Termination is a crucial step in translation, most notably because premature termination can lead to toxic truncated polypeptides. Most interesting is the fact that stop codons are read by a completely different mechanism from that of sense codons. In recent years, rapid progress has been made in the structural biology of complexes of bacterial ribosomes bound to translation termination factors, much of which has been discussed in earlier reviews [1–5]. Here, we present a brief overview of the structures of bacterial translation termination complexes. The first part summarizes what has been learned from crystal structures of complexes containing the class I release factors RF1 and RF2. In the second part, we discuss the results and implications of two recent x-ray structures of complexes of ribosomes bound to the translational GTPase RF3. These structures have provided many insights and a number of surprises. While structures alone do not tell us how these complicated molecular assemblies work, is it nevertheless clear that it will not be possible to understand their mechanisms without detailed structural information.
I. Structures of the Class I Release Factors RF1 and RF2 bound to the ribosome
The discovery of stop codons, the triplet signals in mRNA that specify translation termination, originated from genetic experiments aimed at characterization of nonsense mutations nearly a half century ago [6,7]. The type I release factors RF1 (specific for UAG and UAA) and RF2 (specific for UGA and UAA) were discovered shortly thereafter [8]. Remarkably, clear answers to the key questions surrounding the mechanism of termination have only begun to emerge in the past few years.
Three Key Questions
The unexpected finding that recognition of stop codons is mediated by protein factors, rather than tRNAs, raised three fundamental questions concerning the termination mechanism. First, do the release factors themselves directly recognize the stop codons, or do they indirectly mediate recognition, for example through interactions with ribosomal RNA [9–13]? Second, is the hydrolysis reaction catalyzed by the factors themselves, or is the peptidyl transferase activity of the ribosome subverted in the presence of the factor to use water, rather than aminoacyl-tRNA, as an acceptor [14,15]? A third key question is how the very high accuracy of stop codon-dependent termination (which has an error frequency on the order of 10−5) is maintained in the absence of any proof-reading mechanism. Recent crystal structures of all four possible termination complexes [16–19] have provided clear answers to the first two questions and a solid basis for thinking about the third. The factors and corresponding mRNA stop codons in the four complexes are RF1-UAA (solved at 3.2 Å resolution)[16], RF2-UAA (at 3.0 Å)[17], RF1-UAG (at 3.6 Å)[18] and RF2-UGA (at 3.5 Å)[19].
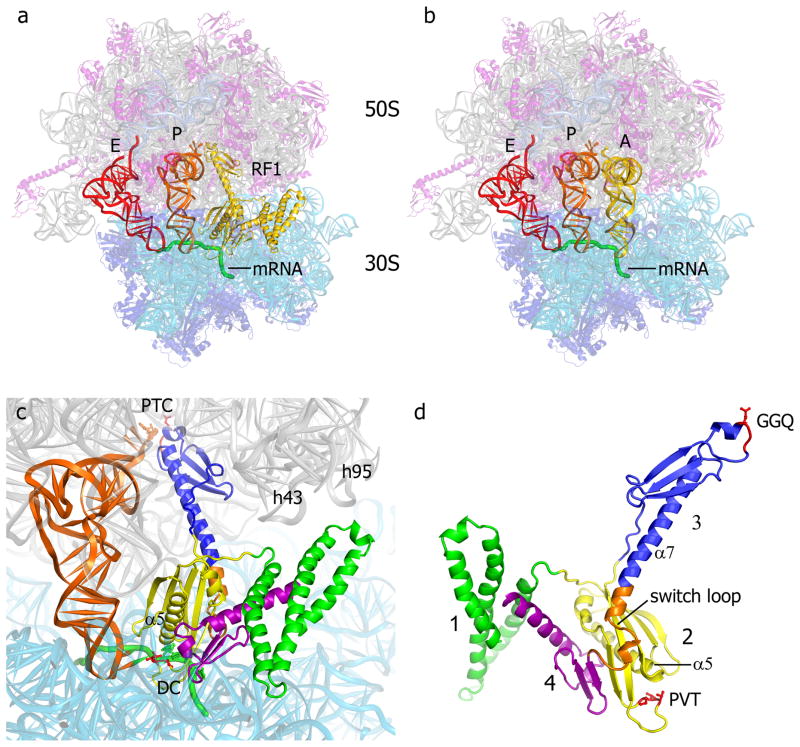
Type I release factors bind to the ribosomal A site, as shown in Figure 1a for RF1 [16]. Domains 2, 3 and 4 of the factor overlap with the space occupied by aminoacyl-tRNA during elongation (Fig. 1a,b,c). The tripeptide sequence PVT in domain 2, implicated in codon recognition by RF1 in domain-swap experiments [20] is found in the 30S subunit decoding site (Fig. 1c,d)[16,18], as is the analogous SPF sequence implicated in codon recognition for RF2 [17,19]. The universally conserved GGQ motif in domain 3, critical for the peptidyl-tRNA hydrolysis activity of release factors [21] is located in the peptidyl transferase center (PTC) next to the 3′ end of the P-site tRNA (Fig. 1c,d). Domain 1 lies adjacent to the A site, near the binding sites for the GTPase factors EF-G, EF-Tu and RF3.
Fig. 1. Binding of RF1 to the 70S ribosome.
(a) Positions of RF1 (yellow), P-site tRNA (orange), E-site tRNA (red) and a mRNA (green) bound to the 70S ribosome [23S rRNA (gray), 5S rRNA (light blue), 16S rRNA (cyan), 50S proteins (magenta) and 30S proteins (dark blue)]. (b) The position of A-site tRNA (yellow) in a 70S elongation complex [54] is similar to that of domains 2, 3 and 4 of RF1. (c) Orientation of RF1 in the A site of the 70S ribosome, along with the P-site tRNA (orange). The peptidyl transferase center (PTC), decoding center (DC) and helices h43 (the L11 stalk) and h95 (the sarcin-ricin loop) of 23S rRNA are indicated. (d) The structure of RF1 in its ribosome-bound conformation. The view is rotated approximately 180° from that shown in panels (a) and (c), for clarity. Domains 1, 2, 3 and 4 are shown in green, yellow, blue and magenta, respectively. The conserved GGQ and PVT motifs are shown in red, and the switch loop (see text) is in orange. From ref. [16].
Stop Codon Recognition
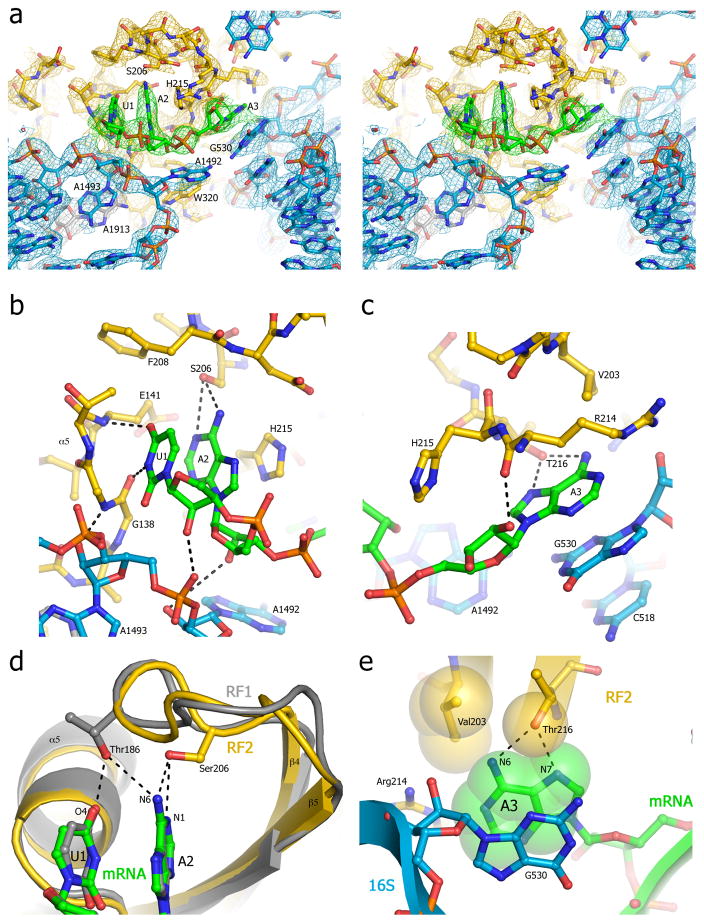
The high-resolution structures clearly show that stop codons are recognized directly by release factors (Fig. 2). The conformation of the stop codons in the A site of termination complexes differs strikingly from that of sense codons [22,23]. The first two bases are stacked on each other, but the plane of the third base lies orthogonal to those of the first two in a separate binding pocket (Fig. 1a). The putative ‘tripeptide anticodon’ motifs PA/VT (for RF1) and SPF (for RF2) do indeed participate in codon recognition; however, only a single amino acid (Thr or Ser, respectively) in each motif is directly involved, and they are primarily responsible for discrimination of the 2nd base of the stop codon (Fig. 2b,d). Thr186 of RF1, positioned between U1 and A2, donates an H-bond to O4 of U1, and so can only accept an H-bond from adenine, whereas Ser206 of RF2 can both donate and accept H-bonds with the Watson-Crick edges of adenine or guanine. To recognize the third base, RF1 uses a combination of Thr194 and Gln181 to discriminate either A or G via their Hoogsteen edges (Fig. 2c). In RF2, there is a Val at the position corresponding to Gln181, restricting the 3rd position to adenine. The first base is universally restricted to U by backbone elements at the N-terminal end of helix α5, where a peptide carbonyl oxygen H-bonds to N3 of the uridine, and a peptide amide NH H-bonds to its O4. This finding explains the failure to obtain mutations that result in altered recognition of the first position of stop codons.
Fig. 2. Recognition of stop codons by RF1 and RF2.
(a) Stereo view of the σA-weighted 3Fo-2Fc electron density map of the stop codon and surrounding elements of RF2 and the ribosome [17]. Electron density is contoured at 1.0 σ for RF2, and at 1.5 σ for rRNA and mRNA, and colored yellow (RF1), green (mRNA) and blue (16S rRNA). (b) Interaction of the hydroxyl group of Ser206 of the SPF motif of RF2 with A2 of the stop codon; (c) Interaction of Thr216 of RF2 with A3 of the stop codon; (d) Comparison of the positions of Thr186 of the PVT motif of RF1 (gray)[16] and Ser206 of the SPF motif of RF2 (yellow)[17], showing their respective modes of recognition of U1 and A2 of the UAA stop codon. (e) Packing of RF2 around A3 of the UAA stop codon. Val203 would be positioned to exclude water from H-bonding to O6 of guanine, helping to discriminate against guanine at position 3. The structure model is represented as Van der Waals surfaces in yellow (RF2), green (mRNA) and blue (16S rRNA). From ref. [17].
Catalysis of Peptidyl-tRNA Hydrolysis
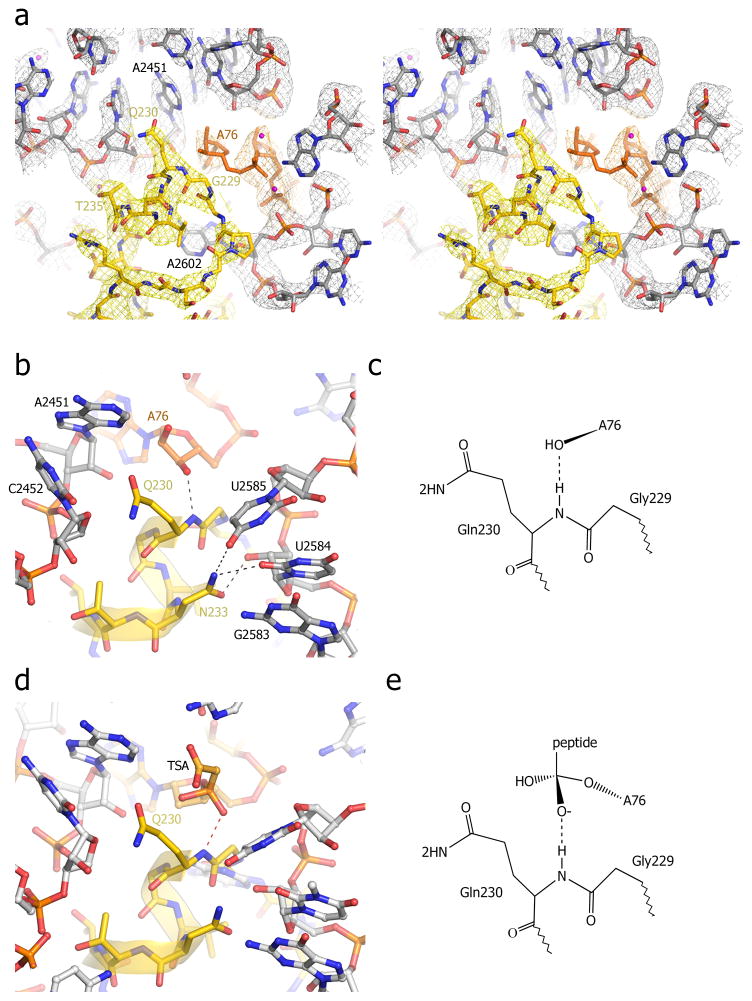
The universally conserved GGQ motif in domain 3 is positioned next to A76 at the 3′ terminus of P-site tRNA (Figs. 1c, 3). In spite of its conservation, mutation of Gln230 in the GGQ sequence to most other amino acids has only a modest effect on catalysis, while mutations of G228 or G229 confer severe decreases in activity [24]. Consistent with this finding, the side-chain of Gln230 is oriented away from the predicted position of the scissile bond. Instead, the backbone NH group of Gln230 is positioned within H-bonding distance of the 3′-hydroxyl of A76. It has been proposed that this backbone NH group plays a dual role in catalysis of the hydrolytic reaction, in stabilization of the tetrahedral transition-state intermediate by H-bonding to the developing oxyanion, and in product stabilization by H-bonding to the 3′-OH of the deacylated tRNA (Fig. 3b–e)[16]. This mechanism was tested by mutation of glutamine 230 to proline, the sole mutation that is able to eliminate the backbone α-NH group [17]. Uniquely among all tested mutations of Gln230, the proline mutation completely abolished catalysis by RF1. The possibility that the effects of the proline substitution were due to a conformational change in the backbone of the GGQ region seems unlikely, since the backbone torsion angles of Gln230 fall well within the range of values commonly observed for proline. Participation of a backbone NH group in stabilization of an oxyanion transititon state has precedent in the mechanisms of esterases [25] and proteases [26] as well as in the Ras-catalyzed hydrolysis of GTP [27].
Fig. 3. Interactions of the universally conserved GGQ motif in the peptidyl transferase center (PTC) of the termination complex.
(a) Stereo view of σA-weighted 3Fo-2Fc electron density map for the PTC. Density for RF1 (yellow), P-tRNA (orange) and 23S rRNA (grey) was contoured at 1.7 σ. (b) Interaction of the backbone amide nitrogen of the universally conserved Gln230 with the 3′-OH of A76 of P-site tRNA. (c) Model for proposed product stabilization of the peptide release reaction by H-bonding between the main-chain amide nitrogen of Gln230 and the 3′-OH of A76. (d) Superposition of the structure of the peptidyl-transferase transition-state analog (TSA, orange) complexed with the 50S subunit (grey)[55] on the structure of the termination complex (this work). The main-chain amide nitrogen of Gln230 is positioned to form a hydrogen bond with the oxyanion of the TSA. (e) Model for potential transition-state stabilization of the peptide release reaction by H-bonding between RF1 and the TSA. From ref. [16].
What is the role of the Gln230 side chain?. In contrast to the comparatively modest effects on the rate of the hydrolytic reaction, there is a striking effect of mutation of Gln230 on the ability of the factor to discriminate against nucleophiles other than water [24]. For the Ser, Ala and Gly mutants, the rate of aminolysis by hydroxylamine was increased by a factor of 15- to 170-fold. Based on their findings, Shaw and Green proposed that the glutamine side chain excludes nucleophiles bulkier than water, and in addition positions water for in-line attack on the substrate. This proposal is consistent with the available structural information, including the structure of a pre-termination-like complex containing a peptidyl-tRNA analogue bound to the P site [28].
Coupling of peptidyl-tRNA hydrolysis to stop codon recognition
Premature hydrolysis of the peptidyl-tRNA ester linkage prior to recognition of an authentic stop codon is of necessity a rare event. It seems likely that this is accomplished by preventing productive docking of the GGQ sequence of domain 3 in the PTC until stop codon recognition is established. In the crystal structures of isolated RF1 and RF2 [29,30], domains 2 and 3 are folded into a compact state that differs dramatically from that of the extended conformations seen in their respective termination complexes [16–19,31–33]. The possibility of interconversion of the compact and extended forms is supported by direct observation of both closed and open conformations of free release factors in solution [34,35]. Comparison of the ribosome-bound structures with those of the free factors suggests that a key event involves rearrangement of a loop (residues 286–300) connecting domains 3 and 4 that we term the “switch loop” (Fig. 1d), which extends helix α7 by two helical turns and directs domain 3 into the PTC (Fig. 1c). The rearranged state of the switch loop appears to be stabilized by extensive hydrogen bonding and packing interactions within a pocket formed by protein S12, the 530 loop of 16S rRNA, the β-sheet of domain 2 and the flipped nucleotides A1492 and A1913, which form the floor of the pocket [16,18]. The extra turns of helix formed at the end of α7 are held in position by contacts with the β-sheet of domain 2 of RF1 and the shifted loop of helix 69 of 23S rRNA.
Rearrangement of the switch loop and fixing the orientation of the extended α7 helix depend on interactions that are only possible following stop codon recognition, which include conformational changes involving A1492 and A1493 of 16S rRNA and A1913 of 23S rRNA. Flipping out of A1493 or failure of A1913 to pack on A1493 in h44 (as seen for sense codon recognition) would not only result in steric clash between these nucleotides and domain 2 of RF1, preventing correct docking of its reading head on the stop codon, but would also prevent formation of the binding pocket for the rearranged switch loop and other contacts needed to position the extended α7 helix, thus docking the GGQ into the PTC. This proposal would account for the stringent dependence of peptidyl-tRNA hydrolysis on stop codon recognition. The crucial role played by A1913 would also explain the deleterious effects on translation termination caused by deletion of helix 69 of 23S rRNA [36].
II. Structures of the Class II Release Factor RF3 bound to the ribosome
The class II release factor RF3 was initially identified by its stimulatory effects on the activity of class I release factors [37]. It is now believed that its role is in dissociating the class I release factors from the ribosome following termination [38,39]. RF3 is of special interest because it is a GTPase structurally related to EF-G, and like EF-G, promotes intersubunit rotation of the ribosome [40,41]. Thus, the crystal structures of its complexes with the ribosome will not only help to understand its own mechanism of action, but may also provide insight into how interactions with the translational GTPases affect the structural dynamics of the ribosome.
RF3-dependent structural changes in the ribosome
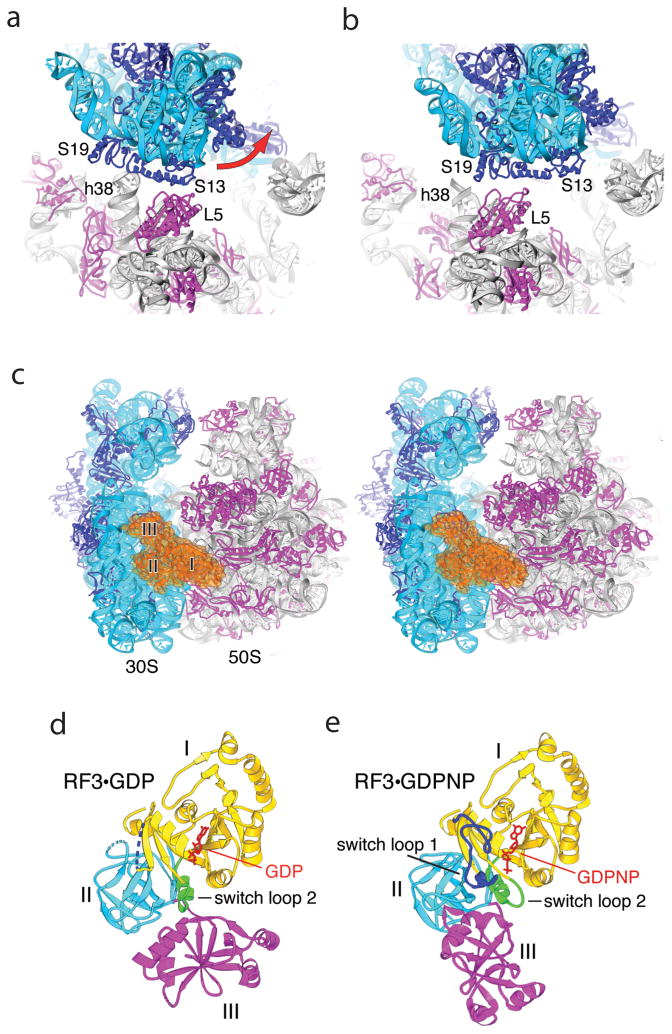
Cryo-EM reconstructions showed that RF3 binds to the ribosome in the same location as observed for EF-G and EF-Tu, and induces intersubunit rotation similar to that of EF-G [40,42]. Recently, two crystal structures of RF3-ribosome complexes have been reported, one of T. thermophilus ribosomes complexed with E. coli RF3·GDPCP [28] and the other of E. coli ribosomes with E. coli RF3·GDPNP [43]. Along with the structure of a complex containing the ribosome recycling factor RRF [44], these represent the first crystal structures showing the 70S ribosome trapped in a rotated state. The RRF and T. thermophilus RF3 structures show a similar intersubunit rotation, relative to the classical state, of about 9°, a 30S head rotation of 3–4° and contain a deacylated tRNA bound in the P/E hybrid state. In contrast, the E. coli RF3 structure has an intersubunit rotation of about 7° and a striking 14° rotation of the 30S head (Fig. 4a,b).
Fig. 4. Effects of binding RF3 to the 70S ribosome.
(a,b) Large-scale rotation of the 30S subunit head induced by RF3 in the E. coli complex [43]. (a) In the classical-state ribosome [17], bridge B1a is formed between protein S13 and helix 38 of 23S rRNA, and B1b between S13 and L5. (b) In the RF3 complex, a large-scale (14°) counter-clockwise rotation of the 30S head results in disruption of bridges B1a and B1b; a new bridge R1b is formed between proteins S13 and S19 from the 30S subunit and L5 from the 50S subunit, involving a 34 Å displacement of the intersubunit contacts. (c) Stereo view of RF3 (orange) bound at the entrance to the interface cavity of the 70S ribosome. (d,e) Comparison of crystal structures of (d) free RF3·GDP [42] and (e) ribosome-bound RF3·GDPNP. A large-scale rotational movement of domain III (magenta) and smaller movements in domains I (yellow) and II (cyan) can be seen. Disordered segments in RF3·GDP are shown as dotted lines. Rearranged segments in RF3·GDPNP containing switch loops I and II are shown in blue and green, respectively. GDP and GDPNP are shown in red. From ref. [43].
Both of the major rotational events result in alteration or disruption of intersubunit bridges. In general, the RNA-rich bridges nearer to the center of rotation at bridge B3 are preserved in the rotated state, made possible by bending, rotation or compression of local elements of rRNA. The peripheral bridges, such as bridge B1a, where displacements of more than 30Å can occur, are disrupted and replaced by new contacts.
Structural changes in RF3
RF3 binds to the ribosome at the entry to the intersubunit cavity (Fig. 4c) in a position that overlaps with that of EF-G and EF-Tu. Domain I contacts the 50S subunit at the sarcin-ricin loop (SRL) of 23S rRNA and protein L6. Domains II and III contact the 30S subunit at helices h5 and h15 of 16S rRNA and protein S12. Compared with the crystal structure of free RF3 solved in its GDP form [42], the ribosome-bound structures of RF3·GDPNP and RF3·GDPCP are dramatically rearranged (Fig. 4d,e). Domain II undergoes a 7° rotation and domain III rotates by 55° [43].
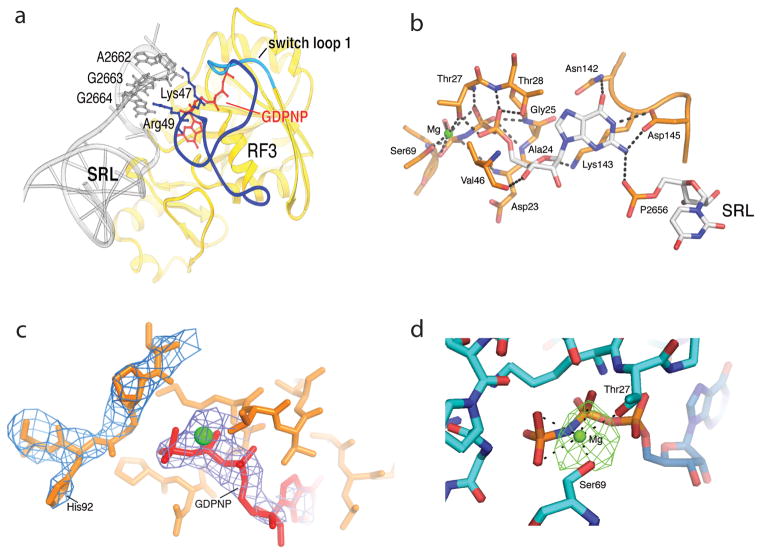
In the GTP-binding domain I, switch loop I, which is conserved among G proteins and disordered in all previous structures of RF3 and EF-G, becomes ordered, forming a cage around the bound GTP analogue (Figs. 5a,b). The histidine residue previously proposed to activate the attacking water molecule in catalysis of the GTPase reactions of all ribosomal G proteins, based on the structure of the EF-Tu·GDPCP-ribosome complex [45], is located more than 8Å from the γ-phosphate, placing it out of range for direct participation in catalysis (Fig. 5c). Electron density for a presumed magnesium ion is found next to the β-γ oxygen, which could play a catalytic role by stabilizing the oxyanion in the transition state (Fig. 5d).
Fig. 5. Ribosome-induced ordering of the GTPase active site of RF3 in the E. coli complex.
(a) Ordering of residues 39–69 of RF3 and re-positioning residues 70–73 (including the switch loop 1 residues 64–72) creates contacts between the G domain of RF3 and the SRL of 23S rRNA. Residues 39–63 are shown in dark blue, the switch loop in cyan, and the rest of RF3 in yellow. (b) An intricate network of H-bonds is formed between the base, ribose and phosphate groups of GDPNP and domain I of RF3, including elements of switch I (residues 64–72), P loop (residues 21–28), NKXD motif (residues142–145) and phosphate 2656 of the SRL of 23S rRNA. A Mg atom (green) is coordinated by Thr27, Ser91 and the β and γ phosphates of GDPNP (see Fig. S9). (c) His92 of RF3 is positioned at a distance of 8 Å from the γ phosphate of GDPNP. The 2Fo-Fc electron density map is shown around His92 and GDPNP, contoured at 1.3 σ and 2.3 σ, respectively. (d) Difference electron density showing Mg bound to GDPNP. An Fo-Fc difference map was calculated using the complete RF3·GDPNP·70S ribosome model, shown here contoured at 4.5 σ. The Mg ion (green) is coordinated with the β- and γ-phosphate oxygens of GDPNP and with the side-chain hydroxyls of Thr27 and Ser 69. From ref. [43].
Implications for the functions of RF3 and EF-G
Following peptidyl-tRNA hydrolysis, RF1/2 remain tightly bound to the ribosome. The observed requirement for deacylation of peptidyl-tRNA for RF3 to effect dissociation of RF1 and RF2 [46] can be explained by allowing P/E hybrid-state formation and therefore intersubunit rotation. The RF3-induced rotated state creates clash between domain IV of RF1/2 and the head of the 30S subunit, and between domain I of RF1/2 and the L11 stalk of 23S rRNA. Thus, both large-scale rotational movements of the ribosome would contribute to destabilization of binding of release factors RF1 or RF2. But these same clashes would interfere with binding of RF3 to a post-termination complex in its GTP state. One possibility is that RF3 binds initially in its GDP form followed by exchange of GTP for GDP [46]. However, RF3·GDP is not observed to bind stably to the termination complex [28,43,47]. An alternative possibility is that the termination complex samples the rotated state, which is stabilized by binding of RF3·GTP, causing dissociation of RF1 or RF2.
One unexplained puzzle in the binding of EF-G to the ribosome is that domain IV of EF-G occupies the same space as A-site tRNA, yet these two ligands must coexist on the ribosome during initial binding of EF-G. In the E. coli structure of the RF3-ribosome complex, domain I of RF3 is rotated by ~45° relative to that of EF-G·GDP or EF-Tu. Docking of EF-G in the RF3 orientation results in positioning of its domain IV near the shoulder of the 30S subunit, relieving potential clash with A-site tRNA [43](Fig. 6a,b). This orientation resembles an earlier cryo-EM reconstruction of an EF-G·ribosome complex trapped in the pre-translocation state in the presence of thiostrepton [48]. Further studies on trapped intermediates containing EF-G will be needed to resolve this confusing aspect of the translocation mechanism.
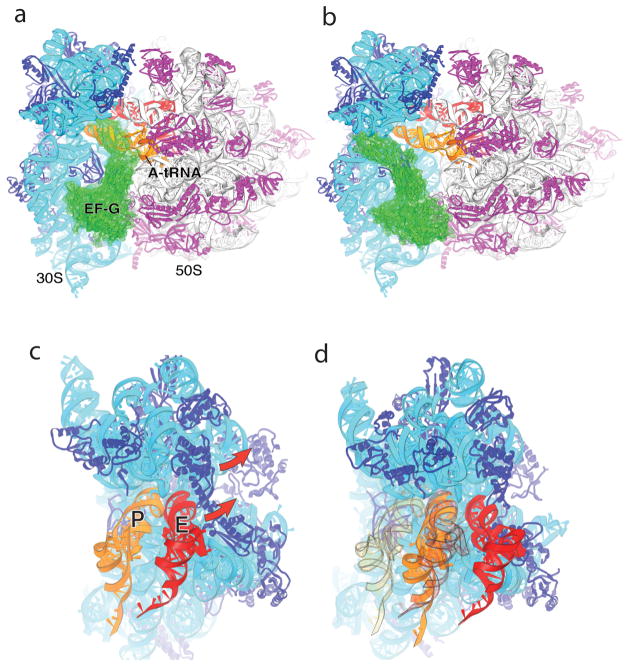
Fig. 6. Implications for EF-G and translocation.
(a) Observed orientation of EF-G (green) bound to the classical-state 70S ribosome·EF-G complex [56], in which tRNA has been modeled into the A site; note the severe steric clash between domain IV of EF-G and A-site tRNA. (b) Orientation of EF-G in the classical-state ribosome, but with its domain I docked on the 50S subunit in the same orientation as observed for RF3·GDPNP in the E. coli structure [43]. Note the absence of steric clash with A-site tRNA or the ribosome. (c,d) P-site ASL contacts in the 30S subunit head are displaced by 23 Å in the RF3 complex, relative to their position in the classical-state complex. Docking of P-site (orange) and E-site (red) tRNAs on the (c) classical-state RF2 and (d) rotated RF3 structures illustrates that the combined 30S head and body rotations seen in the E. coli RF3 structure are sufficient to translocate an ASL from the 30S P site to the E site, while allowing passage of the ASL of P-site tRNA to the E site without clash with the ribosome. The views in (c) and (d) are shown in exactly the same orientation relative to the 50S subunit. In (d), the initial positions of the P- and E-site tRNAs as in (c) are shown in transparent orange and red, respectively. From ref. [43].
Another problem that must be addressed in thinking about the translocation mechanism was recognized by Cate and co-workers [49]. In moving from the 30S P site to the E site, the anticodon stem-loop (ASL) must pass through a channel between the head and platform that is constricted to a width of only 13–14Å in the classical state of the 70S ribosome. Clearly, one or both of these features must move to enable translocation of P-site tRNA, and the latter authors noted that this could be accomplished by rotation of the head of the small subunit as seen in a cryo-EM reconstruction of an 80S ribosome complex [50]. Sufficiently large rotational movements of the head have been observed in cryo-EM [51,52] and crystal [53] structures. Spahn and co-workers [51] have proposed the existence of additional hybrid intermediate states involving head and body rotation in EF-G-catalyzed translocation. In the E. coli RF3-ribosome complex [43], simultaneous large-scale rotations of both the 30S body (7°) and head (14°) are observed. The head rotation opens the constricted channel from 14 to 22Å, sufficient to allow translocation of the ASL. Moreover, the net displacement of the anticodon stem contacts G1338 and A1339 in the 30S subunit head, due to combined body and head rotations, is about 23Å, corresponding almost exactly to the distance traveled by the ASL during movement from the P to the E site (Fig. 6c,d). These findings may have implications for the mechanism of EF-G-dependent translocation.
Acknowledgments
The authors were supported by grants from the NIH, NSF, QB3 and the Agouron Foundation.
References
- 1.Korostelev AA. Structural aspects of translation termination on the ribosome. RNA. 2011;17:1409–1421. doi: 10.1261/rna.2733411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klaholz BP. Molecular recognition and catalysis in translation termination complexes. Trends Biochem Sci. 2011;36:282–292. doi: 10.1016/j.tibs.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Loh PG, Song H. Structural and mechanistic insights into translation termination. Curr Opin Struct Biol. 2010;20:98–103. doi: 10.1016/j.sbi.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Dunkle JA, Cate JH. Ribosome structure and dynamics during translocation and termination. Annu Rev Biophys. 2011;39:227–244. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- 5.Petry S, Weixlbaumer A, Ramakrishnan V. The termination of translation. Curr Opin Struct Biol. 2008;18:70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Brenner S, Stretton AO, Kaplan S. Genetic code: the ‘nonsense’ triplets for chain termination and their suppression. Nature. 1965;206:994–998. doi: 10.1038/206994a0. [DOI] [PubMed] [Google Scholar]
- 7.Weigert MG, Garen A. Base composition of nonsense codons in E. coli. Evidence from amino-acid substitutions at a tryptophan site in alkaline phosphatase. Nature. 1965;206:992–994. doi: 10.1038/206992a0. [DOI] [PubMed] [Google Scholar]
- 8.Capecchi MR. Polypeptide chain termination in vitro: isolation of a release factor. Proc Natl Acad Sci U S A. 1967;58:1144–1151. doi: 10.1073/pnas.58.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murgola EJ, Hijazi KA, Goringer HU, Dahlberg AE. Mutant 16S ribosomal RNA: a codon-specific translational suppressor. Proc Natl Acad Sci U S A. 1988;85:4162–4165. doi: 10.1073/pnas.85.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen ZH, Fox TD. Substitution of an invariant nucleotide at the base of the highly conserved ‘530-loop’ of 15S rRNA causes suppression of yeast mitochondrial ochre mutations. Nucl Acids Res. 1989;17:4535–4539. doi: 10.1093/nar/17.12.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lodmell JS, Dahlberg AE. A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science. 1997;277:1262–1267. doi: 10.1126/science.277.5330.1262. [DOI] [PubMed] [Google Scholar]
- 12.O’Connor M, Brunelli CA, Firpo MA, Gregory ST, Lieberman KR, Lodmell JS, Moine H, Van Ryk DI, Dahlberg AE. Genetic probes of ribosomal RNA function. Biochem Cell Biol. 1995;73:859–868. doi: 10.1139/o95-093. [DOI] [PubMed] [Google Scholar]
- 13.Shine J, Dalgarno L. The 3′-terminal sequence of E coli 16S ribosomal RNA complementarity to nonsense triplets and ribosome binding sites. Proc Nat Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel Z, Zamir A, Elson D. The possible involvement of peptidyl transferase in the termination step of protein biosynthesis. Biochemistry. 1969;8:5161–5168. doi: 10.1021/bi00840a070. [DOI] [PubMed] [Google Scholar]
- 15.Caskey CT, Beaudet AL, Scolnick EM, Rosman M. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc Natl Acad Sci U S A. 1971;68:3163–3167. doi: 10.1073/pnas.68.12.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 17.Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci U S A. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korostelev A, Zhu J, Asahara H, Noller HF. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–2585. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito K, Uno M, Nakamura Y. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 21.Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogle JM, Brodersen DE, Clemons WM, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 23.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 24.Shaw JJ, Green R. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol Cell. 2007;28:458–467. doi: 10.1016/j.molcel.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaeger KE, Dijkstra BW, Reetz MT. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- 26.Wilmouth RC, Edman K, Neutze R, Wright PA, Clifton IJ, Schneider TR, Schofield CJ, Hajdu J. X-ray snapshots of serine protease catalysis reveal a tetrahedral intermediate. Nat Struct Biol. 2001;8:689–694. doi: 10.1038/90401. [DOI] [PubMed] [Google Scholar]
- 27.Maegley KA, Admiraal SJ, Herschlag D. Ras-catalyzed hydrolysis of GTP: a new perspective from model studies. Proc Natl Acad Sci U S A. 1996;93:8160–8166. doi: 10.1073/pnas.93.16.8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin H, Kelley AC, Ramakrishnan V. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci U S A. 2011;108:15798–15803. doi: 10.1073/pnas.1112185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin DH, Brandsen J, Jancarik J, Yokota H, Kim R, Kim SH. Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J Mol Biol. 2004;341:227–239. doi: 10.1016/j.jmb.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 30.Vestergaard B, Van LB, Andersen GR, Nyborg J, Buckingham RH, Kjeldgaard M. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol Cell. 2001;8:1375–1382. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- 31.Petry S, Brodersen DE, Murphy FVt, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 32.Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 33.Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 34.Zoldak G, Redecke L, Svergun DI, Konarev PV, Voertler CS, Dobbek H, Sedlak E, Sprinzl M. Release factors 2 from Escherichia coli and Thermus thermophilus: structural, spectroscopic and microcalorimetric studies. Nucleic Acids Res. 2007;35:1343–1353. doi: 10.1093/nar/gkl696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vestergaard B, Sanyal S, Roessle M, Mora L, Buckingham RH, Kastrup JS, Gajhede M, Svergun DI, Ehrenberg M. The SAXS solution structure of RF1 differs from its crystal structure and is similar to its ribosome bound cryo-EM structure. Mol Cell. 2005;20:929–938. doi: 10.1016/j.molcel.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 36.Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell. 2006;23:865–874. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 37.Goldstein JL, Caskey CT. Peptide chain termination: effect of protein S on ribosomal binding of release factors. Proc Natl Acad Sci U S A. 1970;67:537–543. doi: 10.1073/pnas.67.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M. Release factor RF3 in E. coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grentzmann G, Kelly PJ, Laalami S, Shuda M, Firpo MA, Cenatiempo Y, Kaji A. Release factor RF-3 GTPase activity acts in disassembly of the ribosome termination complex. RNA. 1998;4:973–983. doi: 10.1017/s1355838298971576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klaholz BP, Myasnikov AG, Van Heel M. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature. 2004;427:862–865. doi: 10.1038/nature02332. [DOI] [PubMed] [Google Scholar]
- 41.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of Intersubunit Movement of the Ribosome in Solution Using FRET. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 42.Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 43.Zhou J, Lancaster L, Trakhanov S, Noller HF. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA. 2011;18:230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zavialov AV, Buckingham RH, Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 47.Pel HJ, Moffat JG, Ito K, Nakamura Y, Tate WP. Escherichia coli release factor 3: resolving the paradox of a typical G protein structure and atypical function with guanine nucleotides. RNA. 1998;4:47–54. [PMC free article] [PubMed] [Google Scholar]
- 48.Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 49.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3. 5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 50.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 53.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 54.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5. 5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 55.Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 56.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]