Abstract
To date, the bacterial DNA topoisomerases are one of the major target biomolecules for the discovery of new antibacterial drugs. DNA topoisomerase regulates the topological state of DNA, which is very important for replication, transcription and recombination. The relaxation of negatively supercoiled DNA is catalyzed by bacterial DNA topoisomerase I (topoI) and this reaction requires Mg2+. In this report, we first quantitatively studied the intermolecular interactions between Escherichia coli topoisomerase I (EctopoI) and pBAD/Thio supercoiled plasmid DNA using surface plasmon resonance (SPR) technique. The equilibrium dissociation constant (Kd) for EctopoI-pBAD/Thio interactions is determined to be about 8 nM. We then studied the effect of Mg2+ on the catalysis of EctopoI-pBAD/Thio reaction. A slightly higher equilibrium dissociation constant (~15 nM) was obtained for Mg2+ coordinated EctopoI (Mg2+EctopoI)-pBAD/Thio interactions. In addition, we observed a larger dissociation rate constant (kd) for Mg2+EctopoI-pBAD/Thio interactions (~0.043 s−1), compared to EctopoI-pBAD/Thio interactions (~0.017 s−1). These results suggest that enzyme turnover during plasmid DNA relaxation is enhanced due to the presence of Mg2+ and furthers the understanding of importance of the Mg2+ ion for bacterial topoisomerase I catalytic activity.
Keywords: Surface plasmon resonance, bacterial topoisomerase I, supercoiled plasmid DNA, capture covalent method, equilibrium dissociation constant
1. Introduction
The drug resistance of bacterial pathogens to available antibacterial drugs is a serious public health issue and needs to be addressed. The bacterial topoisomerase I, a DNA topoisomerase I (topoI), is a novel target biomolecule for the discovery of new antibacterial drugs [1]. DNA topoisomerases play important roles on both the supercoiling control of DNA and the resolution of topological barriers during replication, transcription, and recombination [2,3,4,5]. The supercoiling tension caused by translocation of RNA polymerase must be relieved by topoisomerases [6,7]. Topoisomerase I cleaves and rejoins a single DNA strand during topoI-DNA reactions [8], which establishes a transient covalent linkage between these two macromolecules. These complexes can be trapped using topoisomerase inhibitors [2,8].
Topoisomerase I can catalyze interconversion of various topological isomers [9] and this type IA topoisomerase catalytic activity requires Mg2+ [10]. In Escherichia coli, EctopoI removes excess negative supercoils in order to regulate DNA supercoiling [11]. EctopoI is a single polypeptide of 865 amino acids and tyrosine 319 is the active site tyrosine in EctopoI that forms a transient covalent linkage to DNA 5′ phosphoryl group during EctopoI-DNA reaction [12]. We quantitatively studied the interactions for EctopoI-pBAD/Thio supercoiled plasmid DNA (hereafter termed as pBAD/Thio) and Mg2+ bound EctopoI (Mg2+EctopoI)-pBAD/Thio using the surface plasmon resonance (SPR) technique.
SPR is a widely used label free technique to determine equilibrium dissociation constants and the kinetics of bio-molecular interactions [13]. The sensor surface modification for the SPR assay was confirmed by using electrochemical impedance spectroscopy (EIS). The equilibrium dissociation constant (Kd) for EctopoI binding to pBAD/Thio was determined to be about 8 nM. A slightly higher Kd (~15 nM) value was obtained for Mg2+EctopoI-pBAD/Thio interactions. In addition, the dissociation rate constants (kd) for the interactions between the enzymes (EctopoI or Mg2+EtopI) and pBAD/Thio were also derived and a larger dissociation rate constant (kd) was obtained for Mg2+EctopoI-pBAD/Thio interactions. These results can help us further understand the important role of Mg2+ in the interactions between EctopoI and DNA substrate during catalysis [14].
Mycobacterial tuberculosis topoisomerase I (MttopoI), has a different C-terminal DNA binding domain (CTD) that lacks the three Zn2+ binding motifs in the CTD of EctopoI [15]. Binding to pBAD/Thio plasmid DNA by the two enzymes were compared. Under our experimental conditions, weak SPR signals were observed for interactions between MttopoI and pBAD/Thio. Therefore, the Kd value could not be recovered for MttopI-pBAD/Thio interactions.
2. Materials and methods
2.1 Materials
Triethylene glycol mono-11-mercaptoundecyl ether (PEG-thiol), nickel (II) sulfate hexahydrate, sodium, chloride, potassium hexacyanoferrate (III) and ethanolamine HCL were purchased from Sigma-Aldrich, ethanol (200 proof) from Decon Laboratories LLC, 2-{2-[2-(1-mercaptoundec-11-yloxy)-ethoxy]-ethoxy)-ethoxy nitrilotriacetic acid (NTA-thiol) from ProChimia Surfaces, Poland and potassium ferrocyanide trihydrate from Acros Organics. N-hydroxysuccinimide (NHS), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) and GeneJET Plasmid Maxiprep Kit were received from Thermo-Scientific. All other reagents were purchased from VWR international, Randor, PA, USA. Solutions were prepared using deionized (DI) water (~18 MΩ) (Ultra Purelab system, ELGA/Siemens or Milli-Q Direct 8 water system). The polycrystalline gold chips (50 nm Au over 2.5 nm titanium adhesion layers, coated on 18mm × 18mm cover slip glass slides) were purchased from Platypus Technologies, LLC, Madison, WI and each chip was cut into two halves before further processing.
2.2 Methods
2.2.1 Isolation and purification of EctopoI, MttopoI and pBAD/Thio
Recombinant EctopoI and MttopoI were expressed and purified as N-terminal His-tagged proteins according to previously reported procedures [16,17]. For the SPR experiments, the EctopoI was dialyzed against HEPES buffer (10 mM Hepes, pH 7.4 containing 100 mM NaCl and 0.005% (V/V) tween 20 in DI water) overnight at 4°C. The MttopoI was dialyzed against HEPES buffer with 2.5% (V/V) glycerol overnight at 4°C. The glycerol was needed to maintain solubility of MttopoI. The pBAD/Thio supercoiled plasmid DNA was purified using Genejet maxiprep kit (Thermo Scientific) according to the manufacturer’s protocol.
2.2.2 Sensor preparation and characterization
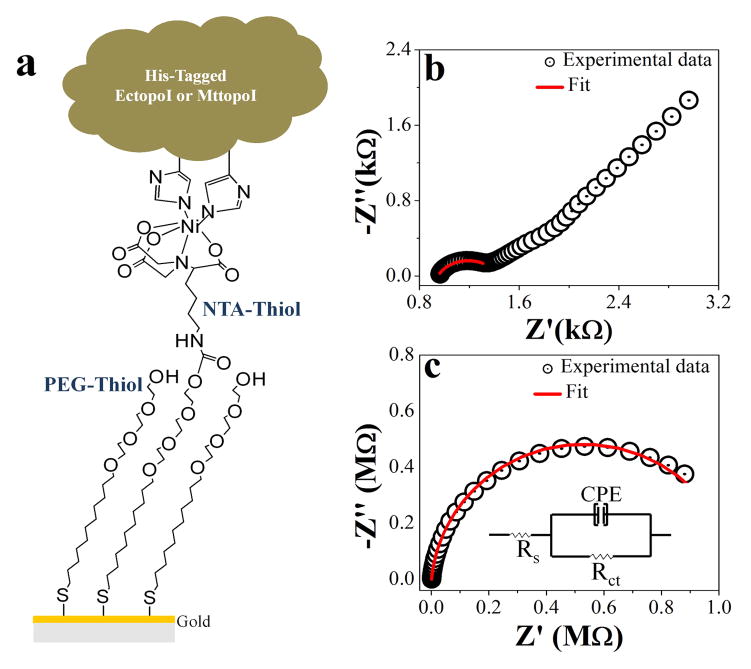
Gold chips were used to prepare SPR sensors. After rinsing in ethanol for 2–3 minutes, the chips were cleaned by oxygen plasma (Plasma Cleaner PDC-001, Harric Plasma, Ithaca, NY) for 40 s at an oxygen pressure of 500–600 mTorr and RF power of 10.2 W. The chips were then annealed with hydrogen flame for 20 s to reduce the surface roughness [18]. To form a self-assembled monolayer (SAM), the hydrogen flamed chip was immediately immersed in mixed thiol solution (1:9 V/V mixture of 1mM NTA-thiol and 1mM PEG-thiol in ethanol respectively) and incubated overnight. The chip was then copiously rinsed with ethanol and DI water to remove physically adsorbed thiol molecules and dried with argon. The formation of SAM on the cleaned gold surface was confirmed by electrochemical impedance spectroscopy (EIS). A detailed explanation of EIS experiments can be found in supplementary information (section S1). Figure 1a shows the scheme of SAM formation and His-tagged EctopoI/MttopoI immobilization. The modified chip was then mounted inside SPR flow cell. A detail of the SPR system similar to the system used here can be found in a previous report [19].
Figure 1.
a) Scheme showing the sensor surface modification with mixed thiols followed by the His-tagged EctopoI/MttopoI immobilization. b) Electrochemical Impedance Spectroscopy (EIS) for the bare gold. c) EIS for SAM modified gold surface, inset: equivalent circuit for EIS data fitting and analysis. A frequency range from 10−1 Hz to 104 Hz was used during EIS measurements. In both Figures b and c, the symbols are experimental data and continuous lines are the CNLS fit (see supplementary information, section S1, for details).
2.2.3 Enzyme immobilization and DNA binding
The sensor surface was activated using a 40 mM nickel (II) sulfate solution prepared in DI water for 2 minutes at a flow rate of 50 μL/min followed by DI water flushing for two minutes. The surface was then equilibrated with the HEPES buffer for 5–10 minutes. His-tagged EctopoI (2μM) or MttopoI (2.5μM) was immobilized on the activated SAM surface at a flow rate of 50 μL/min via the capture covalent method [20]. Sequential treatment of the EctopoI or MttopoI immobilized surface was accomplished with a pulsed injection of NHS-EDC solution (25 mM NHS and 100 mM EDC) in DI water followed by 1 M ethanolamine (pH 8.2) in DI water. This treatment allowed us to achieve the capture covalent immobilization. The experiments with various pBAD/Thio concentrations were accomplished by regeneration of the sensor surface using 1M NaCl. For the experiments involving Mg2+, 0.5 mM MgCl2 (in Tris buffer, pH 8.0) was passed over EctopoI immobilized surface for 5 minutes. All the experiments were carried out at 22°C.
2.2.4 Data analysis
Complex non-linear least square (CNLS) fitting algorithm was used for EIS data using an equivalent circuit model (supplementary information, section S1). Equilibrium data analysis methods were used to analyze the SPR data [21]. The simple nonlinear hyperbolic (SNLH) equation (equation 1 below) was used to fit the equilibrium SPR response plotted vs DNA concentrations,
| (1) |
where, R is the equilibrium response, [DNA] is the analyte (pBAD/Thio) concentration, Kd is the equilibrium dissociation constant, and Rmax is the fitting parameter representing maximum response. The dissociation rate constants were also derived by fitting the SPR dissociation profiles. A detailed explanation of the fitting procedure is given the supplementary information (section S2).
3. Results
3.1 Sensor surface characterization
As shown in Figures 1b&c, the EIS experimental data (symbols) are fitted (continuous lines) using the equivalent circuit (inset, Figure 1c). The parameters obtained from fitting are listed in Table 1. The charge transfer resistance (Rct) for SAM modified gold is significantly higher (~1060 kΩ) compared to bare gold (~0.5 kΩ). This clearly confirms the successful modification of the gold surface. Furthermore, due to the presence of SAM on the gold surface, the interfacial capacitance for modified surface is obviously reduced (313nF) compared to clean gold (1227 nF).
Table 1.
The parameters obtained by SNLH fitting of EIS experimental data to an equivalent circuit (Inset, Figure 2c). The errors are the standard errors of the fitting.
| Surface | C (nF) | Rct (kΩ) | Rs (kΩ) | α |
|---|---|---|---|---|
| Au | 1227±122 | 0.48±0.08 | 0.95±0.04 | 0.75±0.01 |
| Au/SAM | 313±3 | 1060±10 | 0.93±0.08 | 0.94 |
3.2 Enzyme immobilization and pBAD/Thio binding
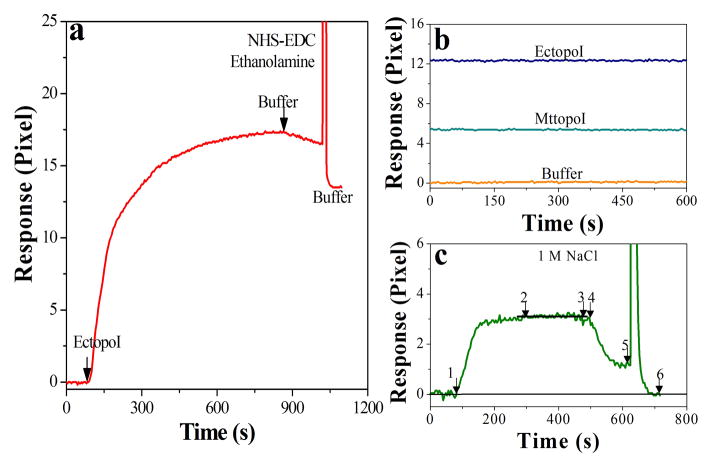
As explained in the methods section, both EctopoI (Figure 2a) and MttopoI (Figure S3, supplementary information) were immobilized onto the Ni2+ chelated NTA surface via the capture covalent method. Figure 2b depicts the sensor surface stability after chemical treatment of the enzyme immobilized surfaces with NHS-EDC followed by ethanolamine. Figure 2c is the representation of pBAD/Thio binding to the enzyme immobilized sensor surfaces, including association, saturation and dissociation of SPR signal. The sensor surface was regenerated using 1 M NaCl solution.
Figure 2.
a) SPR sensorgram for the immobilization of EctopoI via capture covalent method. The arrows show the start of immobilization and buffer wash. b) Representative SPR signals showing stability of sensor surface; with buffer before enzyme immobilization and after capture covalent immobilization of the enzymes. c) Representative SPR profile for enzyme-pBAD/Thio interaction: 1 to 2 association, 2 to 3 saturation, 3 to 4 minor signal shift due to manual changing of valve, 4 to 5 dissociation, and 5 to 6 regeneration.
3.3 pBAD/Thio molecular interaction with EctopoI, Mg2+EctopoI and MttopoI
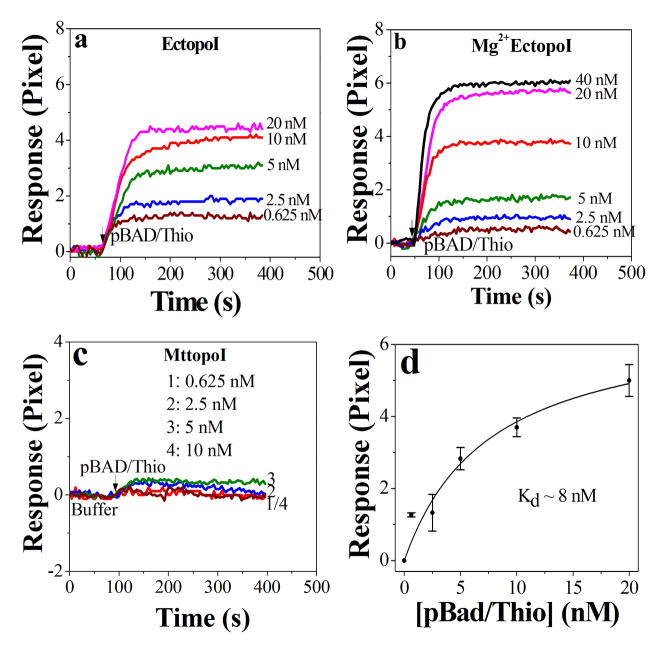
The concentration-dependent pBAD/Thio binding to EctopoI and Mg2+EctopoI are shown in Figure 3a and 3b, respectively. Under our experimental conditions, the pBAD/Thio did not show a considerable response change upon its binding to MttopoI (Figure 3c). The SNLH fitting, using equation 1, to the plot of equilibrium response vs concentration of pBAD/Thio (Figure 3d) provides quantitative information of the interaction; more precisely, the Kd can be derived. The Kd value obtained from the fit is about 8 nM for EctopoI-pBAD/Thio interactions and a slightly higher Kd value (~15 nM) for Mg2+EctopoI-pBAD/Thio interactions. We could not obtain quantitative information for MttopoI-pBAD/Thio interactions.
Figure 3.
SPR sensorgrams for a) pBAD/Thio binding to EctopoI, b) for pBAD/Thio binding to Mg2+ bound EctopoI and c) for pBAD/Thio binding to MttopoI. d) Plot of equilibrium SPR response vs pBAD/Thio concentration. The symbols are average experimental data of three different measurements with error bars as standard deviation and the continuous lines are the simple hyperbolic fit (explained in text).
3.4 Effect of Mg2+on EctopoI-pBAD/Thio molecular interaction
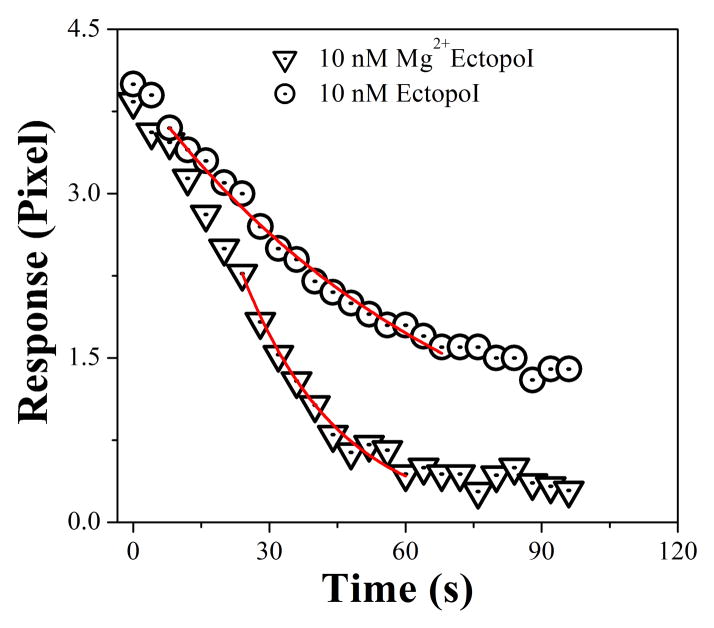
The SPR dissociation signals were fitted using the exponential dissociation rate constant equation (supplementary information, section S2). As shown in Figure 4, stronger dissociation signals were observed for Mg2+EctopoI-pBAD/Thio interactions compared to EctopoI-pBAD/Thio interactions. The derived dissociation rate constant (kd) value for EctopoI-pBAD/Thio interactions is ~0.017 s−1 and for Mg2+EctopoI-pBAD/Thio interactions is ~0.043 s−1. Compared to EctopoI-pBAD/Thio interactions, a larger kd value for Mg2+EctopoI-pBAD/Thio interactions suggests that the rate of enzyme turnover following DNA religation during catalysis of EctopoI-pBAD/Thio relaxation reaction is enhanced in the presence of Mg2+.
Figure 4.
Representative SPR dissociation profiles fitted to exponential dissociation rate equation (equation S2a, supplementary information). The symbols are experimental data and the continuous lines are the best fit to the rate equation, S2a.
4 Discussion
Recognizing EctopoI as an important target biomolecule for the discovery of new antibacterial drugs [1], EctopoI-plasmid DNA interactions were quantitatively studied for the first time (to the best of our knowledge) using SPR, a surface based affinity technique. In the SPR technique, the sensor surface modification plays a crucial role for the minimization of non-specific binding and reproducibility of SPR signals. We carried out EIS measurements to confirm the gold surface modification via the mixed thiols (supplementary information, section S1). The efficient charge (electron) transfer between the redox couple and the working electrode in solution is basically evaluated by charge transfer resistance (Rct) determination. The Rct, obtained either from fitting or crudely from the semicircular diameter (Figures 1b&c) [22], for Au surface is much smaller compared to Au/SAM surface (Table 1). This clearly confirms the successful sensor surface modification. Our data also show that the interfacial capacitance (C) decreases as the surface thickness increases because of the surface modification (Table 1). The successful immobilization of EctopoI or MttopoI on the modified sensor surface was achieved via capture covalent method. Previously reported result shows that the single His-tag has a weak affinity (~1 μM) to the Ni2+-NTA surface [23]. Even immobilized hexahistidine tagged proteins would dissociate upon continuous buffer flushing, and this phenomenon is not suitable for protein-small molecule interaction studies [20]. The immobilized enzyme surface must therefore be stable. This can be achieved by chemical treatment of the enzyme immobilized surface with sequential flow of NHS-EDC and ethanolamine solutions [20] (explained in methods). 30 μL of each solution was injected as a pulse for 20 seconds. Figure 2a and Figure S3 (supplementary information) show the EctopoI and MttopoI immobilization, respectively, onto the sensor surface via capture covalent immobilization. As shown in Figure 2b, the SPR response was found to be stable for 10 minutes before and after the treatment of EctopoI/MttopoI immobilized surfaces and thus guarantees the sensor surface stability and the successful capture covalent immobilization of the enzymes.
Figures 3a shows the interaction of EctopoI-pBAD/Thio in a concentration dependent manner. The EctopoI-pBAD/Thio binding responses saturated quickly; therefore an equilibrium analysis method could easily produce the Kd value via SNLH fitting (Figure 3d). The Kd value for EctopoI-pBAD/Thio interactions was found to be about 8 nM. A slightly higher Kd value (~15 nM) was obtained for Mg2+EctopoI-pBAD/Thio interactions. For the experiments shown in Figures 3a&b, except for 40 nM data, each SPR trace was repeated three times. Due to sample limitation, the 40 nM data was not repeated. After saturated SPR responses were achieved, dissociation SPR profiles were recorded for some experiments. Buffer flushing could dissociate the EctopoI-pBAD/Thio complex considerably. The covalent topoisomerase-DNA complex is transient and reversible in nature [3]. Treatment of the topoisomerase-DNA complex with high salt would promote dissociation of the DNA from enzyme following relegation [24]. After desired SPR signals were recorded, the surface was treated using 1M NaCl (in DI water) via pulsed injection for 2–3 minutes. This could successfully regenerate the surface to the initial (buffer) level and the surface was ready for next pBAD/Thio concentration. Figure 2c shows a typical SPR sensorgram showing association, saturation and dissociation of enzyme-pBAD/Thio interaction followed by regeneration. It should be noted that 10 mM Tris (pH 8.0) buffer was used as running buffer for enzymes-pBAD/Thio interaction experiments. The immobilized enzyme surfaces were equilibrated with the Tris buffer, for approximately 5 minutes, before pBAD/Thio injection. The experiments for each enzyme were performed either on the same chip or on different sensor chips. The variation of chip surfaces resulted in a variation of equilibrium response (lowest to highest) of ~0.6 pixels, on an average.
In order to understand the influence of the topoI C-terminal domain (CTD) sequence on the interactions between topoI and supercoiled plasmid DNA, we also studied the interaction between MttopoI and pBAD/Thio using SPR. There are tetracysteine Zn2+ binding motifs [25] that follow the N-terminal 67kDa transesterification domain [26]. The three tetracysteine motifs are part of a DNA-binding domain at the C-terminus of EctopoI [27]. Unlike EctopoI [28], MttopoI lacks Zn2+ coordination and has evolved to have a different CTD sequence [15]. Under our experimental conditions, we could not see the concentration dependent interaction for MttopoI-pBAD/Thio interactions. As shown in figure 3c, weak SPR signals were observed instead. This result suggests that the tetracysteine Zn2+ binding motifs are required for observing the interaction with plasmid DNA with the SPR protocol described here. Mycobacterium smegmatis topoisomerase I (MstopoI) has a CTD similar to MttopoI that also does not coordinate Zn2+ [15]. Previous reports indicate that MstopoI CTD interacts with DNA during catalysis [29] and is responsible for sequence specific recognition of duplex DNA by MstopoI [30]. Because of its specific interaction with duplex DNA sequence, the Mycobacterium topoisomerase CTD may not be as efficient in promoting high affinity binding to the single-stranded DNA region in supercoiled plasmid DNA as the CTD in EctopoI. Alternatively, the result observed here might be due to the loss of MttopoI activity during protein immobilization. The dialyzed MttopoI sample used for protein immobilization was assayed for relaxation activity and found to be active (Figure S4, supplementary information).
It has been revealed from crystal structure of the 67 kDa N-terminal fragment of EctopoI that there is presence of acidic and basic amino acid residues nearby the active site region [31]. It has also been proposed that Lys-13 and Arg-321 (both basic residues) participate in DNA cleavage [32,33] and three acidic residues Asp-111, ASP-113, and Glu-115 coordinate with Mg2+ [34]. We could not detect the concentration dependent Mg2+ binding to EctopoI, which might be the limiting case of our SPR systems to resolve the detection of very small molecular weight ions/molecules-protein interactions. We also did not analyze our SPR sensorgrams for association rate constant measurements due to rapid association and fast saturation of the SPR signals. Therefore, to better understand the effect of Mg2+ coordination, the SPR dissociation profiles were recorded for the SPR sensorgrams shown in Figures 3a&b. These SPR profiles were fitted to the exponential dissociation rate constant equation. The fitting procedure is explained in detail in the supplementary information (section S2). A larger dissociation rate constant (kd) (~0.043 s−1) was obtained for Mg2+EctopoI-pBAD/Thio compared to EctopoI-pBAD/Thio interactions (~0.017 s−1). This increase in kd value confirms that EctopoI catalytic activity is enhanced with Mg2+. This is consistent with the role of Mg2+ to increase the dissociation rate constants for type IB topoisomerase I-DNA interactions, as reported previously [14]. Such analysis for type IA topoisomerase-DNA interactions has not been previously reported.
Supplementary Material
Research Highlights.
Study of E. coli topoisomerase I interactions with supercoiled DNA quantitatively
Quantitative analysis for the effect of Mg2+ ions on the enzyme-DNA interactions
Higher dissociation rate constant obtained for Mg2+ bound E. coli topoisomerase I
Enzyme turnover during the DNA relaxation would be enhanced in the presence of Mg2+
Acknowledgments
P. Tiwari would like to thank Florida International University (FIU) School of Integrated Science & Humanity, College Arts & Sciences for the research assistantship. This work was supported by NIH grant R01GM054226 (Y.T.) and the start-up funds from FIU (J.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tse-Dinh YC. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res. 2009;37:731–737. doi: 10.1093/nar/gkn936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Champoux JJ. DNA TOPOISOMERASES: Structure, Function, and Mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 4.Corbett KD, Berger JM. STRUCTURE, MOLECULAR MECHANISMS, AND EVOLUTIONARY RELATIONSHIPS IN DNA TOPOISOMERASES. Annu Rev Biophys Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 5.Cheng B, Shukla S, Vasunilashorn S, Mukhopadhyay S, Tse-Dinh YC. Bacterial Cell Killing Mediated by Topoisomerase I DNA Cleavage Activity. J Biol Chem. 2005;280:38489–38495. doi: 10.1074/jbc.M509722200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu HY, Shyy S, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 8.Tsai HP, Lin LW, Lai ZY, Wu JY, Chen CE, Hwang J, Chen CS, Lin CM. Immobilizing topoisomerase I on a surface plasmon resonance biosensor chip to screen for inhibitors. J Biomed Sci. 2010;17:49. doi: 10.1186/1423-0127-17-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JC. DNA Topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 10.Sissi C, Palumbo M. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 2009;37:702–711. doi: 10.1093/nar/gkp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drlica K. Bacterial topoisomerases and the control of DNA supercoiling. Trends Genet. 1990;6:433–437. doi: 10.1016/0168-9525(90)90306-q. [DOI] [PubMed] [Google Scholar]
- 12.Lynn RM, Wang JC. Peptide Sequencing and Site-Directed Mutagenesis Identify Tyrosine-319 as the Active Site Tyrosine of Escherichia coli DNA Topoisomerase I, Proteins. Struct, Funct, Genet. 1989;6:231–239. doi: 10.1002/prot.340060305. [DOI] [PubMed] [Google Scholar]
- 13.Wilson WD. Analyzing Biomolecular Interactions. Science. 2002;295:2103–2105. doi: 10.1126/science.295.5562.2103. [DOI] [PubMed] [Google Scholar]
- 14.Pond CD, Holden JA, Schnabel PC, Barrows LR. Surface plasmon resonance analysis of topoisomerase I-DNA binding: effect of Mg2+ and DNA sequence. Anti-Cancer Drugs. 1997;8:336–344. doi: 10.1097/00001813-199704000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Bhaduri T, Bagui TK, Sikder D, Nagaraja V. DNA Topoisomerase I from Mycobacterium smegmatis: AN ENZYME WITH DISTINCT FEATURES. J Biol Chem. 1998;273:13925–13932. doi: 10.1074/jbc.273.22.13925. [DOI] [PubMed] [Google Scholar]
- 16.Sorokin EP, Cheng B, Rathi S, Aedo SJ, Abrenica MV, Tse-Dinh YC. Inhibition of Mg2+ binding and DNA religation by bacterial topoisomerase I via introduction of an additional positive charge into the active site region. Nucleic Acids Res. 2008;36:4788–4796. doi: 10.1093/nar/gkn460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Annamalai T, Dani N, Cheng B, Tse-Dinh YC. Analysis of DNA relaxation and cleavage activities of recombinant Mycobacterium tuberculosis DNA topoisomerase I from a new expression and purification protocol. BMC Biochem. 2009;10:18. doi: 10.1186/1471-2091-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leopold MC, Bowden EF. Influence of Gold Substrate Topography on the Voltammetry of Cytochrome c Adsorbed on Carboxylic Acid Terminated Self-Assembled Monolayers. Langmuir. 2002;18:2239–2245. [Google Scholar]
- 19.Mulchan NM, Rodriguez M, O’Shea K, Darici Y. Application of a high-resolution SPR technique for monitoring real-time metal/dielectric interactions. Sens Actuators, B-Chem. 2003;88:132–137. [Google Scholar]
- 20.Wear MA, Patterson A, Malone K, Dunsmore C, Turner NJ, Walkinshaw MD. A surface plasmon resonance-based assay for small molecule inhibitors of human cyclophilin A. Anal Biochem. 2005;345:214–226. doi: 10.1016/j.ab.2005.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wammes AEM, Fischer MJE, de Mol NJ, van Eldijk MB, Rutjes FPJT, van Hest JCM, van Delft FL. Site-specific peptide and protein immobilization on surface plasmon resonance chips via strain-promoted cycloaddition. Lab Chip. 2013;13:1863–1867. doi: 10.1039/c3lc41338a. [DOI] [PubMed] [Google Scholar]
- 22.Shan YP, Tiwari PB, Krishnakumar P, Vlassiouk I, Li WZ, Wang XW, Darici Y, Lindsay SM, Wang HD, Smirnov S, He J. Surface modification of graphene nanopores for protein translocation. Nanotechnology. 2013;24:495102. doi: 10.1088/0957-4484/24/49/495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nieba L, Nieba-Axmann SE, Persson A, Hämäläinen M, Edebratt F, Hansson A, Lidholm J, Magnusson K, Karlsson ÅF, Plückthun A. BIACORE Analysis of Histidine-Tagged Proteins Using a Chelating NTA Sensor Chip. Anal Biochem. 1997;252:217–228. doi: 10.1006/abio.1997.2326. [DOI] [PubMed] [Google Scholar]
- 24.Chen SJ, Wang JC. Identification of Active Site Residues in Escherichia coli DNA Topoisomerase I. J Biol Chem. 1998;273:6050–6056. doi: 10.1074/jbc.273.11.6050. [DOI] [PubMed] [Google Scholar]
- 25.Tse-Dinh YC, Beran-Steed RK. Escherichia coli DNA topoisomerase I is a zinc metalloprotein with three repeatitive zinc-binding domains. J Biol Chem. 1998;263:15857–15859. [PubMed] [Google Scholar]
- 26.Ahumada A, Tse-Dinh YC. The role of the Zn(II) binding domain in the mechanism of E. coli DNA topoisomerase I. BMC Biochem. 2002;3:13. doi: 10.1186/1471-2091-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahumada A, Tse-Dinh YC. The Zn(II) Binding Motifs of E. coli DNA Topoisomerase I Is Part of a High-Affinity DNA Binding Domain. Biochem Biophys Res Commun. 1998;251:509–514. doi: 10.1006/bbrc.1998.9500. [DOI] [PubMed] [Google Scholar]
- 28.Yu L, Zhu CX, Tse-Dinh YC, Fesik SW. Solution structure of the C-terminal single-stranded DNA-binding domain of Escherichia coli topoisomerase I. Biochemistry. 1995;34:7622–7628. doi: 10.1021/bi00023a008. [DOI] [PubMed] [Google Scholar]
- 29.Ahmed W, Bhat AG, Leelaram MN, Menon S, Nagaraja V. Carboxyl terminal domain basic amino acids of mycobacterial topoisomerase I bind DNA to promote strand passage. Nucleic Acids Res. 2013;41:7462–7471. doi: 10.1093/nar/gkt506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhaduri T, Sikder D, Nagaraja V. Sequence specific interaction of Mycobacterium smegmatis topoisomerase I with duplex DNA. Nucleic Acids Res. 1998;26:1668–1674. doi: 10.1093/nar/26.7.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lima CD, Wang JC, Mondragon A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 32.Strahs D, Zhu CX, Cheng B, Chen J, Tse-Dinh YC. Experimental and computational investigations of Ser10 and Lys13 in the binding and cleavage of DNA substrates by Escherichia coli DNA topoisomerase I. Nucleic Acids Res. 2006;34:1785–1797. doi: 10.1093/nar/gkl109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feinberg H, Lima CD, Mondragón A. Conformational changes in E. coli DNA topoisomerase I. Nat Struct Biol. 1999;6:918–922. doi: 10.1038/13283. [DOI] [PubMed] [Google Scholar]
- 34.Zhu CX, Tse-Dinh YC. The Acidic Triad Conserved in Type IA DNA Topoisomerases Is Required for Binding of Mg(II) and Subsequent Conformational Change. J Biol Chem. 2000;275:5318–5322. doi: 10.1074/jbc.275.8.5318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.