SUMMARY
Reprogramming differentiated cells into induced pluripotent stem cells (iPSCs) promotes a broad array of cellular changes. Here we show that the let-7 family of microRNAs acts as an inhibitory influence on the reprogramming process through a regulatory pathway involving pro-differentiation factors, including EGR1. Inhibiting let-7 in human cells promotes reprogramming to a comparable extent to c-MYC when combined with OCT4, SOX2, and KLF4, and persistence of let-7 inhibits reprogramming. Inhibiting let-7 during reprogramming leads to an increase in the level of the let-7 target LIN-41/TRIM71, which in turn promotes reprogramming and is important for overcoming the let-7 barrier to reprogramming. Mechanistic studies revealed that LIN-41 regulates a broad array of differentiation genes, and more specifically, inhibits translation of EGR1 through binding its cognate mRNA. Together our findings outline a let-7 based pathway that counteracts the activity of reprogramming factors through promoting the expression of pro-differentiation genes.
INTRODUCTION
Fibroblasts can be reprogrammed into cells remarkably similar to embryonic stem cells (ESCs) by the expression of OCT4, SOX2, and KLF4 (OSK), with or without c-MYC (M) (Maherali et al., 2007; Meissner et al., 2007; Nakagawa et al., 2008; Okita et al., 2007; Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2008; Wernig et al., 2007 Park et al., 2007). Like ESCs, these reprogrammed cells, called induced pluripotent stem cells (iPSCs), can give rise to almost all cellular lineages upon differentiation. While it is known that OSKM induces genome-wide transcriptional changes that result in conversion to iPSCs, less is understood about the downstream events after reprogramming initiation. Furthermore, the efficiency of conversion with OSKM is very low (typically less than 1%). Without M, reprogramming efficiency is even lower (Nakagawa et al., 2008; Wernig et al., 2008). A few barriers contributing to low reprogramming efficiency have been described, including H3K9 methylation (Chen et al., 2013), macroH2A (Gaspar-Maia et al., 2013; Pasque et al., 2012), and upregulation of p53, p21, and p16Ink4a triggered by reprogramming factors (reviewed in Banito and Gil, 2010). Recent reports indicate that MBD3 of the NuRD complex is also a significant barrier to reprogramming (Luo et al., 2013; Rais et al., 2013).
We hypothesized that microRNAs (miRNAs) abundant in fibroblasts but not expressed in iPSCs and ESCs may also be a reprogramming barrier. One candidate was the let-7 family of miRNAs, since it is abundant in differentiated cells and low in pluripotent stem cells (Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008). Supporting this hypothesis, let-7 regulates differentiation in Caenorhabditis elegans, where loss of let-7 results in reiteration of larval cell fates and overexpression results in precocious expression of adult fates (Hunter et al., 2013; Reinhart et al., 2000). Also, let-7 is dowregulated in many types of cancer (reviewed in Boyerinas et al., 2010), consistent with a role in promoting a differentiated state. Therefore, since let-7 has been shown to promote differentiation, we thought it might also be a barrier to reprogramming to pluripotency.
In addition, let-7 is regulated by another heterochronic gene, LIN-28, which has also been shown to promote human reprogramming with the OS+NANOG cocktail of factors (Yu et al., 2007). LIN-28 binds and blocks maturation of the primary and precursor let-7 transcripts (reviewed in Mayr and Heinemann, 2013). LIN-28 is abundantly expressed in pluripotent stem cells and is downregulated as cells differentiate, whereas mature let-7 levels rise as cells differentiate in mice and humans (Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008). let-7 has been implicated in the regulation of reprogramming in mice, as antagonizing let-7 with OSK in mouse embryonic fibroblasts (MEFs) containing an Oct4-GFP reporter induced GFP-positive colonies (Melton et al., 2010). However, the effect of let-7 on human iPSC generation has not been previously examined. In addition, while let-7 targets have been identified in studies of ES cells lacking miRNA processing machinery (Melton et al., 2010), cancer (Johnson et al., 2007; Kumar et al., 2007; Lee and Dutta, 2007; Mayr et al., 2007; Sampson et al., 2007), and development (Johnson et al., 2005; Slack et al., 2000), targets that are important in iPSC reprogramming have not been identified experimentally.
In this study, we found that let-7 is a barrier to human iPSC reprogramming. Combining OSK-transduction with let-7 inhibition in HDFs improved reprogramming efficiency, similar to OSKM, and yielded a larger percentage of colonies with true ESC-like morphology compared to OSKM. Prolonged expression of let-7 blocked reprogramming. Furthermore, we identified the let-7 target LIN-41 (also known as TRIM71 and Mlin41) as a key factor that is necessary to overcome the let-7 barrier to reprogramming. LIN-41 is also a heterochronic gene that has been linked to translational regulation in mammals and C.elegans. Overexpression of LIN-41 in C.elegans results in reiteration of larval fates, and loss of LIN-41 results in precocious differentiation, the opposite effect of let-7 (Reinhart et al., 2000; Slack et al., 2000). We found that LIN-41 regulates expression of genes involved in development and differentiation in its capacity as a reprogramming factor. Finally, we identified the pro-differentiation transcription factor EGR1 (also known as NGFI-A, KROX-24, ZIF268, and TIS8) as a direct target of post-transcriptional regulation by LIN-41 and showed that it also inhibits reprogramming. Thus, we have identified a regulatory pathway downstream of let-7 that acts as a barrier to reprogramming by promoting the expression of pro-differentiation genes.
RESULTS
Inhibiting let-7 Promotes Efficiency and Quality of Human iPSC Reprogramming
Consistent with documented results, we observed that the levels of let-7 miRNAs are high in fibroblasts and low in pluripotent stem cells (Figure S1A and Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2008). To determine if antagonizing let-7 activity promotes reprogramming of human dermal fibroblasts (HDFs) to iPSCs, we transfected let-7 or control antisense inhibitors (inh) during reprogramming with OSK or OSKM. Inhibiting let-7 increased the efficiency of OSK-induced reprogramming by 1–2 orders of magnitude, similar to that observed with OSKM (Figures 1A and S1B). In Oct4-GFP reporter MEFs, let-7 inh was previously found to boost production of colonies by about 4-fold (Melton et al., 2010). Transfecting let-7 inh with OSKM increased reprogramming efficiency by only about 2-fold over control inh, which itself slightly increased reprogramming efficiency, as reported for MEFs (Figure 1A, S1B, and Melton et al., 2010).
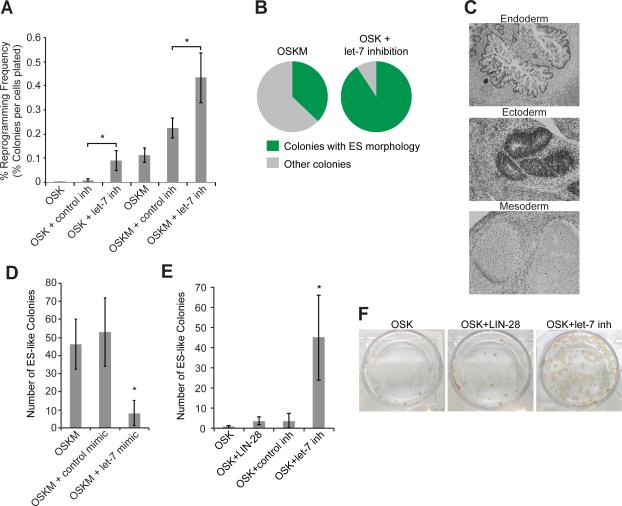
Figure 1. let-7 Inhibition Is Necessary and Sufficient to Promote iPSC Reprogramming.
(A) HDFs were treated with the indicated cocktails. Colonies with hESC-like morphology were counted and stained with TRA-1-60 antibody. Percent efficiency was calculated by dividing by the number of cells reseeded on day 7. Data are represented as mean +/− standard deviation (SD), n=3.
(B) Pie graphs showing results from experiments with HDFs treated with either OSKM (left, n=289 colonies total from 3 experiments) or OSK+let-7 inh (right, n=149 colonies total from 3 experiments). Colonies were counted and scored as having either hESC-like (green) or non-hESC like (gray) morphology. Plates were also stained with Tra-1-60 antibody to confirm the morphological scoring.
(C) Teratomas derived from OSK+let-7 inh reprogramming contain endoderm, ectoderm, and mesoderm.
(D) HDFs were treated with the indicated cocktails, and ES-like colonies were counted per well. Data are represented as mean +/- SD, n=3.
(E) HDFs were treated with the indicated cocktails and ES-like colonies were counted per well. Data are represented as mean +/- SD, n=3.
(F) Representative TRA-1-60 staining of colonies treated with the indicated cocktails.
*p<0.05 See also Figures S1 and S2.
Reprogramming with OSKM produces colonies of which most fail to develop ES-like morphology or become true iPSCs (Figure 1B). In contrast, the vast majority of colonies reprogrammed with OSK+let-7 inh had ES-like morphology and were TRA-1-60-positive (90%, OSK+let-7 inh, versus 40%, OSKM) (Figures 1B, S1C, and data not shown).
Repeated transfections and starting let-7 inhibition early led to the highest number of colonies, which decreased with increasing delay in initiating inhibition (Figure S1D). The greatest improvements in reprogramming efficiency depend on antagonizing let-7 throughout reprogramming (Figure S1D).
We found that inhibiting let-7 during reprogramming slightly increased the number of cells (Figure S1E), consistent with studies showing a role for let-7 in cell cycle regulation (Dong et al., 2010; Johnson et al., 2007; Johnson et al., 2005; Lee and Dutta, 2007; Legesse-Miller et al., 2009; Mayr et al., 2007). However, this minor increase in cell number is unlikely to account for the 1–2 orders of magnitude by which reprogramming was increased due to let-7 inhibition. We conclude that let-7's reprogramming-enhancing effects are most significantly attributed to its direct effects on reprogramming rather than acceleration of cell proliferation, consistent with data showing that let-7 inhibition did not enhance MEF proliferation (Melton et al., 2010).
We picked iPSC colonies derived from the OSK+let-7 inh cocktail and expanded them for further characterization. They expressed pluripotency markers, had normal karyotypes, formed teratomas with all three embryonic germ layers in vivo, and differentiated into derivatives of all three embryonic germ cell lineages in vitro (Figures 1C and S2A-C).
To test if high let-7 levels inhibit reprogramming, we transfected cells with mature let-7 mimic during reprogramming. As expected, overexpressed let-7 resulted in fewer colonies (Figure 1D). As LIN-28 blocks let-7 processing (reviewed in Mayr and Heinemann, 2013), we tested whether adding LIN-28 to OSK during reprogramming would produce results equivalent to reprogramming with OSK+let-7 inh. let-7 inhibition consistently resulted in many more colonies than LIN-28 (Figures 1E and 1F).
The let-7 Target LIN-41 Promotes iPSC Reprogramming
To understand the mechanism by which let-7 inhibition promotes reprogramming, we sought to identify let-7 targets with enhanced expression during reprogramming with OSK+let-7 inh. We tested several known let-7 targets, including HMGA2, CDC34, and LIN-41, as well as RAS- and MYC-family genes (Johnson et al., 2005; Kim et al., 2009; Kumar et al., 2007; Lee and Dutta, 2007; Legesse-Miller et al., 2009; Mayr et al., 2007; Melton et al., 2010; Sampson et al., 2007; Slack et al., 2000). The levels of HMGA2, CDC34, LIN-41, and N-RAS increased upon let-7 inhibition during OSK-induced reprogramming (Figure 2A), but we did not observe significant upregulation of the MYC genes (Figures S3A-C).
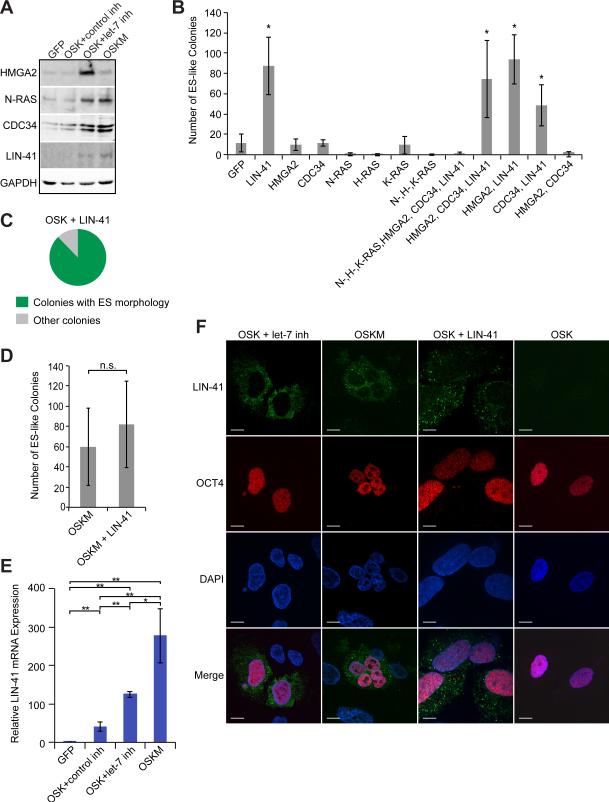
Figure 2. The let-7 Target LIN-41 Promotes Reprogramming with OSK.
(A) Representative western blots of the indicated factors at day 13 after infection with GFP, OSK+control inh, OSK+let-7 inh, or OSKM.
(B) HDFs were treated with OSK+ the indicated factors and scored for the number of ES-like colonies. Data are represented as mean +/- SD, n=3.
(C) Pie graph showing the result from experiments of HDFs treated with OSKL (n= 799 colonies total from 5 experiments). Colonies were counted and scored as having either hESC-like (green) or non-hESC like (gray) morphology. Plates were also stained with Tra-1-60 antibody to confirm the morphological scoring.
(D) HDFs were treated with the indicated factors and ES-like colonies were counted. Data are represented as mean +/- SD, n=10.
(E) qRT-PCR results for LIN-41 after 7 days of treatment with the indicated factors. Data are represented as mean +/- SD, n=3.
(F) Representative immunofluorescence images of cells 8 days post-infection with the indicated factors. Green, LIN-41; Red, OCT-4, blue, DAPI stain for nuclei. Scale bars, 10μm.
*p<0.05, **p<0.01, n.s., not significant See also Figures S1-S5.
We next tested whether HMGA2, CDC34, LIN-41 and the RAS genes alone or in combination could directly promote reprogramming. Expressing a combination of HMGA2, CDC34 and LIN-41 with OSK resulted in more colonies than did OSK alone, while adding RAS-family proteins to this mix inhibited reprogramming (Figure 2B). This is likely because N-RAS and H-RAS inhibited reprogramming (Figure 2B). We found that LIN-41 alone was responsible for the increased number of colonies, while the others were dispensable (Figure 2B). Most colonies obtained with OSK+LIN-41 (OSKL) had ES-like morphology, similar to colonies reprogrammed with OSK+let-7 inh (Figure 2C and Figure S1C). OSKL promoted reprogramming of MEFs but to a lesser extent than it promotes reprogramming of HDFs (Figure S4A). In contrast, expressing LIN-41 with OSKM did not significantly increase reprogramming efficiency, although in most experiments performed with HDFs, the number of colonies was slightly increased (Figures 2D and S4A). LIN-41 expression during reprogramming did not effect cell proliferation (Figure S5A). These data indicate that the let-7 target gene LIN-41 increases OSK-induced reprogramming efficiency. Furthermore, colonies obtained with OSKL were pluripotent, as demonstrated by positive staining with pluripotency makers, in vitro differentiation into the three cellular lineages, ability to form teratomas in vivo, and contribution to chimeric mice (Figures S2A-D and S4B-F).
LIN-41 is abundantly expressed in iPSCs and ESCs but is almost undetectable in fibroblasts (Chang et al., 2012; Rybak et al., 2009). Therefore, we examined by quantitative (q) RT-PCR whether endogenous LIN-41 expression was induced early during reprogramming. At 5 and 7 days post-OSK infection, LIN-41 mRNA levels were upregulated (Figure 2E and S5B). Inhibiting let-7 during OSK-mediated reprogramming increased LIN-41 levels 3-fold compared to OSK alone. By day 7, LIN-41 expression was even further increased when cells were reprogrammed with the OSKM cocktail, to about 5-fold higher levels compared to OSK. At these levels, LIN-41 function may be nearly saturated and may explain why adding LIN-41 to the OSKM cocktail did not substantially increase the number of colonies (Figures 2D). Additionally, LIN-41 upregulation occurs prior to let-7 downregulation (data not shown and Figure S5C). These data suggest that transfecting let-7 inh with OSK helps overcome the let-7 barrier to reprogramming and boosts LIN-41 expression levels toward those achieved by OSKM.
We next sought to determine if the endogenous LIN-41 levels induced by OSK+let-7 inh and OSKM were comparable to the exogenous LIN-41 levels expressed by retrovirus. The LIN-41 retrovirus efficiently expresses LIN-41 protein, as assessed by examining the mixed population of HDFs and reprogramming cells (Figure S5D). Since only a small fraction of the cell population will become iPSCs, we examined LIN-41 expression in individual cells by immunofluorescence (Figure 2F). First, we scored DAPI-stained cells as LIN-41-positive or -negative. As expected, the LIN-41 retrovirus infects ~25% of HDFs, which is 5–7 times more cells than express endogenous LIN-41 due to expressing the right combination of O, S, and K plus either let-7 inh or M (Figure S5E). Next, to determine whether the LIN-41 levels in individual cells were comparable among the OSK+let-7 inh, OSKM, and OSKL cocktails, we quantified fluorescence intensity in individual cells that expressed LIN-41. The level of retroviral LIN-41 was variable, as expected (Figure S5F). We found that many cells transduced with OSK+let-7 inh and OSKM cocktails expressed a level of endogenous LIN-41 that was similar to the level of LIN-41 produced by LIN-41 retrovirus (Figure S5F). These data suggest that LIN-41 levels resulting from let-7 inhibition would likely be sufficient to promote reprogramming in a manner similar to LIN-41 retrovirus.
Multiple Domains of LIN-41 Contribute to Reprogramming Activity
LIN-41 is a member of the RBCC (RING, B-box, Coiled-coil) family of proteins, which contain a RING domain, two B-box domains, a coiled-coil domain, a filamin domain, and six NHL repeats (Figure 3A). To identify LIN-41 domains that facilitate reprogramming, we generated HA-tagged domain deletion mutants (ΔRING, ΔB-boxes, ΔCoiled-coil, ΔFilamin, Δ6xNHL, and NHL-only) and expressed them in HDFs (Figures 3A and 3B). We found that wild-type (wt) LIN-41 and the ΔRING mutant had similar patterns of intracellular localization, though expression of the ΔRING mutant altered fibroblast morphology, imparting a less elongated shape (Figure 3C). Each of the other deletion mutants displayed altered cellular localization patterns (Figure 3C), which may contribute to their differing effects on reprogramming: We found that expression of OSK plus each domain mutant resulted in fewer colonies than OSK+wtLIN-41. ΔRING, Δ6xNHL, and NHL-only mutants produced the fewest colonies (Figure 3D and 3F). When ΔRING was added to OSKM, reprogramming was strongly inhibited (Figures 3E and 3F). Adding the other domain mutants to OSKM did not change the number of colonies (Figure 3E).
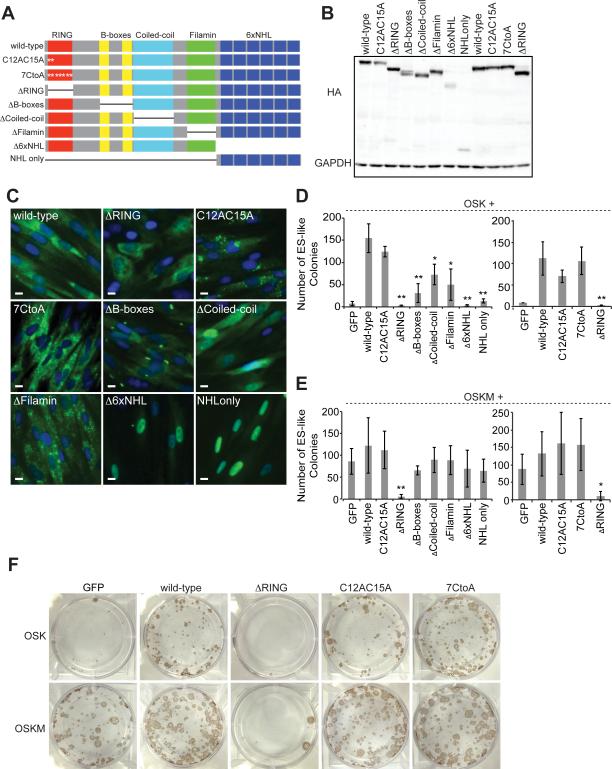
Figure 3. All Domains of LIN-41 Contribute to Reprogramming.
(A) Diagram of the domain structure of LIN-41 and the domain deletion and point mutants constructed. C12AC15A contains alanines in the place of cysteines at positions 12 and 15 of the human LIN-41 open reading frame. 7CtoA contains alanines in place of cysteines at positions 12, 15, 61, 66, 69, 91, and 94. ΔRING lacks amino acids 12-91. ΔB-box lacks amino acids 194-320. ΔCoiled-coil lacks amino acids 328-447. ΔFilamin lacks amino acids 483-583. Δ6xNHL lacks amino acids 593-868. NHL-only contains only an initiating methionine and amino acids 583-868. The white asterisks indicate the position of C to A point mutations.
(B) Representative western blot showing the levels of HA-tagged wtLIN-41 and domain deletion mutants expressed in HDFs.
(C) Representative immunofluorescence images of HA-tagged wtLIN-41 and domain deletion mutants expressed in HDFs. Green, HA tag; blue, DAPI stain for nuclei. Scale bars, 10μm.
(D) HDFs were treated with OSK+ the indicated factors and ES-like colonies were counted. In the right graph, a construct with additional C to A mutations (7CtoA) was tested for reprogramming ability. Data are represented as mean +/- SD, n=3.
(E) The same type of experiment as in D but with OSKM. Data are represented as mean +/- SD, n=3.
(F) Representative TRA-1-60 staining of colonies treated with the indicated cocktails.
*p<0.05, **p<0.01
LIN-41 has been shown to have E3 ubiquitin ligase activity (Chen et al., 2012; Rybak et al., 2008). The RING domain of E3 ubiquitin ligases interacts with E2 ubiquitinconjugating enzymes and ubiquitin, which are critical for proteasome-mediated degradation. Seven cysteines and a histidine residue in the RING domain coordinate the zinc molecules important for maintaining the structure and function of the domain (Deshaies and Joazeiro, 2009; Plechanovova et al., 2012; Rybak et al., 2009). To determine if E3 ubiquitin ligase activity is important for LIN-41-mediated reprogramming, we made cysteine-to-alanine (C to A) point mutations within the RING domain (Figures 3A, 3B, and 3C). Mutating the first two cysteines of this domain disrupts LIN-41's E3 ubiquitin ligase activity (Rybak et al., 2009). Surprisingly, unlike ΔRING, when we expressed OSK with either of the C to A point mutants, we obtained a similar number of colonies as when reprogramming with wtLIN-41 (Figures 3D, 3E, and 3F). These data suggest that LIN-41's function in reprogramming is independent of cysteine-mediated zinc coordination and E3 ubiquitin ligase activity. Constitutive high expression of ΔRING in human ESCs (hESCs) resulted in cell death, suggesting that ΔRING is toxic (data not shown), making it difficult to ascribe a role for the RING domain to reprogramming.
Recent reports implicate LIN-41 in the regulation of multiple signaling pathways, including those mediated by Ago2 (Rybak et al., 2008), FGF (Chen et al., 2012), and mouse ES cell proliferation through the p21/Cdkn1a pathway (Chang et al., 2012). We did not observe changes in the levels of AGO2 or FGF signaling mediators upon LIN-41 and ΔRING expression during reprogramming or LIN-41 knockdown in hESCs (Figures S6A–C). The expression of p21, a negative regulator of reprogramming, is upregulated during OSKM-mediated reprogramming (reviewed in Banito and Gil, 2010). If p21 were downstream of LIN-41 in reprogramming, we would expect LIN-41 and ΔRING to affect p21 levels differentially, as LIN-41 promotes reprogramming while ΔRING inhibits it. While we did observe a reduction in p21 levels upon LIN-41 expression with OSKM, we saw a similar decrease when ΔRING was expressed (Figure S6D). Adding LIN-41 or ΔRING to OSK did not affect p21 levels (Figure S6D). These data suggest p21 is not likely the downstream effector of LIN-41 for reprogramming.
LIN-41 Induction Is Important for Overcoming the let-7 Barrier to iPSC Reprogramming
To determine if LIN-41 activity is important for reprogramming, we knocked down LIN-41 expression during reprogramming by transfecting cells with one of two siRNAs that target LIN-41. Transfecting these siRNAs into hESCs reduced LIN-41 levels but did not affect colony morphology (Figure 4A, 4B, and S6E), suggesting that LIN-41 knockdown does not affect pluripotency. This is consistent with studies of Lin-41 knockout mice, which display defects in neural tube closure and death between embryonic days (E) E8.5–E13.5 (Chen et al., 2012; Maller Schulman et al., 2008). In contrast, we found that knocking down LIN-41 during reprogramming with OSK+let-7 inh resulted in fewer colonies (Figures 4C and 4D). Therefore, endogenous LIN-41 is an important target of let-7 that needs to be upregulated for let-7 inhibition to promote reprogramming.
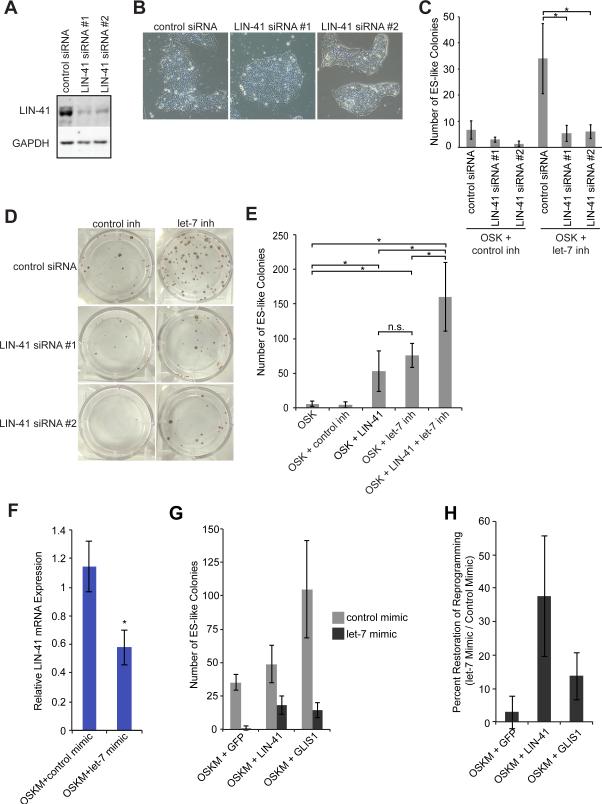
Figure 4. LIN-41 Is Important for Overcoming the let-7 Barrier to Reprogramming.
(A) Representative western blots of H9 hESCs 72 hrs after transfection with control or LIN-41 siRNAs.
(B) Representative images of H9 hESCs 72 hrs after transfection with control or LIN-41 siRNAs.
(C) HDFs were treated with OSK+ control or let-7 inh, transfected with control or LIN-41 siRNAs, and ES-like colonies were counted. Data are represented as mean +/- SD, n=3.
(D) Representative TRA-1-60 staining of colonies treated with OSK+ the indicated factors.
(E) HDFs were treated with the indicated factors and ES-like colonies were counted. Data are represented as mean +/- SD, n=4.
(F) qRT-PCR for LIN-41 after 7 days of treatment with the indicated cocktails. Data are represented as mean +/- SD, n=3.
(G) HDFs were treated with the indicated cocktails and transfected with either control (gray bars) or let-7 (black bars) mimic, and ES-like colonies were counted. Data are represented as mean +/- SD, n=3.
(H) The mean number of colonies obtained in wells transfected with let-7 mimic for each indicated reprogramming cocktail (OSKM+GFP, LIN-41, or GLIS1) was divided by the mean number of colonies obtained in wells transfected with control mimic and the same reprogramming cocktail. Values are percentages ± SD, n=3.
*p<0.05, n.s., not significant.
To test if LIN-41 is the only let-7 target gene important for reprogramming, we compared the efficiency of reprogramming with OSK, OSKL, OSK+let-7 inh, and OSKL+let-7 inh. Retroviral LIN-41 only contains the open reading frame and therefore lacks the let-7 binding sites that regulate endogenous LIN-41 expression. OSKL and OSK+let-7 inh resulted in comparable colony numbers (Figure 4E), while combining OSKL with let-7 inh further enhanced the number of colonies (Figure 4E). Thus, there are likely additional let-7 targets that contribute to reprogramming.
We wanted to determine if LIN-41 expression could overcome the let-7 barrier to reprogramming. First, we confirmed that sustained let-7 levels repress endogenous LIN-41 (Figure 4F). When we reprogrammed with OSKM+GFP or OSKM+LIN-41 in the presence of let-7 mimic, more colonies were obtained with LIN-41 than with GFP (Figures 1D and 4G). As a control we tested GLIS1, a factor that increases OSKM reprogramming efficiency (Figure 4G and Maekawa and Yamanaka, 2011). Overexpressed LIN-41 was more effective at restoring the number of colonies in the presence of let-7 mimic than was GFP or GLIS1 (Figure 4H). Additional let-7 targets must also contribute to overcoming the let-7 barrier to reprogramming, as LIN-41 does not completely restore the number of colonies to that obtained with control mimic. These data indicate that LIN-41 can partially rescue the deficit in reprogramming when let-7 levels are high. Therefore, we have identified LIN-41 as a target of let-7 regulation that is increased during reprogramming, promotes reprogramming, and is important for surmounting the let-7 barrier to reprogramming.
LIN-41 Negatively Regulates EGR1 Expression
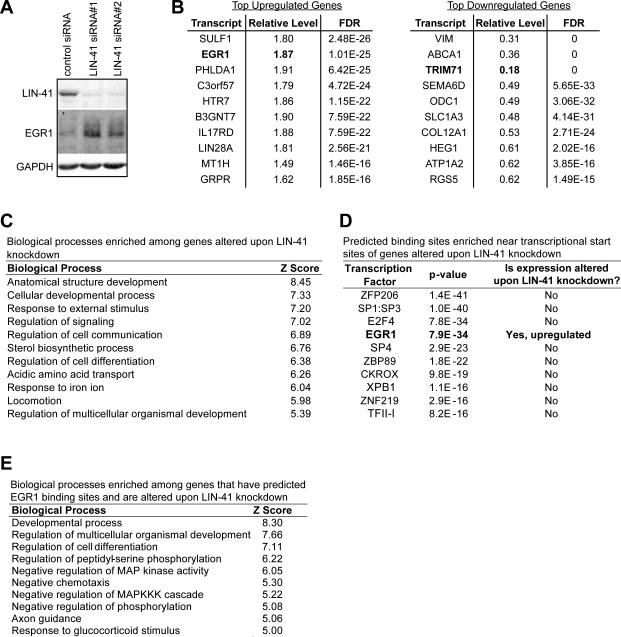
To gain insight into the mechanism by which LIN-41 promotes reprogramming, we first knocked down LIN-41 expression in hESCs by 90% and examined genome-wide transcriptome changes by RNAseq. Expression of over 1,000 genes was altered (Figure 5A, 5B, and Table S1). Gene ontology (GO) analysis suggested that LIN-41 regulates development and differentiation (Figure 5C). We hypothesized that LIN-41 may promote reprogramming by regulating a broadly acting transcription factor. To test this, we used Whole Genome rVISTA (Dubchak et al., 2013) to search for predicted transcription factor (TF) binding sites that are enriched within the set of genes regulated by LIN-41. One TF, EGR1, stood out, as its transcript was also among those most upregulated upon LIN-41 knockdown (Figure 5B and 5D). We found that EGR1 protein expression was also upregulated upon LIN-41 knockdown (Figure 5A).
Figure 5. LIN-41 Knockdown Alters Expression of Genes Involved in Development and Differentiation, Including the Transcription Factor EGR1.
(A) Representative western blots for LIN-41, EGR1, and GAPDH, 72 hrs after transfecting H1 hESCs with control or LIN-41 siRNAs.
(B) List of the most significantly up- (left) and down- (right) regulated transcripts upon LIN-41 knockdown assayed by RNAseq.
(C) List of the top biological processes enriched among genes altered upon LIN-41 knockdown.
(D) List of the top TFs with enriched predicted binding sites among genes altered upon LIN-41 knockdown. Among these TFs, only EGR1 had altered expression when LIN-41 was knocked down.
(E) List of the top biological processes enriched among genes with predicted EGR1 binding sites among genes altered upon LIN-41 knockdown.
GO analysis of the subset of genes with predicted EGR1 binding sites indicated an enrichment of genes involved in development and differentiation, as well as phosphorylation (Figure 5E). EGR1 has been shown to promote differentiation when expressed in embryonal carcinoma cells, which are similar to ESCs, and to regulate differentiation in various contexts (Cao et al., 1990; Carter et al., 2007; Dinkel et al., 1998; Edwards et al., 1991; Harris and Horvitz, 2011; Krishnaraju et al., 1995; Lanoix et al., 1998; Laslo et al., 2006; Le et al., 2005; Lejard et al., 2011; Nguyen et al., 1993; Spaapen et al., 2013; Sukhatme et al., 1988; Topilko et al., 1998; Zhang et al., 2013). Fragola et al. proposed that EGR1 functions as a key TF that maintains the fibroblast transcriptional profile (Fragola et al., 2013). Of the genes we identified with predicted EGR1 binding sites, several have been previously validated by chromatin immunoprecipitation as EGR1 targets in cancer cells by the ENCODE project (Table S2). One of the predicted targets that has also been validated as an EGR1 target is NAB2. NAB2 is not only a target of EGR1 regulation, but also acts as a corepressor or coactivator of EGR1 activity, depending on cellular context (Collins et al., 2006; Kumbrink et al., 2010; Sevetson et al., 2000; Svaren et al., 1996). Both EGR1 and NAB2 are induced by mitogenic stimuli, including serum and purified factors such as FGF (Svaren et al., 1996; reviewed in Gashler and Sukhatme, 1995). During reprogramming, serum and FGF are replenished on a daily basis (serum on reprogramming days 1-8 and FGF thereafter), thus stimulating expression of these differentiation-associated genes (data not shown). Therefore, our data suggest that LIN-41 has a role in overcoming this differentiation barrier.
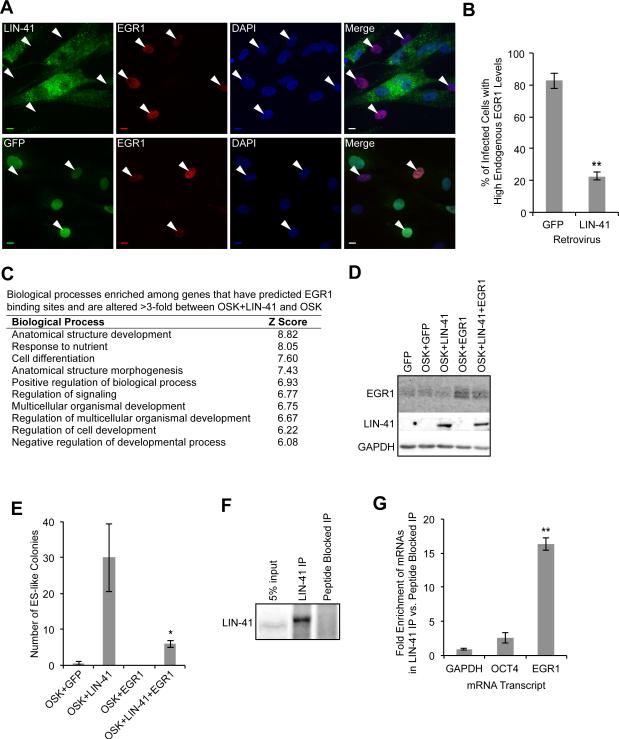
We next examined the effect of LIN-41 on endogenous EGR1 expression on a single-cell basis by infecting cells with HA-LIN-41 or GFP retrovirus, performing immunofluorescence staining with EGR1 and HA antibodies, and quantitating EGR1 fluorescence intensity in individual cells (Figure 6A). We found that LIN-41 expression repressed EGR1 protein expression (Figure 6B). These findings were corroborated by examining EGR1 mRNA expression in isolated TRA-1-60+ reprogramming cells. EGR1 was repressed the most when the OSKL cocktail was used, compared to the OSK, OSK+let-7 inh, and OSKM cocktails (Figure S6F). These data support a recent report showing that EGR1 is downregulated in mouse reprogramming and acquires the repressive histone modification H3K27me3 (Fragola et al., 2013). Supporting our finding that predicted EGR1 binding sites were enriched among genes with altered expression upon LIN-41 knockdown, predicted EGR1 binding sites were also enriched among the genes with a greater than 3-fold difference in expression between the OSKL and OSK cocktails (Figure S6F). In addition, of the top 10 enriched transcription factors, EGR1 was the only factor significantly downregulated in OSKL reprogramming cells. (Figure S6G).
Figure 6. EGR1 Is a Target of LIN-41 that Blocks Reprogramming.
(A) HDFs were infected with HA-LIN-41 or GFP retroviruses and immunostained with anti-HA and anti-EGR1 at 8 days post-infection. Arrowheads indicate EGR1+ cells. Scale bars, 10μm.
(B) We selected random fields of DAPI-stained nuclei and captured images in the blue (DAPI), green (LIN-41 or GFP), or red (EGR1) channels. The level of EGR1 fluorescence intensity was measured using Volocity (PerkinElmer). Cells were scored as having fluorescence intensity above (high) or below (low) a threshold. Values are the mean of the percent of infected cells with high EGR1 expression +/-SD. ≥50 cells were scored for each condition in each experiment, n=3
(C) List of the top biological processes enriched among genes with EGR1 binding sites and a greater than three-fold difference between OSKL and OSK reprogramming cells.
(D) Representative western blots for LIN-41, EGR1, and GAPDH at 7 days post-infection with the indicated cocktails.
(E) HDFs were treated with the indicated factors and ES-like colonies were counted. E Data are represented as mean +/- SD, n=3.
(F) Representative western blot for LIN-41. Beads bound to LIN-41 antibody or LIN-41 antibody pre-incubated with peptide antigen were used in IP experiments with hESC extract.
(G) qRT-PCR was performed with RNA collected from the LIN-41 IPs and peptide blocked IPs. Data are represented as mean +/- SD, n=3.
*p<0.05, **p<0.001 See also Figure S6.
Corroborating our finding with LIN-41 knockdown, biological processes related to development and differentiation were also enriched among genes with predicted EGR1 binding sites in OSKL reprogramming cells (Figure 6C). Based on the above findings, we theorized that EGR1 expression is another barrier that needs to be overcome during reprogramming. To test this, we overexpressed EGR1 with the OSKL cocktail (Figure 6D). This generated fewer colonies than OSKL but more than OSK+GFP (Figure 6E and S6H), indicating that overexpression of EGR1 negates LIN-41's positive effect on reprogramming.
Finally, we wanted to determine if LIN-41 regulates EGR1 expression directly by binding to the EGR1 transcript. We immunoprecipitated (IP'd) endogenous LIN-41 from hESCs using a LIN-41 antibody. We performed side-by-side IPs in which the antibody was either free to bind endogenous LIN-41 or blocked by preincubation with the peptide antigen (Figure 6F). We collected RNA from the IPs and compared enrichment of EGR1 mRNA and control mRNAs GAPDH and OCT4 between the LIN-41 IPs and peptide-blocked IPs. We found that EGR1 mRNA, but not GAPDH or OCT4 mRNA, was enriched when LIN-41 was IP'd (Figure 6G). Collectively, these data suggest that one role of LIN-41 in reprogramming is to lower EGR1 levels and thereby dysregulate genes associated with differentiation.
DISCUSSION
In this study, we found that the let-7 family of miRNAs acts as a barrier to reprogramming via a pathway that promotes the expression of pro-differentiation genes. We found that inhibiting let-7 with the OSK cocktail increases the reprogramming efficiency of HDFs to a level comparable to that seen with OSKM. In addition, we established that let-7 inhibition enhances OSK-mediated reprogramming, at least in part through promoting LIN-41 expression. Exogenous LIN-41 expression promotes reprogramming with OSK, while knocking down endogenous LIN-41 expression reduces the formation of iPSC colonies. Furthermore, we found that LIN-41 expression is upregulated during reprogramming with OSK+let-7 inh, as well as with OSKM, indicating that antagonizing let-7 helps to increase LIN-41 levels and consequently, the reprogramming power of the otherwise inefficient OSK cocktail. LIN-41 can also partially overcome the negative effect of let-7 expression on reprogramming. Finally, we found that EGR1 mRNA is bound and negatively regulated by LIN-41 and acts to block reprogramming. Analysis of the genes with predicted EGR1 binding sites and altered expression upon LIN-41 knockdown or LIN-41 expression during reprogramming link LIN-41 to regulation of development and differentiation. Therefore, we have characterized a pathway in which antagonizing let-7 results in upregulation of let-7 targets including LIN-41, which in turn inhibits expression of pro-differentiation factors such as EGR1.
LIN-41 is a conserved target of let-7 regulation (Lin et al., 2007; O'Farrell et al., 2008; Rybak et al., 2009; Schulman et al., 2005; Slack et al., 2000). lin-41 and let-7 were identified in C. elegans as heterochronic genes, whereby overexpression of let-7 or deletion of lin-41 resulted in precocious differentiation into adult cell fates, and deletion of let-7 or overexpression of lin-41 led to the reiteration of larval cell fates (Reinhart et al., 2000; Slack et al., 2000). We demonstrate that the let-7/LIN-41 pathway also regulates iPSC reprogramming. We found that LIN-28, another heterochronic gene, did not phenocopy let-7 inh and LIN-41 in promoting reprogramming with OSK. However, LIN-28 has been shown to play a role in reprogramming (Hanna et al., 2009; Yu et al., 2007), suggesting that a let-7-independent function of LIN-28 may be involved (reviewed in Mayr and Heinemann, 2013).
Another interesting link to the heterochronic pathway is our finding that LIN-41 regulates EGR1 expression and that EGR1 blocks reprogramming. The C. elegans heterochronic gene MAB-10 is an ortholog to the EGR1 co-factors NAB1 and NAB2 (Harris and Horvitz, 2011). MAB-10 interacts with another heterochronic gene, LIN-29, via a LIN-29 domain that is conserved in EGR proteins (Harris and Horvitz, 2011). The timing of LIN-29 expression is regulated by LIN-41, although the mechanism by which LIN-41 regulates LIN-29 remains unknown (Slack et al., 2000). In mammals, EGR1 has been shown to regulate differentiation and development in several contexts (Cao et al., 1990; Carter et al., 2007; Dinkel et al., 1998; Edwards et al., 1991; Krishnaraju et al., 1995; Laslo et al., 2006; Le et al., 2005; Lejard et al., 2011; Nguyen et al., 1993; Spaapen et al., 2013; Sukhatme et al., 1988; Topilko et al., 1998; Zhang et al., 2013), and expression of EGR1 in P19 embryonal carcinoma cells resulted in spontaneous differentiation (Lanoix et al., 1998). EGR1 is an early growth response gene that is induced by mitogenic stimuli, including serum and purified factors such as FGF, EGF, and TGFβ (reviewed in Gashler and Sukhatme, 1995). As EGR1 is expressed in HDFs and induced by such stimuli present in the cell culture medium, it is logical that EGR1 expression would need to be downregulated for reprogramming to occur.
Future studies to address the mechanism by which LIN-41 regulates translation and to understand how it recognizes particular transcripts will help to further elucidate the role of LIN-41 in regulating differentiation pathways.
EXPERIMENTAL PROCEDURES
Cell Culture and Reprogramming
Cells were maintained using standard methods (described in the Supplemental Experimental Procedures [SEP]). HDFs from Cell Applications were used in this study (lots 1429, 1323, and 1503). Reprogramming was carried out with retroviruses as described (Takahashi et al., 2007). 7 days post-infection, the cells were trypsinized, counted, and reseeded onto SNL feeders at 2×104 or 5×104 per well for reprogramming with or without c-MYC, respectively. Cells were transfected with miRNA inh (20nM, Dharmacon, control inh [IN-001005-01] or let-7c inh [IH-300477-05]) (Robertson et al., 2010) on days 1, 6, 12, 18 and 24 unless otherwise indicated. Cells were transfected with siRNAs (20nM) every 3 days starting on day 2.
Western Blotting
1° antibodies are listed in Table S3. Li-cor 2° antibodies were used and blots were scanned using an Odyssey Fc.
Knockdowns
siRNAs from Ambion (TRIM71: s43598 and s43599, Negative Control 1: 4390844) were transfected using Lipofectamine RNAimax (Life Technologies) at 20nM final concentration for reprogramming or 50 nM for knockdowns in hESCs.
Immunofluorescence
1° antibodies are listed in Table S3. Alexa Fluor 2° antibodies (Life Technologies) were used at a 1:200 dilution. The staining protocol is described in the SEP.
LIN-41 IP
LIN-41 IPs were performed using hESC extract and LIN-41 monoclonal antibody (peptide antigen: CVRAHQRVRLTKDHYIER; developed in collaboration with Epitomics). Dynabeads with captured anti-LIN-41 were either left free to bind LIN-41 or first blocked with 3X-LIN-41 peptide (NH2-RVRLTKDHYIERRVRLTKDHYIERRVRLTKDHYIERCOOH) to block the LIN-41 antibody binding sites. RNA was collected an analyzed by qRT-PCR. Additional details are in the SEP.
qRT-PCR
Trizol-extracted RNA was reverse-transcribed using Superscript III (Life Technologies) and random priming. Taqman assays were performed (probes listed in Table S3). Gene expression was normalized to GAPDH.
Transcriptome Analyses
TRA1-60+ cells on reprogramming day 11 were isolated and analyzed as described previously (Tanabe et al., 2013). Gene expression upon LIN-41 knockdown was analyzed by Illumina HiSeq 2000 and as described in the SEP. We analyzed genes with differential expression (FDR<0.05) between the control siRNA samples (n=3) and the LIN-41 siRNA samples (n=6) using GO-Elite (http://www.genmapp.org/go_elite/; (Zambon et al., 2012). We performed a similar analysis between TRA-1-60+ OSKL vs. OSK reprogramming cells. We used Whole Genome rVISTA (Dubchak et al., 2013) to identify enriched predicted TF binding sites among these gene sets.
Cloning
The LIN-41 cDNA was obtained from Thermo (clone 610064) and the EGR1 cDNA from GeneCopoeia (clone GC-0600487). These and the LIN-41 domain and point mutants were cloned into the retroviral expression vector pMXs. Oligos and cloning methods are described in Table S3 and the SEP.
Statistical Analysis
Values are means ± standard deviation, unless otherwise indicated. Significance was determined with Student's t-tests.
Accession Numbers
The RNAseq and microarray data reported in this paper have been deposited to NCBI GEO with the accession numbers GSE52133 and GSE52052.
Supplementary Material
HIGHLIGHTS.
Inhibition of let-7 promotes reprogramming to human iPSCs
The let-7 target LIN-41 also promotes reprogramming
LIN-41 negatively regulates pro-differentiation genes including EGR1
LIN-41 binds the EGR1 mRNA to inhibit translation
ACKNOWLEDGEMENTS
We would like to thank the Yamanaka and Srivastava lab members for helpful discussions, Alex Williams of the Gladstone Bioinformatics Core for RNAseq analysis, Yanxia Hao of the Gladstone Genomics Core for RNAseq library preparation, the Gladstone Histology Core for teratoma sectioning, Anna Lisa Lucido and Gary Howard for editorial review, Bethany Taylor and Karena Essex for administrative support, Pengzhi Yu for providing E8 medium, and F. Gregory Wulczyn for providing a LIN-41 antibody. K.A.W. was supported by an NIH fellowship (F32GM88988-02) and is a California Institute for Regenerative Medicine scholar. These studies were also made possible by funding from the L.K. Whittier Foundation (S.Y. and D.S.), the Roddenberry Foundation (S.Y. and D.S.), the Gladstone Institutes, and NHLBI/NIH (U01-HL100406, U01-HL098179) (D.S. and S.Y.). The Gladstone Institutes received support from a National Center for Research Resources Grant RR18928-01. S.Y. is a member without salary of the scientific advisory boards of iPierian, iPS Academia Japan, Megakaryon Corporation, and HEALIOS K. K. Japan; D.S. is a member of the scientific advisory boards of iPierian, Inc., Berkeley Lights, and RegeneRx Pharmaceuticals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Banito A, Gil J. Induced pluripotent stem cells and senescence: learning the biology to improve the technology. EMBO reports. 2010;11:353–359. doi: 10.1038/embor.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocrine-related cancer. 2010;17:F19–36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- Cao XM, Koski RA, Gashler A, McKiernan M, Morris CF, Gaffney R, Hay RV, Sukhatme VP. Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Molecular and cellular biology. 1990;10:1931–1939. doi: 10.1128/mcb.10.5.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JH, Lefebvre JM, Wiest DL, Tourtellotte WG. Redundant role for early growth response transcriptional regulators in thymocyte differentiation and survival. J Immunol. 2007;178:6796–6805. doi: 10.4049/jimmunol.178.11.6796. [DOI] [PubMed] [Google Scholar]
- Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nature communications. 2012;3:923. doi: 10.1038/ncomms1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lai F, Niswander L. The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes & development. 2012;26:803–815. doi: 10.1101/gad.187641.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Chen J, Wu Y, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nature genetics. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Collins S, Wolfraim LA, Drake CG, Horton MR, Powell JD. Cutting Edge: TCR-induced NAB2 enhances T cell function by coactivating IL-2 transcription. J Immunol. 2006;177:8301–8305. doi: 10.4049/jimmunol.177.12.8301. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annual review of biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dinkel A, Warnatz K, Ledermann B, Rolink A, Zipfel PF, Burki K, Eibel H. The transcription factor early growth response 1 (Egr-1) advances differentiation of pre-B and immature B cells. The Journal of experimental medicine. 1998;188:2215–2224. doi: 10.1084/jem.188.12.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Meng P, Wang T, Qin W, Qin W, Wang F, Yuan J, Chen Z, Yang A, Wang H. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PloS one. 2010;5:e10147. doi: 10.1371/journal.pone.0010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak I, Munoz M, Poliakov A, Salomonis N, Minovitsky S, Bodmer R, Zambon AC. Whole-Genome rVISTA: A Tool to Determine Enrichment of Transcription Factor Binding Sites in Gene Promoters from Transcriptomic Data. Bioinformatics. 2013;29:2059–61. doi: 10.1093/bioinformatics/btt318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SA, Darland T, Sosnowski R, Samuels M, Adamson ED. The transcription factor, Egr-1, is rapidly modulated in response to retinoic acid in P19 embryonal carcinoma cells. Developmental biology. 1991;148:165–173. doi: 10.1016/0012-1606(91)90327-y. [DOI] [PubMed] [Google Scholar]
- Fragola G, Germain PL, Laise P, Cuomo A, Blasimme A, Gross F, Signaroldi E, Bucci G, Sommer C, Pruneri G, et al. Cell reprogramming requires silencing of a core subset of polycomb targets. PLoS genetics. 2013;9:e1003292. doi: 10.1371/journal.pgen.1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gashler A, Sukhatme VP. Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Progress in nucleic acid research and molecular biology. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A, Qadeer ZA, Hasson D, Ratnakumar K, Leu NA, Leroy G, Liu S, Costanzi C, Valle-Garcia D, Schaniel C, et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nature communications. 2013;4:1565. doi: 10.1038/ncomms2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DT, Horvitz HR. MAB-10/NAB acts with LIN-29/EGR to regulate terminal differentiation and the transition from larva to adult in C. elegans. Development. 2011;138:4051–4062. doi: 10.1242/dev.065417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter SE, Finnegan EF, Zisoulis DG, Lovci MT, Melnik-Martinez KV, Yeo GW, Pasquinelli AE. Functional genomic analysis of the let-7 regulatory network in Caenorhabditis elegans. PLoS genetics. 2013;9:e1003353. doi: 10.1371/journal.pgen.1003353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer research. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes & development. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaraju K, Nguyen HQ, Liebermann DA, Hoffman B. The zinc finger transcription factor Egr-1 potentiates macrophage differentiation of hematopoietic cells. Molecular and cellular biology. 1995;15:5499–5507. doi: 10.1128/mcb.15.10.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nature genetics. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Kumbrink J, Kirsch KH, Johnson JP. EGR1, EGR2, and EGR3 activate the expression of their coregulator NAB2 establishing a negative feedback loop in cells of neuroectodermal and epithelial origin. Journal of cellular biochemistry. 2010;111:207–217. doi: 10.1002/jcb.22690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoix J, Mullick A, He Y, Bravo R, Skup D. Wild-type egr1/Krox24 promotes and dominant-negative mutants inhibit, pluripotent differentiation of p19 embryonal carcinoma cells. Oncogene. 1998;17:2495–2504. doi: 10.1038/sj.onc.1202166. [DOI] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Le N, Nagarajan R, Wang JY, Svaren J, LaPash C, Araki T, Schmidt RE, Milbrandt J. Nab proteins are essential for peripheral nervous system myelination. Nature neuroscience. 2005;8:932–940. doi: 10.1038/nn1490. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes & development. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legesse-Miller A, Elemento O, Pfau SJ, Forman JJ, Tavazoie S, Coller HA. let-7 Overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. The Journal of biological chemistry. 2009;284:6605–6609. doi: 10.1074/jbc.C900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejard V, Blais F, Guerquin MJ, Bonnet A, Bonnin MA, Havis E, Malbouyres M, Bidaud CB, Maro G, Gilardi-Hebenstreit P, et al. EGR1 and EGR2 involvement in vertebrate tendon differentiation. The Journal of biological chemistry. 2011;286:5855–5867. doi: 10.1074/jbc.M110.153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Hsieh LC, Kuo MW, Yu J, Kuo HH, Lo WL, Lin RJ, Yu AL, Li WH. Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Molecular biology and evolution. 2007;24:2525–2534. doi: 10.1093/molbev/msm195. [DOI] [PubMed] [Google Scholar]
- Luo M, Ling T, Xie W, Sun H, Zhou Y, Zhu Q, Shen M, Zong L, Lyu G, Zhao Y, et al. NuRD blocks reprogramming of mouse somatic cells into pluripotent stem cells. Stem Cells. 2013;31:1278–1286. doi: 10.1002/stem.1374. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Yamanaka S. Glis1, a unique pro-reprogramming factor, may facilitate clinical applications of iPSC technology. Cell Cycle. 2011;10:3613–3614. doi: 10.4161/cc.10.21.17834. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell stem cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, Slack FJ. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle. 2008;7:3935–3942. doi: 10.4161/cc.7.24.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr F, Heinemann U. Mechanisms of Lin28-mediated miRNA and mRNA regulation--a structural and functional perspective. International journal of molecular sciences. 2013;14:16532–16553. doi: 10.3390/ijms140816532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nature biotechnology. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature biotechnology. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HQ, Hoffman-Liebermann B, Liebermann DA. The zinc finger transcription factor Egr-1 is essential for and restricts differentiation along the macrophage lineage. Cell. 1993;72:197–209. doi: 10.1016/0092-8674(93)90660-i. [DOI] [PubMed] [Google Scholar]
- O'Farrell F, Esfahani SS, Engstrom Y, Kylsten P. Regulation of the Drosophila lin-41 homologue dappled by let-7 reveals conservation of a regulatory mechanism within the LIN-41 subclade. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237:196–208. doi: 10.1002/dvdy.21396. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Pasque V, Radzisheuskaya A, Gillich A, Halley-Stott RP, Panamarova M, Zernicka-Goetz M, Surani MA, Silva JC. Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. Journal of cell science. 2012;125:6094–6104. doi: 10.1242/jcs.113019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–20. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2007;451:141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Robertson B, Dalby AB, Karpilow J, Khvorova A, Leake D, Vermeulen A. Specificity and functionality of microRNA inhibitors. Silence. 2010;1:10. doi: 10.1186/1758-907X-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nature cell biology. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature cell biology. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer research. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 2005;234:1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevetson BR, Svaren J, Milbrandt J. A novel activation function for NAB proteins in EGR-dependent transcription of the luteinizing hormone beta gene. The Journal of biological chemistry. 2000;275:9749–9757. doi: 10.1074/jbc.275.13.9749. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Molecular cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Spaapen F, van den Akker GG, Caron MM, Prickaerts P, Rofel C, Dahlmans VE, Surtel DA, Paulis Y, Schweizer F, Welting TJ, et al. The immediate early gene product EGR1 and polycomb group proteins interact in epigenetic programming during chondrogenesis. PloS one. 2013;8:e58083. doi: 10.1371/journal.pone.0058083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhatme VP, Cao XM, Chang LC, Tsai-Morris CH, Stamenkovich D, Ferreira PC, Cohen DR, Edwards SA, Shows TB, Curran T, et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Molecular and cellular biology. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Nakamura M, Narita M, Takahashi K, Yamanaka S. Maturation, not initiation, is the major roadblock during reprogramming toward pluripotency from human fibroblasts. Proc Natl Acad Sci USA. 2013;110:12172–9. doi: 10.1073/pnas.1310291110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilko P, Schneider-Maunoury S, Levi G, Trembleau A, Gourdji D, Driancourt MA, Rao CV, Charnay P. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol Endocrinol. 1998;12:107–122. doi: 10.1210/mend.12.1.0049. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell stem cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell stem cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zambon AC, Gaj S, Ho I, Hanspers K, Vranizan K, Evelo CT, Conklin BR, Pico AR, Salomonis N. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28:2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cho J, Ptak D, Leung YF. The role of egr1 in early zebrafish retinogenesis. PloS one. 2013;8:e56108. doi: 10.1371/journal.pone.0056108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.