SUMMARY
The transcription factor Krüppel-like factor 4 (KLF4) is an important regulator of cell fate decision, including cell cycle regulation, apoptosis, and stem cell renewal, and plays an ambivalent role in tumorigenesis as a tissue specific tumor suppressor or oncogene. Here we report that the Von Hippel-Lindau gene product, pVHL, physically interacts with KLF4 and regulates its rapid turnover observed in both differentiated and stem cells. We provide mechanistic insights into KLF4 degradation and show that pVHL depletion in colorectal cancer cells leads to cell cycle arrest concomitant with increased transcription of the KLF4-dependent p21 gene. Finally, immunohistochemical staining revealed elevated pVHL and reduced KLF4 levels in colon cancer tissues. We therefore propose that unexpectedly pVHL, via the degradation of KLF4, is a facilitating factor in colorectal tumorigenesis.
INTRODUCTION
The ubiquitin-proteasome system plays an important role in controlling the level, activity and location of various cellular proteins. Particularly, many transcription factors are unstable proteins and their rate of destruction is regulated by specific E3 ligases. Examples of proteolytic regulation of transcription factors involved in carcinogenesis include the targeting of tumor suppressor p53 by MDM2 and the oncoprotein Myc by Skp2. Another well documented example is the degradation of hypoxia inducible factor (HIF) by the von Hippel-Lindau (pVHL) tumor suppressor (Kaelin, 2007).
Krüppel-like factor 4 (KLF4, GKLF) is a transcription factor involved in cellular responses to a variety of environmental and intracellular stress signals such as DNA damage, oxidative stress, and inflammation (Evans and Liu, 2008). KLF4 activation determines the cell fate by activating or inhibiting a network of genes involved in cellular functions as diverse as cell cycle regulation, stem cell renewal, adhesion, apoptosis, or metabolism (Rowland and Peeper, 2006). Several of these KLF4 regulated pathways affect checkpoints in cancer development. Surprisingly, KLF4 appears to function as an oncogene or tumor suppressor depending on the tissue type. Although several KLF4 downstream-targets have been well characterized, it is unknown why elevated KLF4 levels promote tumorigenesis in the skin, but inhibit cancer formation in the colon. Indeed, the mechanisms of KLF4 activation itself are largely unexplored. The expression of KLF4 is strongly induced after genotoxic stress, such as γ–irradiation (Ghaleb et al., 2007), and serum starvation (Chen et al., 2005). Following the observation that KLF4 is a protein with a high turnover rate, we attempted to identify proteins that regulate it post-translationally and here report the interaction of KLF4 with pVHL.
Mutations in the VHL gene have been linked to a variety of tumors including clear cell renal carcinoma, retinal and cerebellar hemangioblastoma, and phaeochromocytoma (Kaelin, 2007). The VHL protein is the substrate-recognizing subunit of the CBCVHL E3 ubiquitin ligase complex consisting of pVHL, the elongins B and C, Cullin 2, and the RING-H2 finger protein Rbx1. CBCVHL binds and degrades the α subunits of the heterodimeric hypoxia-inducible transcription factors HIF1 and HIF2 (Maxwell et al., 1999). Recognition of HIF1 and HIF2 by pVHL is contingent on the post-translational modification of conserved HIF proline residues located within LXXLAP motifs by specific prolyl-hydroxylases (PHDs), a process involving molecular oxygen (reviewed in (Kaelin, 2008)). Under normoxic or athmospheric oxygen levels, HIF α subunits are efficiently modified by PHDs, ubiquitylated by CBCVHL, and rapidly degraded by the 26S proteasome. However, under hypoxic conditions proline hydroxylation is inhibited and HIF-α subunits are stabilized, leading to their dimerization with constitutively stable HIF-β subunits and activation of hypoxia-responsive genes. Absence or dysfunctional pVHL leads to impaired downregulation of HIF under normoxic conditions and constitutive expression of HIF target genes, likely a key pathogenic event in VHL disease as evidenced by xenograft assays and immunostaining of patient tissues (Kaelin, 2008). However, increasing evidence indicate that pVHL activity is not limited to regulation of HIFs. Other substrates of its ubiquitin ligase activity include atypicial protein kinase Cλ, pVHL-interacting deubiquitinating enzymes VDU1 and VDU2, and two subunits of RNA polymerase II (Rpb1 and Rpb7). Unrelated to its ligase activity, pVHL was shown to bind and stabilize p53 and Jade-1, to regulate microtubule stability, assembly of extracellular matrix and intercellular junctions, and primary cilium maintenance (Nyhan et al., 2008), and to promote proper spindle orientation and chromosomal stability (Thoma et al., 2009). In addition, pVHL was found to play a HIF-independent role in cell cycle regulation. Vhl−/− mouse embryonic fibroblasts (MEFs) and fibrosarcomas derived thereof grow slower than the corresponding Vhl+/+ cells (Mack et al., 2005) and acute loss of Vhl triggered a senescence-like growth arrest in MEFs that was independent of p53 and HIF (Young et al., 2008). Furthermore, p21 and p27 levels are elevated in embryonic stem cells and MEFs derived from Vhl−/− mice (Mack et al., 2005) and inactivation of the Vhl locus in MEFs leads to an increase in p27 levels (Young et al., 2008). Intriguingly, both cyclin-dependent kinase inhibitors, p21 and p27, are reported downstream targets of KLF4 (Rowland and Peeper, 2006; Wei et al., 2008).
In this study, we report that pVHL interacts directly with and degrades KLF4 via the ubiquitin-proteasome pathway. Furthermore, VHL deficiency in colon cancer cells leads to KLF4 upregulation and concomitant increase of p21 transcription as well as to cell cycle arrest. Our findings suggest that KLF4 is a downstream mediator of HIF-independent roles of VHL. As such KLF4 provides a new link in the pathway leading to growth arrest triggered by the loss of VHL observed in certain cells. In such cells, characterized by a requirement of KLF4 for proliferative control, pVHL overexpression could contribute to tumor formation, at least at the early stage of tumorigenesis. Indeed, we find that pVHL upregulation associated with a decrease in KLF4 levels is a common feature in colon cancers. Mechanistically, we identified the KLF4 amino-terminus as particularly important for the regulation of KLF4 turnover by being the site of ubiquitylation and pVHL binding. Our findings provide the basis for future studies on how KLF4 degradation is regulated and how VHL affects KLF4 functions in other pathways such as stem cell renewal and apoptosis.
RESULTS
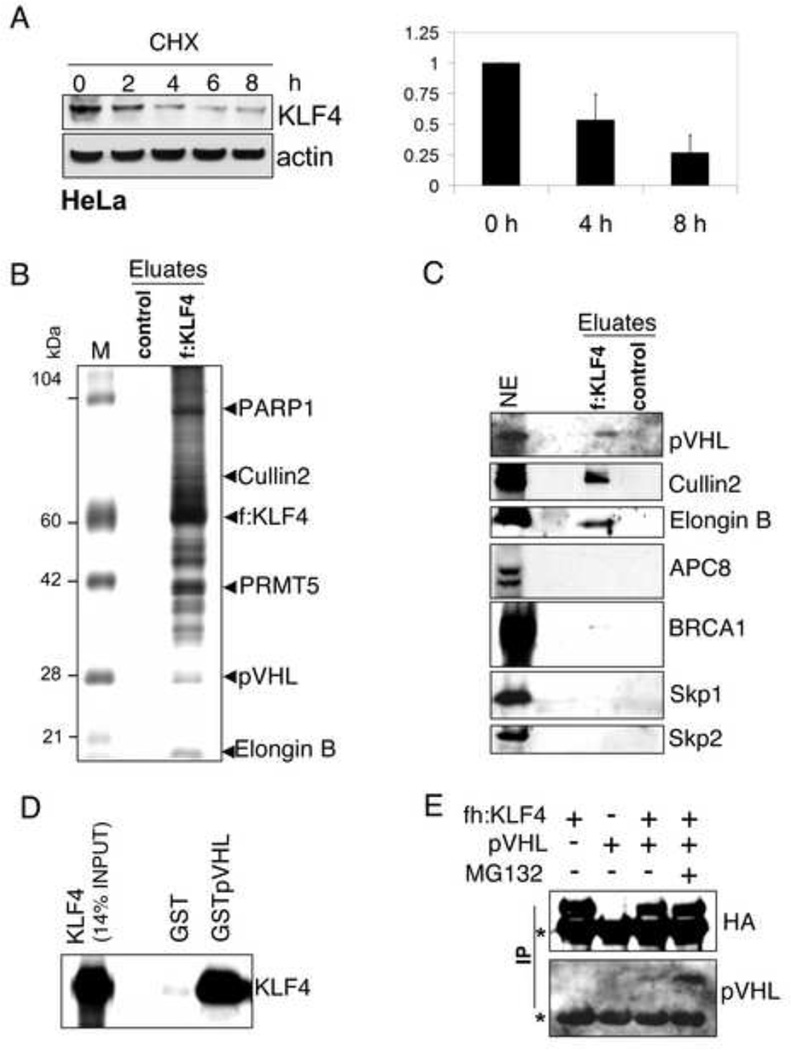
The transcription factor Krüppel-like Factor 4 (KLF4) plays an ambivalent role in tumorigenesis. It can act both as a tissue specific tumor suppressor or oncogene, likely due to its ability to induce cell cycle arrest and/or block apoptosis. Recently, KLF4 has also drawn a lot of attention due to its role as key factor in stem cell renewal and induction of pluripotency in differentiated cells. In all cases, tight regulation of KLF4 levels is pivotal for cell fate decisions. Indeed, expression of KLF4 is strongly induced after genotoxic stress, such as γ–irradiation (Ghaleb et al., 2007), and serum starvation (Chen et al., 2005). Because KLF4 was previously reported to be highly unstable in HCT116 (Chen et al., 2005), we tested whether the same was true for HeLa cells. Under homeostatic conditions KLF4 has a high turnover rate in HeLa cells as shown by an immunoblot after inhibiting protein synthesis (Fig. 1A). HeLa S cells were treated with cycloheximide for 0, 2, 4, 6, and 8 hours and KLF4 levels were assessed by immunoblots for KLF4. KLF4 was rapidly degraded with a half-life of around 4 hours, suggesting that post-translational regulation plays an important part in determining cellular KLF4 levels.
Figure 1. KLF4 interacts with pVHL.
(A) Short half-life of KLF4 in HeLa cells. Equal protein amounts of whole cell lysates from HeLa cells treated for the indicated time with the protein synthesis inhibitor cycloheximide (CHX) were probed for KLF4 by immunoblotting. (B) Proteins co-precipitating with KLF4. Anti-FLAG (M2 agarose) immunoprecipitates from nuclear extracts of HeLa cells (control) or HeLa cells stably transfected with a retrovirus expressing FLAG-tagged KLF4 were peptide eluded and analyzed by SDS-PAGE with Coomassie staining. M, size marker, f: FLAG-tag. (C) pVHL, Cullin2, and elongin B co-precipitate with KLF4. The immunoprecipitate from the HeLa cell line that stably expresses FLAG-tagged KLF4 was probed for various proteins associated with ubiquitin ligase complexes. NE, nuclear extract. (D) Interaction of in vitro translated radiolabeled KLF4 (detected by SDS-PAGE and autoradiography) with GST-fused pVHL. (E) Co-precipitation of pVHL with FLAG-HA-tagged KLF4 from MDAMB231 cells. Anti-FLAG (M2 agarose) immunoprecipitates (IP) were analyzed by immunoblot with antibodies indicated on the right. Asterisks denote signals from the light and heavy chains of M2. fh: FLAG-HA-tag.
pVHL co-precipitates with KLF4
To identify proteins that might regulate KLF4 degradation we established HeLa cell lines stably expressing FLAG-tagged KLF4. Cytosolic and nuclear extracts were prepared from HeLa cells and HeLa cells expressing tagged KLF4. Both, tagged and endogenous KLF4 showed similar degradation kinetics and cellular distribution (data not shown), with the majority found in the nuclear fraction in agreement with previous findings (McConnell et al., 2007). Tagged KLF4 and interacting partners were precipitated with M2-(anti-FLAG)-agarose from nuclear extracts and analyzed by SDS-PAGE and Coomassie blue staining. HeLa cells served as control (Fig. 1B). Prominent bands were excised and sent for mass spectrometric sequencing leading to the identification of several proteins with a potential role as transcription cofactors (data not shown). The smear above 100 KDa was mainly due to post-translationally modified KLF4. The immunoprecipitate was also subjected to immunoblotting with a collection of antibodies against E3 ligases or subunits of ubiquitin-ligase complexes. Fig. 1C shows that, unlike several other tested E3 ligases, the van Hippel-Lindau gene product pVHL was specifically precipitated by tagged KLF4. pVHL is the substrate recognizing subunit of a multiprotein ubiquitin-ligase complex that also contains Cullin2, the Elongins B and C, and Rbx1. We tested for Cullin2 and Elongin B and found that they also co-precipitated with KLF4, although it seems with lower efficiency than pVHL, probably because not all KLF4-bound pVHL is complexed and/or Cullin2 and Elongin B also associate with other proteins to form complexes. This suggests that the observed binding of pVHL with KLF4 is direct. VHL has two isoforms migrating as approximately 28 and 18 kDa proteins and of roughly equal cellular abundance. Interestingly, only the long form of pVHL binds KLF4 (Suppl. Fig. S1A), indicating that the pVHL fragment lacking in the 18 kDa form is directly binding KLF4 or important for the required pVHL folding. Endogenous pVHL also co-precipitated with endogenous KLF4 from the nuclear extract of HCT116 cells (Suppl. Fig. S1B).
KLF4 directly interacts with pVHL
To test whether KLF4 and pVHL interact directly, GST and GST-fused full-length pVHL proteins (Suppl. Fig. S1C) were independently bound to glutathione-Sepharose and incubated with in vitro translated KLF4. KLF4 bound to pVHL, but not to the control GST (Fig. 1D). In a similar assay, also His-tagged KLF4 purified from bacteria bound GST-pVHL, but not GST (Suppl. Fig. S1E). Furthermore, we tested intracellular interactions by transient co-expression of FLAG-HA-tagged KLF4 with untagged pVHL in MDAMB231 cells. Derived whole cell lysates were incubated with M2 (anti-FLAG) agarose and immunoprecipitates were analyzed by immunoblot. pVHL was co-immunoprecipitated with KLF4, albeit more efficiently when KLF4 expressing cells were treated with the ubiquitin-proteasome inhibitor MG132 before harvesting (Fig. 1E). The interaction of pVHL with KLF4 is therefore direct and not specific to HeLa.
KLF4 is post-translationally regulated by VHL
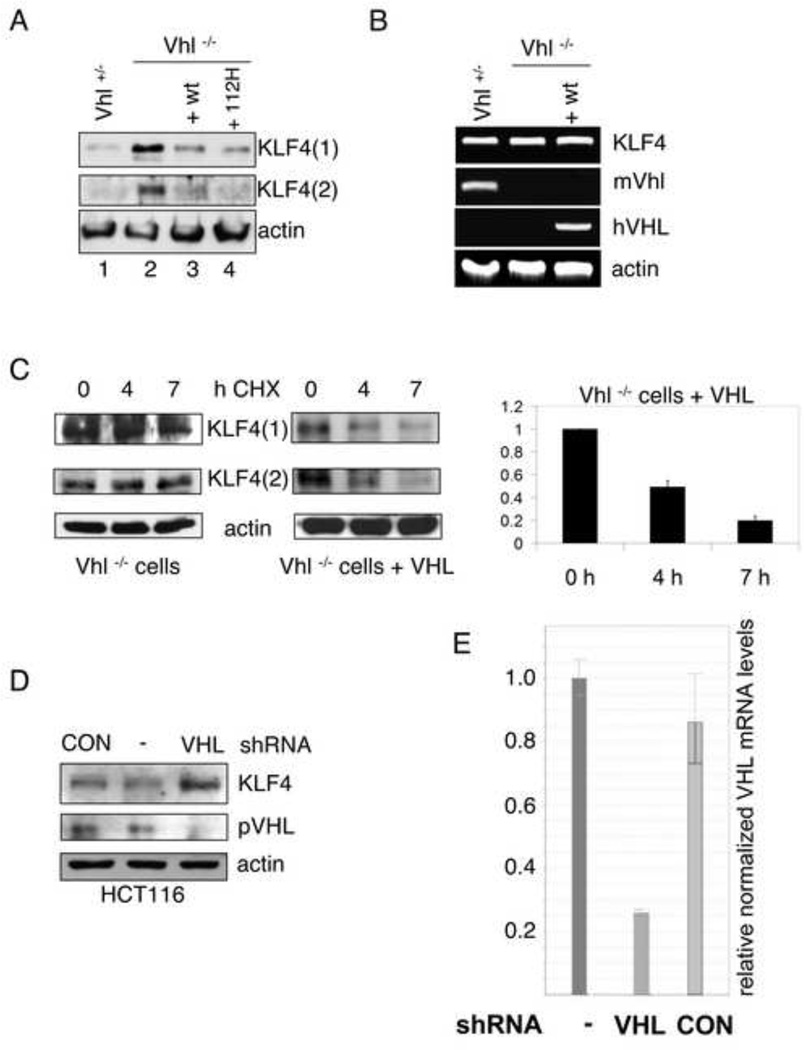
To test whether pVHL controls KLF4 turnover, we wanted to look at VHL-negative cells. Because VHL loss is embryonic lethal and because we did not want to use cancer cells (such as VHL-negative clear cell carcinomas), we resorted to embryonic stem cells (ESCs) derived from mice lacking Vhl described earlier (Rathmell et al., 2004). Interestingly, these cells have elevated levels of the cdk inhibitors p21 and p27 (Mack et al., 2005) compared to wild type or Vhl hemizygous mice. We examined KLF4 protein levels in ESCs from Vhl+/− and Vhl−/− mice as well as in Vhl−/− ESCs reconstituted with human wild type (VHLwt) or mutant VHL (VHL112H) cDNA (Rathmell et al., 2004). Immunoblots using two different antibodies against KLF4 show highly elevated levels of KLF4 in Vhl−/− compared to Vhl+/− ESCs (Fig. 2A, compare lanes 1 and 2). Murine ESCs expressing human wild type or Y112H mutant pVHL had KLF4 levels similar to Vhl+/− ESCs (Fig. 2A), indicating that murine KLF4 is a downstream target of both murine and human pVHL. Furthermore, deletion but not Y112H mutation of VHL leads to KLF4 upregulation. Tyrosine 112 lies in the beta domain of VHL (Stebbins et al., 1999), in the pocket bound by the carboxy-terminal oxygen-dependent degradation motif of HIF1α (Hon et al., 2002). Y112H is a frequent mutation found in VHL patients and was proposed to impair the ability of pVHL to bind and degrade human HIF1α. The fact that Y112H mutant pVHL was able to lower KLF4 levels suggests that VHL-dependent regulation of KLF4 is HIF-independent and/or mechanistically different from the regulation of HIF by pVHL.
Figure 2. VHL regulates the turnover of KLF4.
(A) Elevated KLF4 levels in Vhl negative embryonic stem cells (ESCs). Whole cell extracts of ESCs from mice expressing pVhl (Vhl+/−) or from Vhl knockout mice (Vhl−/−) complemented with wild type (+ wt) or mutant (+ 112H) human pVHL, were tested for KLF4 expression by immunoblot with two different antibodies against KLF4 (1, goat derived; 2, rabbit derived). (B) RT-PCR analysis of KLF4 and VHL (with species specific primers) mRNAs of ESCs from mice with different VHL background. Total RNA of ESCs was purified and cDNA was prepared. Ethidium bromide stains of PCR products are shown. β-actin levels served as internal controls. (C) VHL-dependent half-life of KLF4. Equal protein amounts of whole cell lysates from Vhl knockout ESCs without (left panel) or with reconstituted pVHL expression (right panel), treated for the indicated time with the protein synthesis-inhibitor cycloheximide (CHX), were probed for KLF4 by immunoblot. Two antibodies against KLF4 were used like in (A). Exposure times were adjusted to show trends in the relative KLF4 amounts. (D) VHL knockdown leads to elevated KLF4 levels. HCT116 cells were infected with lentiviruses with an empty cassette (CON) or stably expressing shRNA against VHL. Whole cell lysates of puromycin-selected batches or wild type HCT116 (−) were tested for KLF4 and pVHL expression. (E) VHL mRNA levels of HCT116 after shRNA treatment. Total RNA of infected or uninfected cells was purified and cDNA was prepared. VHL mRNA levels were quantified by real time PCR. β-actin levels served as internal controls. Error bars represent standard deviations.
Vhl+/−, Vhl−/− and VHLwt ESCs have similar KLF4 mRNA levels (Fig. 2B), suggesting that KLF4 levels were not altered due to increased transcription or differences in RNA stability. To further corroborate that KLF4 levels are post-translationally regulated, the stability of the KLF4 protein was tested by measuring protein levels after blocking protein synthesis with cycloheximide (CHX). KLF4 was rapidly degraded in VHLwt ESCs, but had a much longer half-life in ESCs lacking VHL (Fig. 2C). To test whether VHL-mediated regulation of KLF4 is also a feature of human and differentiated cells, we used lentiviruses expressing shRNA to knockdown VHL in HCT116 cells. A VHL-specific, but not control (empty) shRNA vector, led to an approximately 75% reduction in VHL levels (Fig. 2E; see also Suppl. fig. S2 and Experimental Procedures) and a strong induction of KLF4 levels (Fig. 2D). These findings suggest that VHL-dependent regulation of KLF4 is a common feature of murine and human, stem and differentiated cells.
KLF4 has a short half-life in various cancer cells
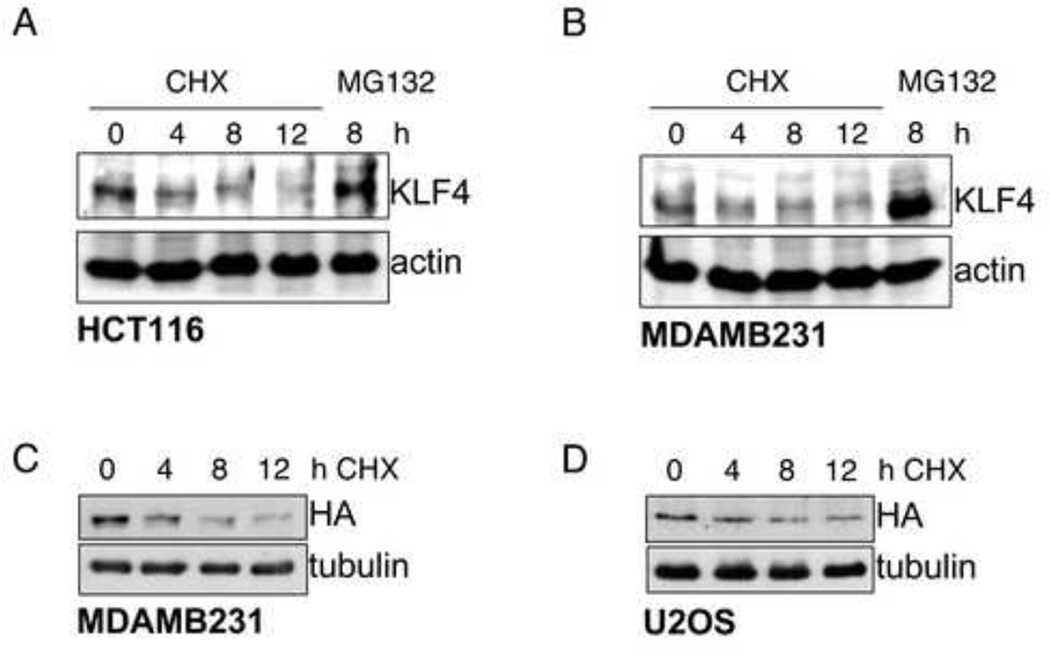
KLF4 has been proposed to function as an oncogene or tumor suppressor depending on the tissue type. Thus, KLF4 levels promote tumorigenesis in the skin, but inhibit cancer formation in the stomach and colon. Furthermore, KLF4 over-expression represents a common feature of breast cancers and probably constitutes an early event in their development (Evans and Liu, 2008). We compared KLF4 levels in cancer cell lines derived from colon (HCT116), lung (U2OS), and breast (HCC1937, MCF7, MDAMB231). KLF4 levels varied greatly with MDAMB231 having high and U2OS low levels of KLF4 (Suppl. Fig. 3A). Yet, treatment of HCT116 and MDAMB231 with cycloheximide showed that turnover rates of KLF4 were rapid also in these cells with relatively high protein levels (Figures 3A and 3B). Conversely, KLF4 levels increased drastically after treatment with MG132, a proteasome inhibitor (Figures 3A and 3B, compare first and last lanes). Furthermore, levels of exogenously expressed tagged KLF4 also rapidly declined in transfected MDAMB231, U2OS, and HCT116, and MCF7 cells after addition of cycloheximide (Figures 3C and 3D; data not shown). These results suggest that KLF4 is kept at cellular steady state levels by a balance of synthesis and ubiquitin/proteasome-mediated turnover in all tested cells, albeit at cell specific levels.
Figure 3. KLF4 has a high turnover rate in several cell types.
(A), (B) KLF4 has a high turn over rate in HCT116 and MDAMB231. Equal protein amounts of whole cell lysates from HCT116 (A) or MDAMB231 (B) treated for the indicated time with the protein synthesis inhibitor cycloheximide (CHX) or the proteasome inhibitor MG132 were probed for endogenous KLF4 by immunoblot. (C, D) Transfected KLF4 has a high turn over rate in MDAMB231 and U2OS. MDAMB231 (C) or U2OS (D) cells were transfected with a FLAG-HA-tagged KLF4 expression vector under the control of a CMV promoter. Equal protein amounts of whole cell lysates from MDAMB231 or U2OS treated for the indicated time with cycloheximide were probed for exogenous KLF4 by immunoblot with an antibody against the hemagglutinin (HA) tag.
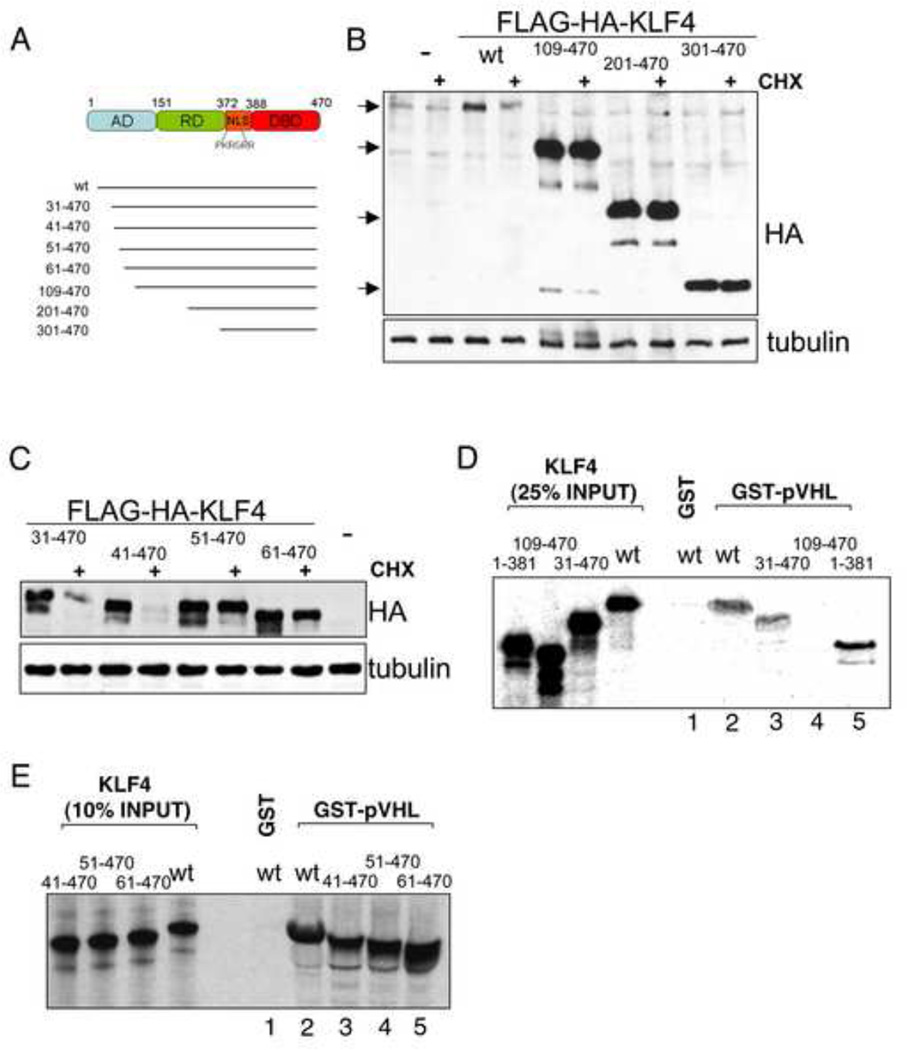
An amino-terminal region of KLF4 is required for its degradation
KLF4 functions as a gene specific activator or repressor. The approximately 55 kDa protein contains an amino-terminal activation domain, shown to interact with the histone acetyltransferases p300 and CBP, a central repressive domain and a carboxy-terminal DNA-binding domain consisting of three zinc fingers. Two carboxy-terminal nuclear localization signals target KLF4 to the nucleus (Evans and Liu, 2008). To identify the regions of KLF4 responsible for its degradation, we generated several FLAG-HA-tagged deletion mutants (Fig. 4A). All deletion mutants retain the nuclear localization signal, avoiding artefacts due to a potential difference in cellular localization. Deletion of the amino-terminal 108 amino acids led to increased stability of KLF4 following cycloheximide treatment (Fig. 4B), suggesting that the amino-terminus contained the ubiquitylation sites and/or the sites of interaction with the ubiquitin ligase. Using additional deletion mutants, a region regulating the degradation of KLF4 could be narrowed to the amino-terminal 50 residues, since KLF4(41–470) was rapidly degraded, but KLF4(51–470) was stable (Fig. 4C). Interestingly, the amino-terminus of KLF4 contains two lysines, K23 and K43, that could function as acceptor sites for ubiquitin (Suppl. fig. S4C).
Figure 4. The KLF4 amino-terminus is important for degradation and interaction with pVHL.
(A) Schematic of human KLF4 with previously identified domains as well as deletion mutants used in the present study. AD, activation domain. RD, repression domain. NLS, nuclear localization signal. DBD, DNA-binding domain. (B) Deletion of KLF4 amino acids 1–108 stabilizes KLF4. Vectors for FLAG-HA-tagged wild type (wt) or deletion mutants of KLF4 were transfected into U2OS. 2 days after transfection cells were mock or cycloheximide (CHX) treated for 6 hours. Equal protein amounts of whole cell lysates were probed for KLF4 mutant levels by immunoblot with an antibody against the hemagglutinin (HA) tag. (C) Fine mapping of the amino-terminal KLF4 region required for degradation. The indicated deletion mutants were tested like in B. (D, E) An amino-terminal region of KLF4 is required for its interaction with KLF4. Beads with GST alone or GST-fused pVHL were incubated with various in vitro translated deletion mutants of KLF4 and bound proteins were analyzed by SDS-PAGE plus autoradiography. Deletion of amino acids 41–50 that stabilize KLF4 (C) does not affect pVHL binding (E).
To determine regions of KLF4 necessary for pVHL binding, in vitro translated deletion-mutants of KLF4 were incubated with glutathione-Sepharose bound GST-pVHL. An analysis of bound proteins showed that amino acids 31–108 of KLF4 were essential for pVHL binding since deletion of the amino-terminal 30 KLF4 residues did not affect binding to GST-pVHL, whereas deletion of residues 1–108 abolished it (Fig. 4D). These findings are in agreement with the previously determined region responsible for KLF4 turnover and provide strong evidence for KLF4 being a direct substrate of the ubiquitin ligase pVHL. We further dissected the interaction site of KLF4 with pVHL to see whether the stabilization of KLF4 by the deletion of residues 41–50 is due to a disruption of pVHL binding to KLF4. Deletion of up to 60 amino-terminal KLF4 residues though had no affect on its interaction with pVHL (Fig. 4E, compare lanes 2 to 5), indicating that pVHL binding is mediated by amino acids 61–108 of KLF4 and raising the possibility that residues 41–50 contain an important ubiquitin acceptor site.
KLF4 degradation is mediated by VHL
To further test the relevance of KLF4 interaction with pVHL for KLF4 stability, we co-expressed FLAG-HA-tagged KLF4 with increasing amounts of FLAG-HA-tagged pVHL in U2OS cells. An immunoblot of the cell lysates showed that KLF4 was degraded by pVHL in a dose-dependent manner (Fig. 5A). Importantly, deletion of amino-terminal 108 residues, shown to be important for the interaction with pVHL (Fig. 4D) and to lead to stabilization of KLF4 after cycloheximide treatment (Fig. 4C), also led to the inability of pVHL to target KLF4 for destruction (Fig. 5B). These results support the hypothesis that pVHL interaction with KLF4 is important for KLF4 degradation and, in combination with the data showing increased endogenous KLF4 levels after VHL deletion or reduction, strongly suggest that pVHL is a major regulator of KLF4.
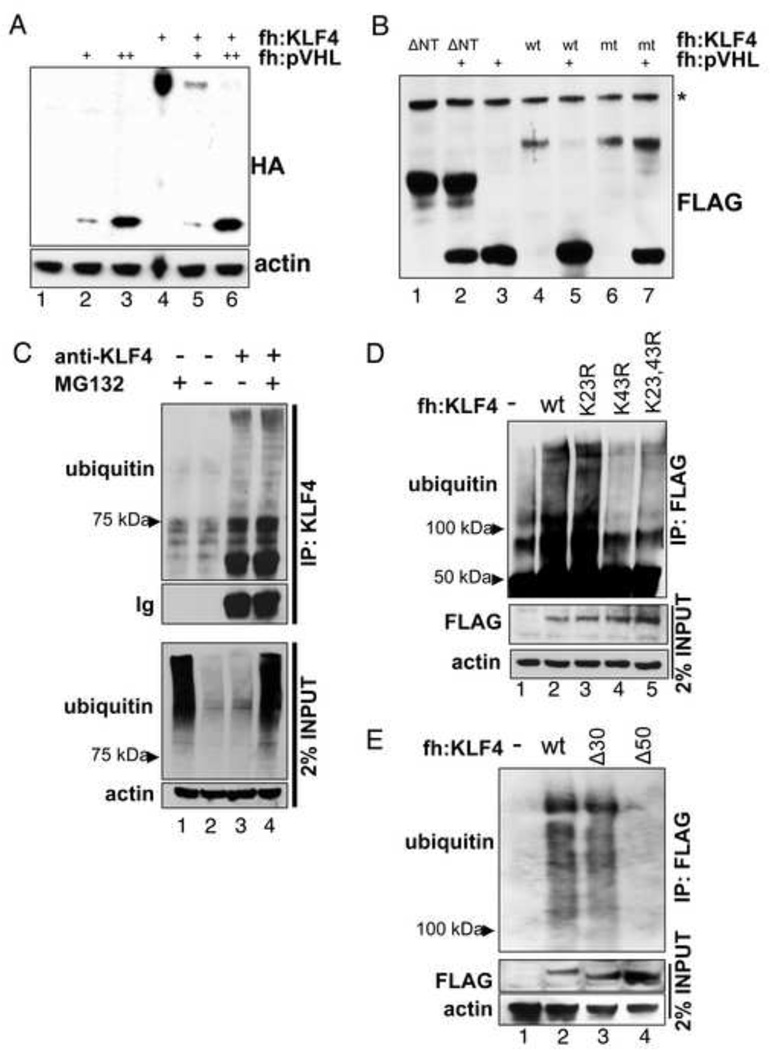
Figure 5. KLF4 is ubiquitylated and targeted for degradation by pVHL.
(A) Dose-dependent degradation of KLF4 by pVHL. An expression vector for FLAG-HA-tagged KLF4 was co-transfected with empty vector or increasing amounts of FLAG-HA-tagged pVHL vector. Equal protein amounts of whole cell lysates were probed for KLF4 and pVHL levels by immunoblot with an antibody against the hemagglutinin (HA) tag. fh: FLAG-HA-tag. (B) An amino-terminal deletion mutant and a point mutant of KLF4 are not degraded by pVHL. KLF4 degradation of KLF(109–470) (ΔNT) or of the KLF4 double mutant K23R,K43R (mt) by pVHL was tested like in A. *, unspecific band. (C) Ubiquitylation of KLF4. Endogenous KLF4 was immunoprecipitated from HCT116 and tested for ubiquitylation by immunoblot. Where indicated cells were treated with MG132. (D, E) Certain point or deletion mutations affect KLF4 ubiquitylation. Cells were transiently transfected with an expression vector for FLAG-HA-tagged wild type or KLF4 with amino-terminal point or deletion mutations. Cells were treated with MG132 for 4 hours before harvesting. M2 (anti-FLAG) immunoprecipitates were probed for ubiquitylated KLF4 by immunoblot with an antibody against ubiquitin. 2% lysate input were tested as control. Δ30: KLF4(31–470); Δ50: KLF4(51–470).
pVHL mediated degradation of its substrates is mediated by the ubiquitin-proteasome pathway. We immunoprecipitated endogenous KLF4 with an antibody against KLF4 and probed the immunoblot for ubiquitin. KLF4 showed a poly-ubiquitylation pattern characteristic for proteins targeted for proteasome dependent degradation (Fig. 5C). Treatment of cells with the proteasome inhibitor MG132 increased high molecular weight ubiquitin signals (Fig. C, lanes 3 and 4).
The mechanistic role of the KLF4 amino-terminus in regulation by pVHL
Our studies with KLF4 deletion mutants indicate that the KLF4 amino-terminus plays a special role in the regulation of KLF4: it contains the pVHL-interaction site (Fig. 4D) and deletion of amino acids 40 to 50 stabilizes the protein (Fig. 4C). HIF1α and HIF2α contain prolines within LAP (leucine-alanine-proline) consensus sequences that are hydroxylated by prolyl-hydroxylases and subsequently bound by pVHL (Ivan et al., 2001; Kaelin, 2008). Intriguingly, the KLF4 amino-terminus contains a proline at residue 81 in a region that also would fit the description of the HIF degron (see Suppl. Fig. S4A). Mutation of proline 81 to alanine though did not abolish pVHL-mediated degradation of KLF4 (Suppl. Fig. S4B), suggesting that the mechanism of KLF4 degradation by pVHL differs from HIF destruction. Interestingly, the amino-terminus of KLF4 contains two lysines, K23 and K43, within the KLF4 activation domain. We next wanted to test whether these could function as acceptor sites for ubiquitin. A previous proteomic screen for ubiquitylated proteins had identified KLF4 K23 as an ubiquitylation site (Tan et al., 2008). We therefore introduced a charge conserving K23R mutation in our tagged KLF4 vector and transfected HCT116 cells with a plasmid expressing FLAG-HA-tagged wild type or mutant KLF4. The cells were treated with MG132 before harvesting and precipitation of KLF4 from whole cell lysates with M2-(anti-FLAG)-agarose. Ubiquitylation was analyzed by immunoblotting. A characteristic smear for ubiquitylated KLF4 above 100 KDa was observed in precipitates of cells expressing tagged wild type KLF4 (Fig. 5D, compare lanes 2 to 5). An immunoblot for ubiquitin of purified mutant KLF4 revealed no significant change in ubiquitylation of the K23R mutant compared to wild type (Fig. 5D, lanes 2 versus 3). Mutation of lysine K43 alone or in combination with K23R though nearly completely abolished KLF4 ubiquitylation (Fig. 5D, lane 4 and 5) indicating that K43 is a major receptor residue for ubiquitin and likely the cause for the stabilizing effect of residues 40 to 50 deletion. To further corroborate the importance of K43 for KLF4 ubiquitylation, we used the same approach to examine the effect of deletion mutants in the amino-terminus on KLF4 ubiquitylation. Deletion of residues 1–30 did not significantly influence the degree of KLF4 ubiquitylation, whereas the deletion of an additional 20 residues (to KLF4 51–470) drastically reduced the signal for precipitated ubiquitin chains (Fig. 5E), establishing K43 as the main KLF4 acceptor site for poly-ubiquitin chains. Intriguingly, mutation of the amino-terminal KLF4 lysines K23 and K43 protects KLF4 from pVHL mediated degradation (Fig. 5B).
Antimitotic affect of VHL knockdown in HCT116 cells
Previous reports have associated elevated KLF4 levels in HCT116 cells with growth arrest (Yoon et al., 2003), likely due to the ability of KLF4 to induce the cdk inhibitor p21 and to inhibit the cyclin D1 promoters (Evans and Liu, 2008). We therefore compared HCT116 cells with normal or pVHL levels reduced by stable shRNA mediated knockdown (see Figures 2D and E). To investigate the impact of VHL status and the concomitant change in KLF4 levels on the CDK inhibitor p21 transcription, we quantified p21 mRNA levels by real time PCR. p21 mRNA levels were more than three times higher in VHL knockdown HCT116 cells compared to wild type HCT116 (Fig. 6A), in agreement with higher p21 levels found in VHL-knockout murine embryonic stem cells (Mack et al., 2005).
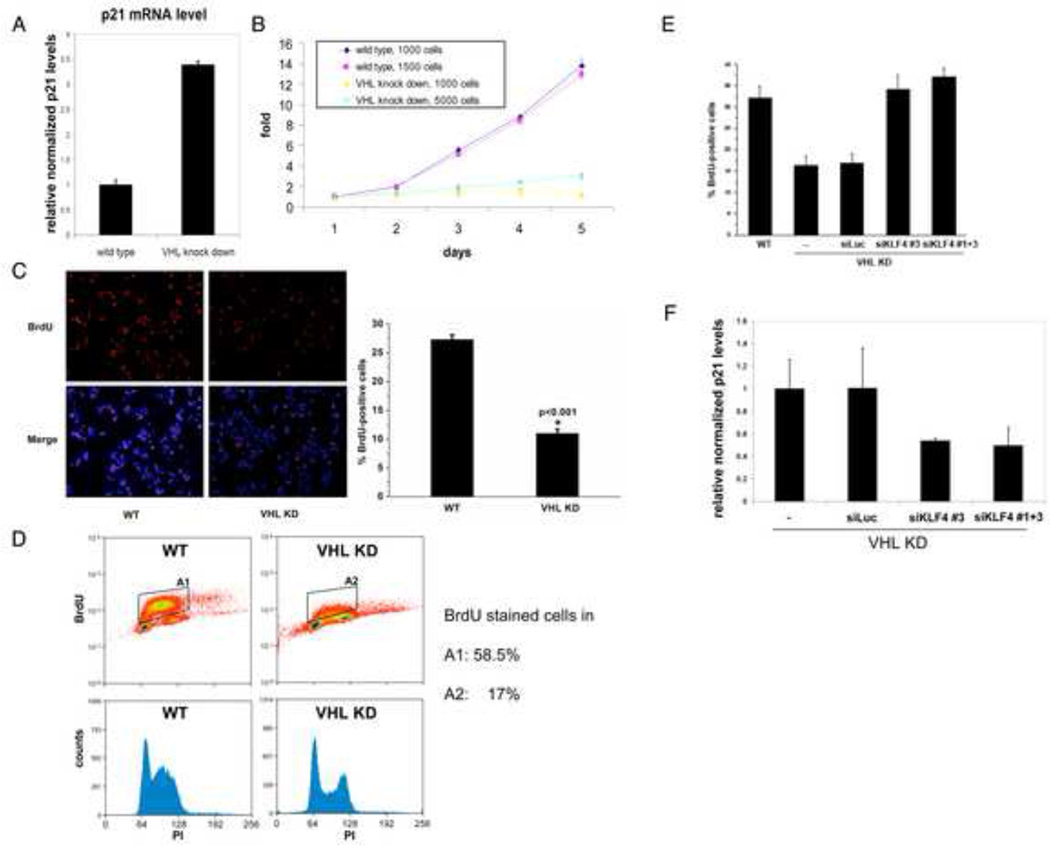
Figure 6. Characterization of HCT116 cells with VHL knockdown.
(A) p21 levels are elevated in VHL-knockdown HCT116 cells. Total RNA from HCT116 cells with wild type or knockdown levels of VHL was purified and cDNA was prepared. p21 mRNA levels were quantified by real time PCR and normalized to β-actin levels. (B) Cell proliferation of HCT116 is impaired by VHL knockdown. Indicated numbers of cells were plated and MTS assays were used to measure the amount of viable cells over 5 days. The MTS values at day one were arbitrarily set at 1. (C) Reduced cell cycle entry of HCT116 by VHL knockdown. HCT116 cells with wild type (WT) or knockdown levels of VHL (VHL KD) were treated with BrdU for 45 minutes and stained for BrdU (upper, left panel) and DAPI. A merged picture is shown in the lower left panel. A diagram of quantitated staining is shown in the right panel. (D) Cell cycle profile of wild type and VHL knock down HCT116. HCT116 cells with wild type or knockdown levels of VHL were BrdU and propidium iodide stained and analysed by FACS. VHL knockdown leads to a strong decrease in cells in S phase. (E) KLF4 silencing in VHL knockdown cells leads to increased cell proliferation. HCT116 cells with knockdown levels of VHL (VHL KD) were transfected with siRNA against KLF4 or luciferase and treated with BrdU (left panel) like in (C). Wild type (WT) HCT116 served as comparison for the BrdU staining. (F) p21 levels are reduced in VHL-knockdown HCT116 cells after KLF4 silencing. Total RNA from HCT116 cells with knockdown levels of VHL was purified after transfection with siRNA against KLF4 or luciferase and cDNA was prepared. p21 mRNA levels were quantified by real time PCR and normalized to β-actin levels. Error bars represent standard deviations.
Measuring cell proliferation using an MTS assay, we found that HCT116 with wild type VHL levels have a doubling time of less than 1 day in the exponential proliferation phase. HCT116 cells with knockdown levels of VHL showed a much reduced proliferation rate with doubling times of more than 48 hours, and sparsely seeded cells were particularly growth impaired (Fig. 6B). Finally, to confirm that the observed colorimetric values of formazan production in the MTS assay reflected VHL induced changes in cell cycle, we also measured bromodeoxyuridine (BrdU) incorporation by immunocytochemical staining and FACS. HCT116 with VHL knockdown have a strongly reduced population staining for BrdU compared to wild type cells (Fig. 6C, 6D), mirroring the observed respective proliferation rates.
In order to see whether the antimitotic effect of VHL knockdown is KLF4 dependent we silenced KLF4 in VHL knockdown cells by transient transfection with siRNA against KLF4. KLF4 silencing decreased p21 levels (Fig. 6F) and enabled VHL knockdown cells to reenter the cell cycle as evidenced by increased BrdU staining (Fig.6E).
Increased levels of pVHL in colon cancer cells correlate with reduced KLF4 levels
pVHL is a well documented tumor suppressor and the loss of VHL plays an important role particular in the formation of renal cancers (Kaelin, 2007). Our observations as well as previous findings that VHL deficiency in mouse models impairs tumor growth (Mack et al., 2005) and acute loss of VHL can lead to senescence (Young et al., 2008) allows for the possibility that under certain conditions pVHL could function as tumor facilitating factor. Such circumstances could be found in cells where KLF4 plays a role as tumor suppressor, such as gastric and colorectal cells. Indeed, our analysis of a published Affymetrix microarray data set from 5 normal and 100 cancer colon tissues (Kaiser et al., 2007), revealed that none of the assayed colon cancer cells showed loss of the VHL gene or even reduced VHL transcription. Indeed, VHL mRNA levels in the colon cancer cells were significantly upregulated, although on average by a small percentage (Fig. 7A). Because RNA levels often are a poor indicator for actual protein expression, we used histochemical staining to look at the expression of both pVHL and KLF4 in colon cancer and adjacent tissue samples. As expected given the well documented role of KLF4 as tumor suppressor in colorectal and gastric cancers, KLF4 levels were significantly decreased in colon cancer tissues. Intriguingly, VHL protein levels were much higher in colon cancer than in adjacent normal tissues (Fig. 7B). A representative stain is shown in Fig. 7C. We speculate that in these colon cancer samples pVHL also retained its function, at least in regard to its ability to target KLF4 for degradation. Because the tumor-suppressive potential of VHL is in great part attributable to the role of its downstream-target HIF in angiogenesis, we predict that a tumor-facilitating effect of pVHL overexpression might be a feature of early tumorigenesis in tissues where KLF4 plays an important antimitotic role.
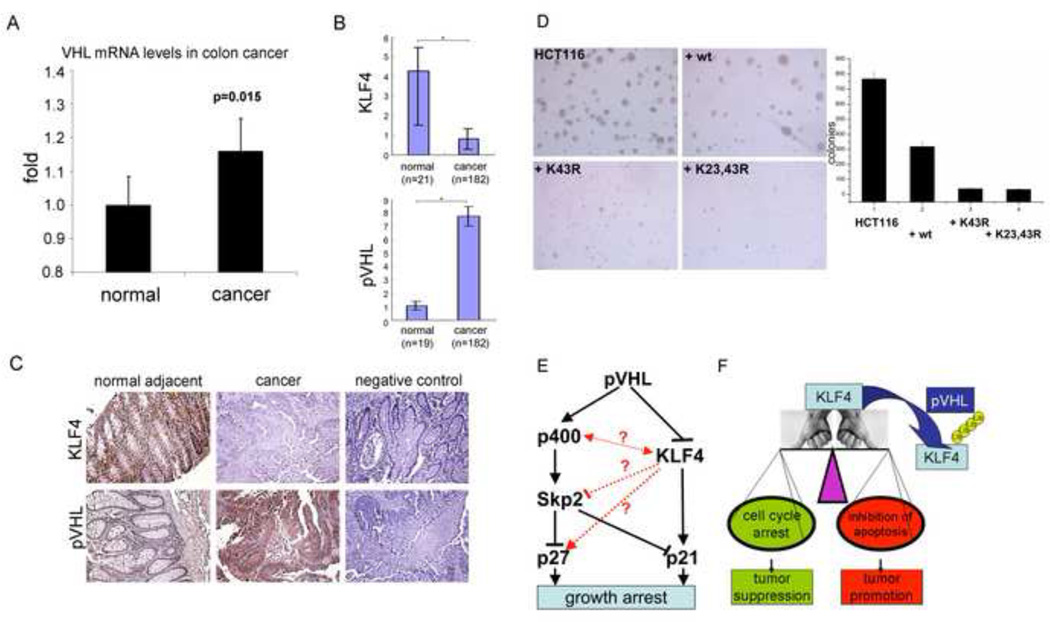
Figure 7. The role of KLF4 and VHL in colon cancer.
(A) mRNA levels of VHL in colon cancer. Published Affymetrix microarray data of 5 normal and 100 cancer colon tissues were analyzed for VHL mRNA levels. None of the analyzed cancer samples showed loss of the VHL gene or decreased VHL RNA levels. (B), (C) Immunohistochemical analysis of KLF4 and pVHL expression in colon cancer tissues. Sections from colon cancer and adjacent tissues were analyzed by immunostaining against KLF4 and pVHL. Staining without primary antibody served as negative control. A quantification of tissue staining, as described in Experimental Procedures, is shown in (B). A representative sample is shown in (C). *, p<0.05. (D) Transformation suppression by wild type and mutant KLF4. HCT116 cells were infected with lentiviruses expressing wild type or KLF4 mutants and tested for colony formation in soft agar in anchorage-independent growth assays. Wild type KLF4 greatly reduced colony formation, but KLF4 with mutant ubiquitylation sites increased the inhibitory potential of KLF4 even further. (E) Model of parallel pathways signaling from pVHL to p21 and p27 and possible cross talks between them. (F) Diagram of the importance of KLF4 in cell fate decision and the regulation of KLF4 by pVHL. Since KLF4 is thought to function as tumor suppressor or oncogene depending on the balance between its impact on cell cycle arrest or inhibition of apoptosis, so would its degradation by pVHL lead to either tumor promotion or suppression. Error bars represent standard deviations.
KLF4 turnover can affect tumor formation
To see whether KLF4 stability affects colon cancer tumorigenesis, we infected HCT116 cells with lentiviruses expressing wild type or KLF4 mutants. These cells were then tested for colony formation in soft agar as anchorage-independent growth assays. Representative images of stained plates as well as a quantification are shown in Fig. 7D (for raw data and immunoblots of KLF4 levels see Supplementary Figures 7A and 7B). Wild type KLF4 greatly reduced colony formation (Fig. 7D, compare upper images and quantification), but KLF4 with mutant ubiquitylation sites increased the inhibitory potential of KLF4 much further (compare upper right image and lower images). While the inhibitory effect of exogenous wild type KLF4 was around 2.5 fold, K43R and K23,43R KLF4 mutants decreased colony formation over 10 fold. These findings confirm that KLF4 has a strong tumor suppressor role in colon cancer and show that the ubiquitylation sites of KLF4, and by inference the turnover rate of KLF4, can influence tumor progression in colon cancer cells.
DISCUSSION
Previous studies have shown that pVHL plays a role in murine cell cycle control independent of its regulation of the hypoxia inducible factors (Mack et al., 2005; Young et al., 2008). The cyclin-dependent kinase inhibitors p21 and p27 were found to mediate growth arrest after VHL loss in a mechanistically unidentified pathway involving Skp2 and p400 (Fig. 7E, left side), but in a p53 (and HIF) independent fashion. The work presented here establishes the transcription factor KLF4 as a target of the E3 ligase pVHL in murine and human cells and provides evidence that the degradation of KLF4 by pVHL plays an important role in VHL-dependent growth control. KLF4 was directly bound by pVHL and studies in which pVHL was either overexpressed or depleted have documented a role of VHL in KLF4 degradation and in determining the proliferation rate of human colorectal cancer cells. These studies indicate that KLF4, an activator of the p21 gene, is an important mediator of the reported VHL dependent cell cycle regulation (Fig. 7E, right side).
Parallel pathways or interconnected regulatory network?
Studies on the impact of acute pVHL loss in MEFs led to the proposal of a VHL/p400/Skp2/p27(p21) pathway (Young et al., 2008). Our current analysis shows that pVHL, KLF4, and p21 form a cascade also inducing cell cycle delay. But are the two pathways parallel or interconnected? Are they cell specific? Loss of p400 has been shown to increase both p21 mRNA and protein levels and induce p21-dependent senescence in several human cell lines. Intriguingly, p400 is a component of the human TIP60/NuA4 HAT complex. TIP60 was found to directly interact with KLF4 and to act as co-repressor of KLF4 in the regulation of the HDC promoter activity (Ai et al., 2007). Regarding Skp2 and p27, it was observed that KLF4 overexpression in human pancreatic adenocarcinoma cells not only led to increased mRNA levels of p27 and p21, but also to reduced Skp2 protein levels (Wei et al., 2008). The mechanism and universality of Skp2 down-regulation by KLF4 are presently unknown, but could constitute another interesting connection between players downstream of VHL (Fig. 7E).
Degradation of KLF4 and its impact on cancer
KLF4 was proposed to be a tumor suppressor based on the observation that ectopic expression of KLF4 in several cell types results in slow growth and that KLF4 plays a role in maintaining the integrity of the G1/S and G2/M checkpoints following DNA damage. Several studies have shown down-regulation or mutation of KLF4 in human tumors, especially of the colon and stomach, and over-expression of KLF4 in a human colon cancer cell line reduces in vivo tumorigenicity (McConnell et al., 2007). Furthermore, gastric-tissue-specific deletion of KLF4 in mice induces gastric hyperplasia and polyps (Katz et al., 2005). This anti-proliferative activity of KLF4 is congruent with its ability to transcriptionally activate, at least in certain cells, the cell cycle inhibitors p21, p27, and p57 and to repress various cyclins and ornithine decarboxylase (Evans and Liu, 2008).
In contrast, elevated levels of KLF4 have been reported in up to 70% of primary mammary cancers and KLF4 over-expression at the stage of ductal carcinomas suggests that this represents an early event in breast cancer progression. Nuclear localization of KLF4 was found to be a prognostic factor indicating an aggressive phenotype in breast cancers. Furthermore, depletion of KLF4 from breast cancer cells was shown to cause p53-dependent apoptosis, in agreement with an anti-apoptotic function of KLF4 observed also in other cell types (Rowland et al., 2005). KLF4 over-expression was also linked to the development of squamous cell carcinoma and unbiased genetic screens have identified KLF4 as a potential oncogene (Rowland and Peeper, 2006). Thus, depending on the type of cancer involved, KLF4 can play a repressive or an activating role in tumorigenesis. At present the molecular mechanisms underlying this ambiguity are poorly understood. It was proposed that the genetic background, particularly concerning genes controlling p21, determines the outcome of KLF4 signaling (Rowland et al., 2005). The pathways of KLF4 and of the tumor suppressor p53 are interlinked and both genes regulate growth arrest and apoptosis. Yet, in contrast to p53, KLF4 seems to negatively regulate apoptosis, a fact that might at least partially be responsible for its oncogenic potential (Rowland and Peeper, 2006). The key role of KLF4 in this complex intersection of pro- and anti-proliferative signalling network emphasizes the importance of understanding the factors and modifications that control KLF4.
Von Hippel-Lindau syndrome is characterized by the dominantly inheredited predisposition to develop highly vascularized tumors (Kaelin, 2007). Mouse models demonstrate that homozygous disruption of VHL results in embryonic lethality and tumorigenesis in VHL patients is associated with the loss or inactivation of the wild type allele following Knudson’s “two-hit” hypothesis (Kaelin, 2007). Loss of VHL function is also observed in sporadic cases of clear-cell renal carcinomas (Kaelin, 2008) and a decrease of VHL levels has been correlated with breast cancers.
Although most studies correlate VHL function as tumor suppressor to its ability to degrade HIF under normoxic conditions, several aspects point to a more complex role of pVHL in tumorigenesis: Other substrates of its ubiquitin ligase activity have been identified (see introduction) and besides the already discussed growth inhibitory effect of VHL loss on proliferation (Mack et al., 2005; Mack et al., 2003) VHL was shown to play a role in microtubule stability, assembly of extracellular matrix and intercellular junctions, primary cilium maintenance (reviewed in (Nyhan et al., 2008)), spindle orientation and chromosomal stability (Thoma et al., 2009).
Our findings that KLF4 is a high turn over protein in several human cell types and is targeted for proteasome-dependent degradation by pVHL open the possibility to a better understanding of HIF-independent effects of VHL functional status on tumorigenesis and the pleiotropic phenotype of VHL mutations. While it is not yet understood how the forces that KLF4 exerts on the balance between its ability to promote (via inhibition of apoptosis) or inhibit (via cell cycle arrest) tumorigenesis are regulated, their effect on the cell fate decision could be mitigated or exacerbated by pVHL levels (Fig. 7F). Thus pVHL could conversely repress or facilitate tumor formation and, besides its well documented role as tumor suppressor in certain tissues, play a deleterious role in other tissues given the appropriate cellular environment. Because pVHL’s best documented tumor suppressive action is via HIF, particularly due to the downstream regulation of VEGF-induced angiogenesis, we envision that circumstances where pVHL promotes tumor formation is at an early stage and in cells where KLF4 is an important inducer of cell cycle arrest after genotoxic stress. Interestingly, it has been proposed that loss of KLF4 in colorectal cancer is an early event (Evans and Liu, 2008) and high levels of pVHL were reported in early stages of colorectal cancers, whereas an angiogenic switch in cancer progression was suggested to select for downregulation of pVHL at later stages (Giles et al., 2006).
Regulation of KLF4 degradation
Although the knockdown of VHL in several cell lines led to an increase in KLF4 levels, we do not see a strict inverse correlation between VHL and KLF4 when comparing different cell lines (data not shown). This indicates that KLF4 degradation by VHL is tightly regulated. Possible mechanisms include post-translational modification of KLF4 or compartimentalization. Several substrates of ubiquitin ligases have been found to be regulated by modification of their interaction or ubiquitylation sites. Such modifications include phosphorylation, acetylation, or hydroxylation. The mechanisms governing KLF4 degradation by pVHL are of interest not only for the role of KLF4 in cancer, but also in differentiation and stem cell renewal.
EXPERIMENTAL PROCEDURES
Nuclear extract preparation and complex purification from a HeLa S cell line stably expressing KLF4
Hela S cells were infected with a retrovirus encoding FLAG-tagged KLF4. Stable tagged KLF4 expressing cell lines were established from individual clones after selection with puromycin. Nuclear extract from HeLa was prepared as described (Dignam et al., 1983). KLF4 interacting proteins were purified from the nuclear extract of a HeLa cell line that stably expresses FLAG-tagged KLF4 by immunopurification on M2 agarose (Sigma) and washing thrice with BC buffer (10% glycerol, 20 mM Tris-HCl pH 7.9, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM DTT, 0.05% NP40) containing 300 mM KCl and once with BC buffer containing 100 mM KCl. The complex was eluted with FLAG peptide in BC buffer containing 100 mM KCl.
RT-PCR assays
RNA was isolated from cells with TRIZOL® (Invitrogen) and reverse transcribed with SuperscriptTM II RT (Invitrogen) according to the manufacturer’s protocol. RTPCR results were run on agarose gels and stained with ethidium bromide. Alternatively, cDNA was used for quantitative PCR analysis on a StepOnePlus thermocycler (Applied Biosystems).
Interaction assays
GST-fused pVHL was expressed in the Escherichia coli strain XA90. Wild type KLF4 and deletion mutants were labelled with 35S-methionine using TNT reticulate lysates, following the standard protocol provided by Promega. In vitro translated proteins were incubated with resin bound GST-pVHL (10µg) by rotating at 4°C for 4 hours in BC buffer containing 100 mM KCl and 0.1 µg/µl BSA. After washes with BC buffer containing 150 mM KCl, the resin was directly boiled in SDS loading buffer before subjecting to SDS-PAGE. Gels were incubated in AmplifyTM (Amersham Pharmacia) before drying and autoradiography to enhance the signal. For in vivo interaction assays, FLAG-HA-tagged KLF4 was co-expressed with pVHL in MDAMB231 cells. 24 h after transfection the cells were washed and crosslinked with 1 mM dimethyl 3,3’-dithiobispropionimidate (DTBP) in PBS by shaking for 20 minutes at room temperature. After washes with PBS, the cells were lysed in stop-lysis-wash (SLW) buffer (50 mM Tris-HCl, pH7.5, 200 mM NaCl, 5 mM EDTA, 0.5% NP40, 0.5 mM PMSF) and sonicated. The lysate was cleared by centrifugation and incubated with M2 agarose for 8 h. After washes with SLW buffer, the beads were boiled in reducing SDS buffer for elution and reversal of the crosslink. Eluates were examined by immunoblotting. (The secondary antibodies recognize the bands of the M2 heavy and short chains as well.)
Stability, degradation, and ubiquitylation assays
Untransfected or transiently transfected cells were treated with 100µg/ml of the protein synthesis inhibitor cycloheximide (Sigma-Aldrich) to measure, respectively, the stability of endogenous or exogenous levels of wild type or mutant KLF4. Unless otherwise stated, cells were harvested 6 hours after addition of the drug. In the case of exogenous KLF4, cycloheximide was added 2 days after transfection. The proteasome dependent degradation was assessed by addition of 30µM MG132 for the indicated amount of time. For VHL dependent degradation assays HCT116 or U2OS cells were transfected with 2 µg KLF4 expression vector and either 1 or 2 µg VHL expression and/or empty vector. For ubiquitylation assays cells were treated with 30µM MG132 for 4 hours were indicated. In the case of experiments with tagged KLF4, cells were transfected with 10 µg FLAG-HA-tagged KLF4 vector or empty vector. Whole cell lysates were incubated with anti-KLF4 antibody bound to Prot A/G agarose or M2 agarose for 4 h and washed thrice with BC buffer containing 300 mM KCl and once with BC buffer containing 100 mM KCl. The beads were boiled in reducing SDS and eluates were examined by immunoblotting against ubiquitin.
Cell culture conditions, immunohistochemical staining, cell cycle analyses, soft agar colony formation assays, and antibodies are described in Supplementary Experimental procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs. Wade Harper and Jianping Jin for kindly providing the TAP purification vector. We also thank Hyun Kim for his help to establish the stable HeLa cell line and especially Dr. Zhuan Zhou for his help with the soft agar and endogenous interaction assays, Drs. Wei Qian and Serah Choi for their help with FACS analysis, and Lucas Santana dos Santos for his help to analyze published affymetrix data. This work is supported by grant CA154695 from the National Institute of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ai W, Zheng H, Yang X, Liu Y, Wang TC. Tip60 functions as a potential corepressor of KLF4 in regulation of HDC promoter activity. Nucleic Acids Res. 2007;35:6137–6149. doi: 10.1093/nar/gkm656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Wang X, Zhou Y, Offner G, Tseng CC. Destabilization of Kruppel-like factor 4 protein in response to serum stimulation involves the ubiquitin-proteasome pathway. Cancer Res. 2005;65:10394–10400. doi: 10.1158/0008-5472.CAN-05-2059. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PM, Liu C. Roles of Krupel-like factor 4 in normal homeostasis, cancer and stem cells. Acta Biochim Biophys Sin (Shanghai) 2008;40:554–564. doi: 10.1111/j.1745-7270.2008.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaleb AM, Katz JP, Kaestner KH, Du JX, Yang VW. Kruppel-like factor 4 exhibits antiapoptotic activity following gamma-radiation-induced DNA damage. Oncogene. 2007;26:2365–2373. doi: 10.1038/sj.onc.1210022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles RH, Lolkema MP, Snijckers CM, Belderbos M, van der Groep P, Mans DA, van Beest M, van Noort M, Goldschmeding R, van Diest PJ, et al. Interplay between VHL/HIF1alpha and Wnt/beta-catenin pathways during colorectal tumorigenesis. Oncogene. 2006;25:3065–3070. doi: 10.1038/sj.onc.1209330. [DOI] [PubMed] [Google Scholar]
- Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Kaelin WG. Von Hippel-Lindau disease. Annu Rev Pathol. 2007;2:145–173. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG, Furth EE, Kaestner KH. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–945. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Mack FA, Patel JH, Biju MP, Haase VH, Simon MC. Decreased growth of Vhl−/− fibrosarcomas is associated with elevated levels of cyclin kinase inhibitors p21 and p27. Mol Cell Biol. 2005;25:4565–4578. doi: 10.1128/MCB.25.11.4565-4578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack FA, Rathmell WK, Arsham AM, Gnarra J, Keith B, Simon MC. Loss of pVHL is sufficient to cause HIF dysregulation in primary cells but does not promote tumor growth. Cancer Cell. 2003;3:75–88. doi: 10.1016/s1535-6108(02)00240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyhan MJ, O'Sullivan GC, McKenna SL. Role of the VHL (von Hippel-Lindau) gene in renal cancer: a multifunctional tumour suppressor. Biochem Soc Trans. 2008;36:472–478. doi: 10.1042/BST0360472. [DOI] [PubMed] [Google Scholar]
- Rathmell WK, Hickey MM, Bezman NA, Chmielecki CA, Carraway NC, Simon MC. In vitro and in vivo models analyzing von Hippel-Lindau disease-specific mutations. Cancer Res. 2004;64:8595–8603. doi: 10.1158/0008-5472.CAN-04-1430. [DOI] [PubMed] [Google Scholar]
- Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Kaelin WG, Jr, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- Tan F, Lu L, Cai Y, Wang J, Xie Y, Wang L, Gong Y, Xu BE, Wu J, Luo Y, et al. Proteomic analysis of ubiquitinated proteins in normal hepatocyte cell line Chang liver cells. Proteomics. 2008;8:2885–2896. doi: 10.1002/pmic.200700887. [DOI] [PubMed] [Google Scholar]
- Thoma CR, Toso A, Gutbrodt KL, Reggi SP, Frew IJ, Schraml P, Hergovich A, Moch H, Meraldi P, Krek W. VHL loss causes spindle misorientation and chromosome instability. Nat Cell Biol. 2009;11:994–1001. doi: 10.1038/ncb1912. [DOI] [PubMed] [Google Scholar]
- Wei D, Kanai M, Jia Z, Le X, Xie K. Kruppel-like factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer Res. 2008;68:4631–4639. doi: 10.1158/0008-5472.CAN-07-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AP, Schlisio S, Minamishima YA, Zhang Q, Li L, Grisanzio C, Signoretti S, Kaelin WG., Jr VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nat Cell Biol. 2008;10:361–369. doi: 10.1038/ncb1699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.