar]
- Opresko LK, Wiley HS. Receptor-mediated endocytosis in Xenopus oocytes. I. Characterization of the vitellogenin receptor system. The Journal of biological chemistry. 1987;262:4109–4115. [PubMed] [Google Scholar]
- Pangrsic T, Reisinger E, Moser T. Otoferlin: a multi-C(2) domain protein essential for hearing. Trends Neurosci. 2012;35:671–680. doi: 10.1016/j.tins.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Patel P, Harris R, Geddes SM, Strehle EM, Watson JD, Bashir R, Bushby K, Driscoll PC, Keep NH. Solution structure of the inner DysF domain of myoferlin and implications for limb girdle muscular dystrophy type 2b. Journal of molecular biology. 2008;379:981–990. doi: 10.1016/j.jmb.2008.04.046. [DOI] [PubMed] [Google Scholar]
- Robinson JM, Ackerman WEt, Behrendt NJ, Vandre DD. While dysferlin and myoferlin are coexpressed in the human placenta, only dysferlin expression is responsive to trophoblast fusion in model systems. Biol Reprod. 2009;81:33–39. doi: 10.1095/biolreprod.108.074591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Yu C, Leung C, Trane A, Lau M, Utokaparch S, Shaheen F, Sheibani N, Bernatchez P. A new role for the muscle repair protein dysferlin in endothelial cell adhesion and angiogenesis. ArteriosclerThromb Vase Biol. 2010;30:2196–2204. doi: 10.1161/ATVBAHA.110.208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MK, Wakimoto BT. Complex regulation and multiple developmental functions of misfire, the Drosophila melanogaster ferlin gene. BMC developmental biology. 2007;7:21. doi: 10.1186/1471-213X-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, Coffman JA, Dean M, Elphick MR, Ettensohn CA, Foltz KR, Hamdoun A, Hynes RO, Klein WH, Marzluff W, McClay DR, Morris RL, Mushegian A, Rast JP, Smith LC, Thorndyke MC, Vacquier VD, Wessel GM, Wray G, Zhang L, Elsik CG, Ermolaeva O, Hlavina W, Hofmann G, Kitts P, Landrum MJ, Mackey AJ, Maglott D, Panopoulou G, Poustka AJ, Pruitt K, Sapojnikov V, Song X, Souvorov A, Solovyev V, Wei Z, Whittaker CA, Worley K, Durbin KJ, Shen Y, Fedrigo O, Garfield D, Haygood R, Primus A, Satija R, Severson T, Gonzalez-Garay ML, Jackson AR, Milosavljevic A, Tong M, Killian CE, Livingston BT, Wilt FH, Adams N, Belle R, Carbonneau S, Cheung R, Cormier P, Cosson B, Croce J, Fernandez-Guerra A, Geneviere AM, Goel M, Kelkar H, Morales J, Mulner-Lorillon O, Robertson AJ, Goldstone JV, Cole B, Epel D, Gold B, Hahn ME, Howard-Ashby M, Scally M, Stegeman JJ, Allgood EL, Cool J, Judkins KM, McCafferty SS, Musante AM, Obar RA, Rawson AP, Rossetti BJ, Gibbons IR, Hoffman MP, Leone A, Istrail S, Materna SC, Samanta MP, Stole V, Tongprasit W, Tu Q, Bergeron KF, Brandhorst BP, Whittle J, Berney K, Bottjer DJ, Calestani C, Peterson K, Chow E, Yuan QA, Elhaik E, Graur D, Reese JT, Bosdet I, Heesun S, Marra MA, Schein J, Anderson MK, Brockton V, Buckley KM, Cohen AH, Fugmann SD, Hibino T, Loza-Coll M, Majeske AJ, Messier C, Nair SV, Pancer Z, Terwilliger DP, Agca C, Arboleda E, Chen N, Churcher AM, Hallbook F, Humphrey GW, Idris MM, Kiyama T, Liang S, Mellott D, Mu X, Murray G, Olinski RP, Raible F, Rowe M, Taylor JS, Tessmar-Raible K, Wang D, Wilson KH, Yaguchi S, Gaasterland T, Galindo BE, Gunaratne HJ, Juliano C, Kinukawa M, Moy GW, Neill AT, Nomura M, Raisch M, Reade A, Roux MM, Song JL, Su YH, Townley IK, Voronina E, Wong JL, Amore G, Branno M, Brown ER, Cavalieri V, Duboc V, Duloquin L, Flytzanis C, Gache C, Lapraz F, Lepage T, Locascio A, Martinez P, Matassi G, Matranga V, Range R, Rizzo F, Rottinger E, Beane W, Bradham C, Byrum C, Glenn T, Hussain S, Manning G, Miranda E, Thomason R, Walton K, Wikramanayke A, Wu SY, Xu R, Brown CT, Chen L, Gray RF, Lee PY, Nam J, Oliveri P, Smith J, Muzny D, Bell S, Chacko J, Cree A, Curry S, Davis C, Dinh H, Dugan-Rocha S, Fowler J, Gill R, Hamilton C, Hernandez J, Hines S, Hume J, Jackson L, Jolivet A, Kovar C, Lee S, Lewis L, Miner G, Morgan M, Nazareth LV, Okwuonu G, Parker D, Pu LL, Thorn R, Wright R. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub E, Fiziev P, Rosenthal A, Hinzmann B. Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. BioEssays : news and reviews in molecular, cellular and developmental biology. 2004;26:567–581. doi: 10.1002/bies.20032. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends in cell biology. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Washington NL, Ward S. FER-1 regulates Ca2+ -mediated membrane fusion during C. elegans spermatogenesis. J Cell Sci. 2006;119:2552–2562. doi: 10.1242/jcs.02980. [DOI] [PubMed] [Google Scholar]
- Wessel GM, Reich AM, Klatsky PC. Use of sea stars to study basic reproductive processes. Syst Biol Reprod Med. 2010;56:236–245. doi: 10.3109/19396361003674879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Pallikkuth S, Hou Z, Mignery GA, Robia SL, Han R. Dysferlin forms a dimer mediated by the C2 domains and the transmembrane domain in vitro and in living cells. PloS one. 2011;6:e27884. doi: 10.1371/journal.pone.0027884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga S, Grati M, Cohen-Salmon M, El-Amraoui A, Mustapha M, Salem N, El-Zir E, Loiselet J, Petit C. A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nature genetics. 1999;21:363–369. doi: 10.1038/7693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.