Abstract
Metastatic melanoma is a highly lethal disease notorious for its aggressive clinical course and eventual resistance to existing therapies. Currently we possess a limited understanding of the genetic events driving melanoma progression, and much effort is focused on identifying pro-metastatic aberrations or perturbed signaling networks that constitute new therapeutic targets. In this study, we validate and assess the mechanism by which homeobox transcription factor A1 (HOXA1), a pro-invasion oncogene previously identified in a metastasis screen by our group, contributes to melanoma progression. Transcriptome and pathway profiling analyses of cells expressing HOXA1 reveals up-regulation of factors involved in diverse cytokine pathways that include the TGFβ signaling axis, which we further demonstrate to be required for HOXA1-mediated cell invasion in melanoma cells. Transcriptome profiling also shows HOXA1’s ability to potently down-regulate expression of microphthalmia-associated transcription factor (MITF) and other genes required for melanocyte differentiation, suggesting a mechanism by which HOXA1 expression de-differentiates cells into a pro-invasive cell state concomitant with TGFβ activation. Our analysis of publicly available datasets indicate that the HOXA1-induced gene signature successfully categorizes melanoma specimens based on their metastatic potential and, importantly, is capable of stratifying melanoma patient risk for metastasis based on expression in primary tumors. Together, these validation data and mechanistic insights suggest that patients whose primary tumors express HOXA1 are among a high-risk metastasis subgroup that should be considered for anti-TGFβ therapy in adjuvant settings. Moreover, further analysis of HOXA1 target genes in melanoma may reveal new pathways or targets amenable to therapeutic intervention.
Keywords: HOXA1, melanoma, metastasis, MITF, TGFβ
INTRODUCTION
Metastasis is responsible for greater than 90% of cancer-related deaths (1) and is primarily thought to occur through a complex progression of interrelated steps by which primary tumor cells acquire the capacity to invade adjacent tissue, enter and survive in circulation, extravasate and proliferate at distant organs sites (2). This multifaceted process requires that cells acquire a wide range of biological capabilities in order to overcome numerous barriers to dissemination and growth in foreign microenvironments.
While much evidence supports this multi-step transit to metastasis, other data indicate that tumors may also be pre-ordained with early, metastasis-promoting genetic events (3). This deterministic model is supported by the finding that gene expression data derived from primary tumors can often predict metastasis (4) in addition to the fact that alterations found in metastases can be traced back to their subclonal presence in early primary lesions (5). There is a great need to identify such early metastasis-promoting events or their activated pathways, particularly those with potential to serve as new therapeutic targets. Moreover, it is equally as important to continue efforts toward developing early cancer detection strategies and intratumoral biomarkers that predict metastatic risk. This is especially true for notoriously aggressive cancers such as melanoma, whose current staging system is based on a measure of the vertical tumor growth termed Breslow thickness (6) in addition to other factors that include mitotic index, lymph node involvement and skin ulceration. Patients diagnosed with metastatic melanoma have an abysmal median survival of 6–9 months and a survival rate of only 10–20% due to melanoma’s aggressive behavior and eventual resistance to all therapies (7). Patients diagnosed with thin (<1mm) melanoma have a high survival rate following tumor excision, and the majority of these individuals will have no evidence of metastasis at diagnosis. However, approximately 5–10% of melanoma patients with low-staged lesions (i.e., Stage I/II) will die of recurrence and metastatic disease within 10 years of diagnosis despite surgical removal of the primary tumor (8). This is further evidenced by a retrospective study of 9,129 fatal melanoma cases spanning 1988–2006 that discovered equivalent numbers of patients diagnosed with thin (<1mm; 2,472 cases) and thick (>4mm; 2,041 cases) melanomas died from their disease (9). These statistics suggest that there is a high-risk melanoma subpopulation that is not identified by the current standard pathological and clinical staging system and illustrates the need for new molecular-based risk assessment strategies and novel targeted therapeutics to manage this aggressive malignant disease.
Given the pressing need to identify genetic mediators of melanoma metastasis and better prognostic indicators, we devised an oncogenomics-guided screening strategy to identify genes capable of driving cancer cell invasion and metastasis (10). This approach leveraged a multi-level oncogenomics comparison founded on (1) genetically engineered mouse (GEM) models of melanoma with differing metastatic potential and (2) genomics data derived from human melanoma. These methods revealed a conserved list of 360 genes correlating with metastatic potential, functional screening of which identified 18 genes whose ectopic expression could significantly enhance cell invasion. Among those 18 genes, our initial study focused on validating the ACP5 phosphatase, expression of which we found to not only enhance melanoma cell invasion but also drive in vivo tumorigenesis and metastasis by a mechanism involving modulation of the phosphorylation status of proteins comprising the focal adhesion complex.
In this study, we validate the top performing pro-invasion candidate identified by our initial screening approach, homeobox transcription factor A1 (HOXA1). We demonstrate that HOXA1 exhibits pro-invasive and oncogenic activities across several melanoma cell systems in a manner dependent HOXA1’s functional DNA binding domain. Transcriptome profiling comparisons reveal HOXA1’s marked influence on the expression of genes involved in diverse cytokine signaling pathways, and cell-based studies support a mechanism by which HOXA1 hyperactivates the TGFβ signaling pathway to elicit HOXA1-mediated cell invasion. Finally, we provide evidence that HOXA1 potently down-regulates genes involved in melanocyte differentiation, suggesting that HOXA1 expression de-differentiates cells into a state of higher metastatic potential. Importantly, the HOXA1 gene signature successfully stratifies melanoma patients into two subgroups with significant differences in metastasis-free survival based on expression in primary tumor specimens, suggesting that this prognostic signature may provide insight into new means by which to identify at risk patients and potentially reveal new targets for therapeutic intervention.
RESULTS
Functional validation of HOXA1 as a pro-invasion oncogene in melanoma
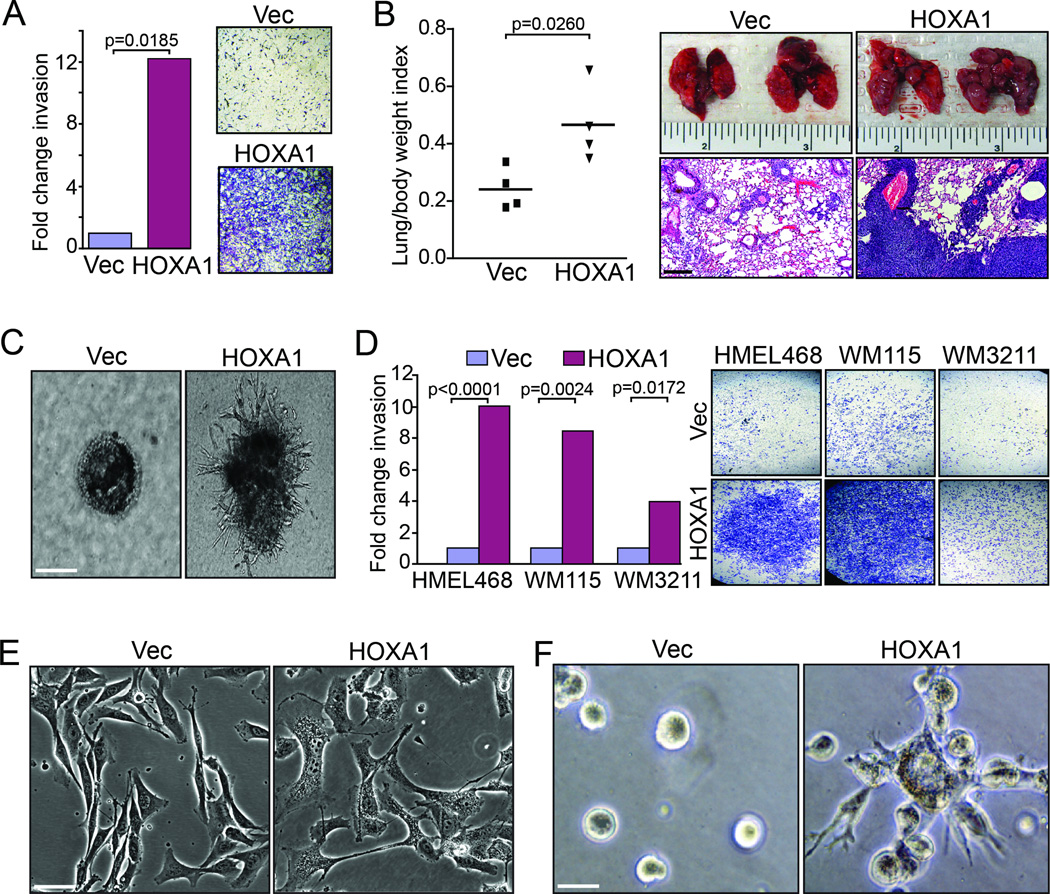
In a recent effort to identify early genetic drivers of melanoma metastasis (10), we devised a comparative oncogenomics strategy employing use of genomics data derived from human and GEM melanoma tumors originating from: (1) An inducible H-RASG12V-driven model (iHRAS) that develops non-metastatic tumors (11) and (2) a similarly engineered model for induction of the receptor tyrosine kinase MET (iMET) that initiates metastatic melanomas (10). Gene candidates identified by this in silico strategy were enlisted into a functional screen for drivers of in vitro cell invasion, an effort that identified HOXA1 as the top scoring pro-invasion gene (10). To begin our validation efforts on HOXA1 in this study, we sought to examine HOXA1 activity in Ink4a/Arf−/− mouse-derived melanocytes transduced with H-RASG12V (hereafter referred to as M3HRAS cells (12)) given that our candidate gene list was derived from genomic comparisons of non-metastatic iHRAS and metastatic iMet GEM tumors. Non-metastatic M3HRAS cells stably expressing HOXA1 exhibited increased invasion through Matrigel by 12-fold compared to vector control cells in transwell invasion assays (Fig. 1A; p=0.0185). Even more striking was HOXA1’s ability to enhance macroscopic lung nodule formation by M3HRAS cells following intravenous injection into NOD-SCID mouse tail veins, a surrogate assay for metastasis (Fig. 1B). Consistent with these pro-invasion and metastatic phenotypes, expression of HOXA1 in M3HRAS cells induced an invasive cell morphology characterized by invasive, stellate protrusions when plated in Matrigel matrix compared to M3HRAS vector control cells that proliferated as individual colonies devoid of invasive structures (Fig. 1C). Together, these data demonstrate the pro-invasive activity of HOXA1 in an H-RASG12V cells thereby supporting its role of as a pro-metastasis gene identified from our initial comparison of iHRAS and iMet tumors.
Figure 1. HOXA1 drives cell invasion and metastasis in weakly metastatic melanoma models.
(A–C) M3HRAS cells over-expressing vector control (Vec) or HOXA1 were assayed for (A) in vitro invasion using transwell invasion chambers (B) lung seeding capacity, a correlate of metastatic activity, following intravenous injection into mouse tail veins and (C) invasive morphology in 3D Matrigel colony assays. Images in (A) show representative invasion readouts and (B) H&E stained lung sections harvested from experimental animals. P-values calculated by t-test. Scale bars = 100 µm, (B); 50 µm, (C). (D) Human cell lines HMEL468, WM115 and WM3211 over-expressing vector control or HOXA1 were assayed for in vitro invasion using transwell invasion chambers. Representative invasion readouts shown at right. P-value calculated by t-test. (E–F) WM115 cells expressing vector control or HOXA1 propagated in (E) 2D tissue culture and (F) 3D Matrigel colony formation assays. Scale bars = 100 µm, (E); 50 µm, (F).
Given HOXA1’s robust activities in the murine M3HRAS cell line, we sought to validate HOXA1’s activity in human melanoma cell line models. As demonstrated previously (10), expressing HOXA1 in engineered TERT immortalized melanocytes expressing BRAFV600E (PMEL/hTERT/CDK4(R24C)/p53DD (13) hereafter referred to as “HMEL468”) enhanced in vitro cell invasion approximately 10-fold over vector-expressing control cells (Fig. 1D; p<0.0001). Likewise, HOXA1 greatly enhanced cell invasion over vector control cells when expressed in weakly invasive cell lines that include WM115 (Fig. 1D; p=0.0024) and to a lesser extent in WM3211 cells (Fig. 1D; p=0.0172). The phenotypic effect of expressing HOXA1 appeared most dramatic on WM115, as cells transduced with HOXA1 underwent a dramatic change in morphology (Fig. 1E) denoted by a marked increase in cell spreading, membrane ruffling (Movie S1 and S2), and invasive protrusions when plated in Matrigel matrix (Fig. 1F) with no obvious changes in cell proliferation (Fig. S1) compared to vector-expressing control cells.
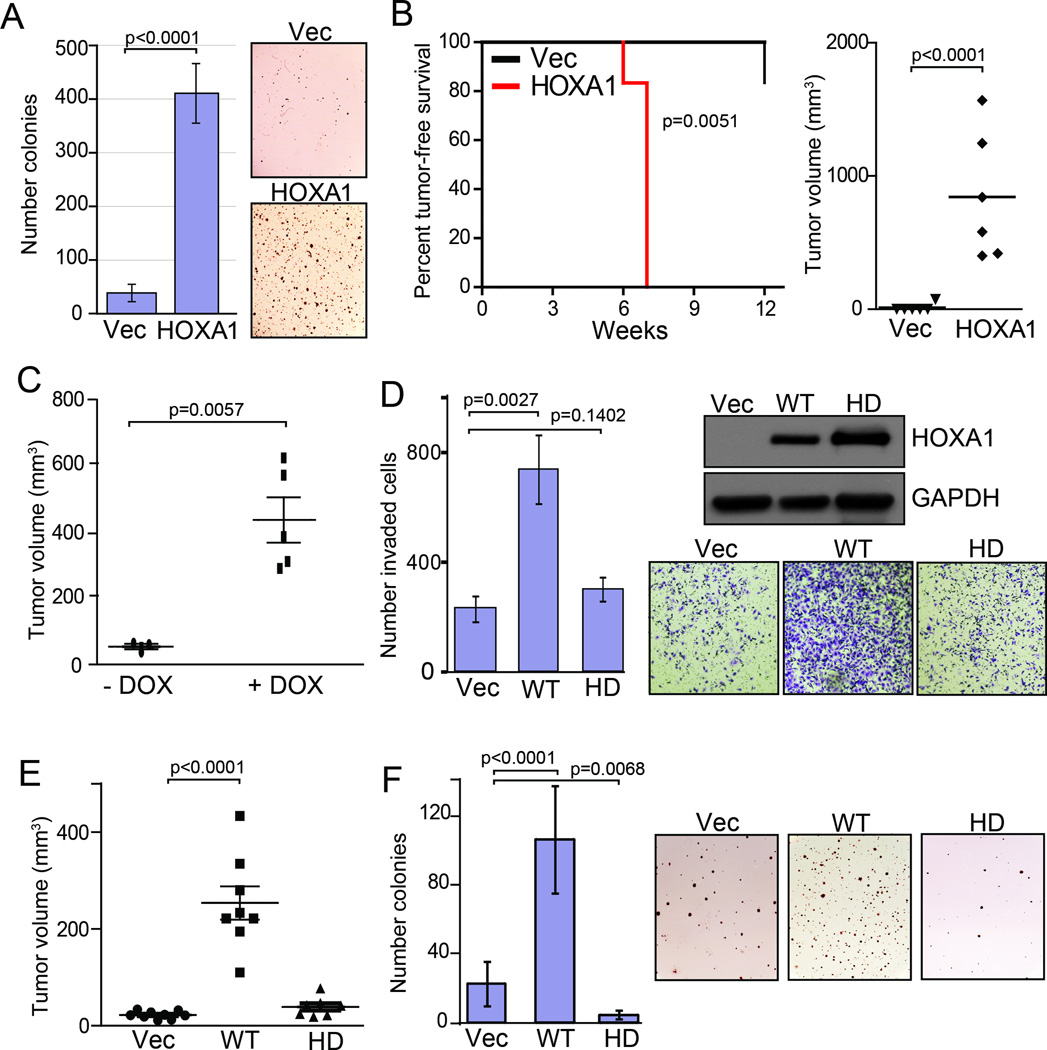
We showed previously that HOXA1 displays a pattern of progression-correlated expression across the benign-to-malignant transition, exhibits oncogenic activity when expressed with BRAFV600E in immortalized melanocytes and is required to maintain anchorage-independence growth (10). Given HOXA1‘s pronounced phenotypic effects on the weakly tumorigenic WM115 melanoma cell line (Fig. 1D–F), we next examined HOXA1’s ability to enhance WM115 colony formation in anchorage-independent growth assays. As expected, expressing HOXA1 markedly enhanced WM115 colony formation 10.6-fold compared to vector-expressing cells when plated in soft agar assays (Fig. 2A; p<0.0001). Consistent with this in vitro study, we next examined HOXA1‘s ability to enhance tumor growth by implanting WM115 cells expressing HOXA1 or vector control into the flanks of athymic mice. HOXA1 expression led to a significant increase in xenograft tumor incidence (p=0.0051) and growth (p<0.0001) compared to control cells that largely failed to form palpable tumors within the time course of these studies (Fig. 2B). To confirm this finding, we constructed a WM115 cell line engineered with a doxycycline inducible HOXA1 expression construct, whose activation with doxycycline following cell implantation led to an increase in tumor growth compared to a control mouse cohort maintained off of doxycycline (Fig. 2C; p=0.0057). These validation data support a pro-tumorigenic role for HOXA1 in melanoma, and together with the progression correlation data and our previous finding that HOXA1 can cooperatively transform immortalized primary melanocytes expressing BRAFV600D (10), suggests that HOXA1 may be selected for early during transformation where it also drives tumor metastasis.
Figure 2. HOXA1-mediated oncogenicity requires a functional DNA binding domain.
(A–B) WM115 cells expressing vector control (Vec) or HOXA1 were assessed for (A) colony formation in anchorage-independent growth assays and (B) xenograft tumor growth in athymic mice. Kaplan-Meier tumor-free survival (left) and endpoint tumor size (right) are shown for panel (B). (C) Endpoint size analysis of WM115 xenograft tumors induced by a doxycycline (DOX) inducible HOXA1 construct. (D–F) WM115 cells stably expressing vector control (Vec), HOXA1 (WT) or HOXA1 mutated at its DNA binding domain (HD) were examined for (D) in vitro invasion activity using transwell invasion chambers, (E) xenograft tumor growth and (F) anchorage-independent growth. The immunoblot shown in (D) reflects the level of WT HOXA1 and HOXA1-HD protein expressed in cells used for experiments shown in (D–F). Error bars indicated +/− s.d.; All p-values calculated by t-test except for Fig. 1B, left (p-value calculated by log rank).
To begin investigating the mechanism by which HOXA1 influences cell invasion and tumor growth, we mutated homeodomain residues glutamine 50 and asparagine 51 to alanine (QN>AA) which has been shown by others to impair DNA binding activity of HOXA1 (14). In contrast to WM115 cells expressing wild-type HOXA1, cells stably expressing the homeodomain dead (HD) mutant failed to increase cell invasion compared to WM115 control cells despite being expressed at similar levels (Fig. 2D). Similarly, WM115 cells expressing HOXA1-HD failed to recapitulate HOXA1-mediated increases in in vivo tumor growth following implantation into athymic mice (Fig. 2E). Mutating the DNA binding domain completely attenuated HOXA1’s ability to promote anchorage independent growth in soft agar (Fig. 2F; WT vs. HD, p<0.0001), and comparison of vector- and HOXA1-HD-expressing cells suggest that HOXA1-HD exhibits a dominant negative effect in this assay (p=0.0068). Together, these data suggest that HOXA1 requires its native ability to bind DNA to elicit invasive and oncogenic effects, likely due to its transcription factor activity.
Transcriptome analysis links HOXA1 to cytokine signaling pathways
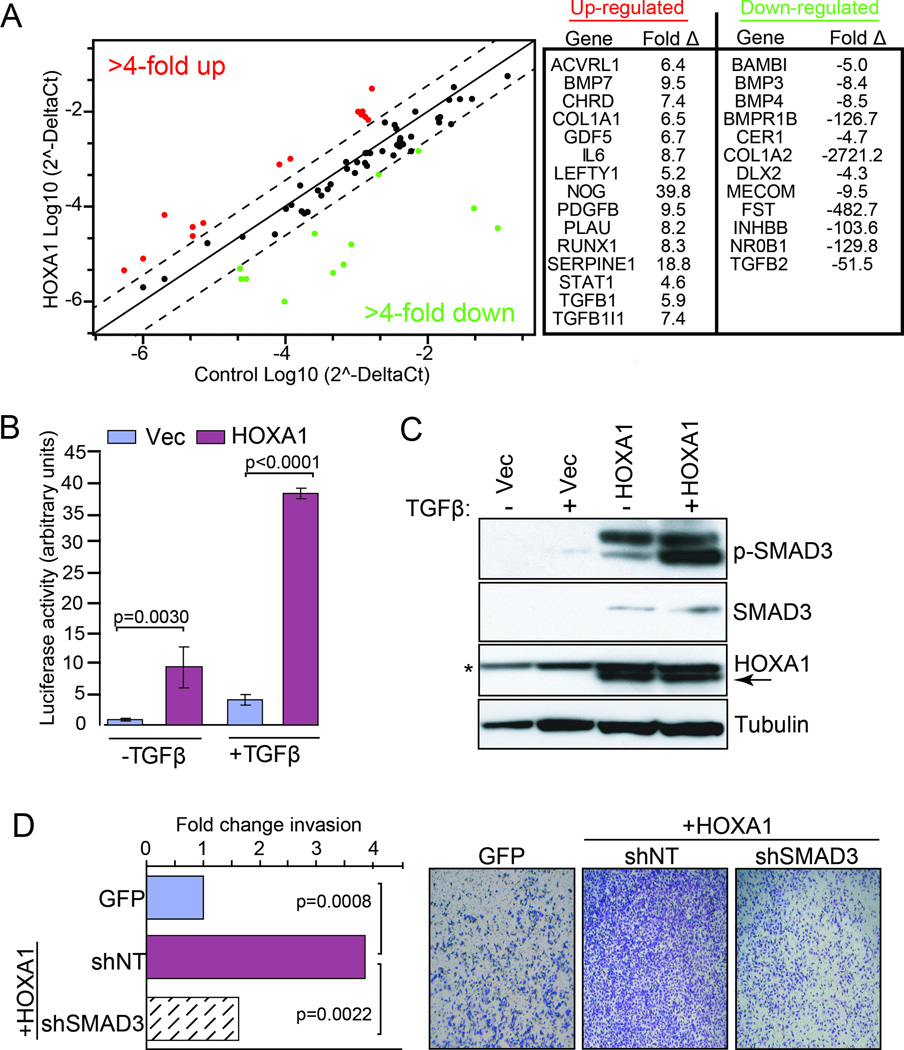
Our finding that HOXA1 requires its native ability to bind DNA to elicit its pro-invasion and oncogenic activities combined with its known role as a transcription factor led us to examine the HOXA1 transcriptome to gain insight into the protein’s mode-of-action in melanoma. Given the strong phenotypes exhibited by WM115 cells following enforced expression of HOXA1, we chose to first profile WM115 cells stably expressing HOXA1 or vector control using Affymetrix gene chips. Analysis for differential gene expression revealed 852 up- and 882 down-regulated genes (p<0.01, >3-fold change, average intensity >25) in cells constitutively expressing HOXA1 (Table S1), which is consistent with HOX genes ability to both activate and repress gene transcription (15). Functional annotation clustering via David Bioinformatics Resources revealed “regulation of cell migration“ (p=5.67E-10) as the top scoring functional group (Table S2). A large portion of the differentially expressed genes (249 up- and 192 down-regulated) exhibited a greater than 10-fold change in expression in WM115-HOXA1 cells compared to vector control cells (Table S1). Closer analysis of the up-regulated gene set (>10-fold change) indicated significant enrichment for “secreted factors” (p=5.5E-26), “extracellular region” (p=7.7E-19), “chemotaxis” (p=3.2E-10) and “cytokine activity” (p=2.7E-09) among the top functional groups (Table S2). Knowledge-based pathway analysis using Ingenuity Pathway Analysis (IPA) revealed a top scoring network (Fig. S2) whose node centered on transforming growth factor beta 1 (TGFβ1), a cytokine that regulates diverse cellular processes that include cell growth, movement, differentiation and apoptosis (16).
Given the known role for the TGFβ pathway in regulating cancer progression processes, we used WM115 cells to perform a focused PCR profiling array to analyze HOXA1-induced expression changes of 84 genes associated with the TGFβ/BMP signaling pathway. These assays confirmed that expression of HOXA1 significantly modulates expression of multiple genes encoding components that signal through this pathway that include receptors (e.g. PDGFB, 9.5×; BMPR1B, −126.7×), ligands (e.g. BMP7, 9.5×; TGFB1, 5.9×; GDF5, 6.7×) and other molecules (e.g., SERPINE1/PAI-1, 18.8×, CER1, −4,7×) positioned throughout the TGFβ signaling axis (Fig. 3A). As a more directed measure of HOXA1’s influence on TGFβ signaling, we transfected control and HOXA1-expressing WM115 cells with a TGFβ-responsive reporter construct (p3TP-Lux; (17)). As shown in Figure 3B, expression of HOXA1 enhanced basal reporter activity (11.0-fold, p=0.0030) in the absence of TGFβ and evoked a 9.3-fold increase in response to TGFβ ligand compared to control cells (p<0.0001). Correspondingly, levels of phosphorylated SMAD3 (p-SMAD3), which is the activated form of this protein and is required for signaling through the TGFβ pathway, were elevated in cells expressing HOXA1 and increased further after stimulation with TGFβ ligand thus corroborating active TGFβ signaling (Fig. 3C). Notably, total SMAD3 levels were also elevated in HOXA1-expressing cells (Fig. 3C), which is consistent with SMAD3‘s 4-fold up-regulation observed from our transcriptome analysis (Table S1). To determine whether HOXA1-mediated invasion is dependent on the TGFβ pathway, we treated control- and HOXA1-expressing cells with RNAi against SMAD3. Depletion of SMAD3 decreased HOXA1-mediated invasion (Fig. 3D), which further indicates that HOXA1 requires the TGFβ pathway to fully elicit its pro-invasion phenotype.
Figure 3. HOXA1 promotes invasion through enhanced TGFβ signaling.
(A) Focused TGFβ/BMP PCR profiling array on cDNA isolated from WM115 cells stably expressing vector control or HOXA1. Shown at right are genes up-regulated (red) and down-regulated (green) greater than 4-fold in HOXA1-expressing cells compared to vector control. (B) WM115 cells expressing vector control (Vec) or HOXA1 were transfected with the TGFβ-inducible p3TP-Lux luciferase reporter construct, followed by treatment with or without TGFβ to assess responsiveness. Error bars indicate +/− s.d.; p-values calculated by t-test. (C) Whole cell lysates from WM115 cells stably expressing vector control or HOXA1 were propagated in 1% serum with or without TGFβ for immunoblot analysis using the indicated antibodies. P-SMAD3 = phosphorylated SMAD3, Ser423+Ser425. * = denotes tubulin band. (D) WM115 cells expressing vector control or HOXA1 were treated with or without SMAD3 shRNA (shSMAD3) or non-targeting shRNA (shNT) and loaded onto transwell invasion chambers. P-values calculated by t-test.
HOXA1 regulates expression genes controlling melanocyte differentiation
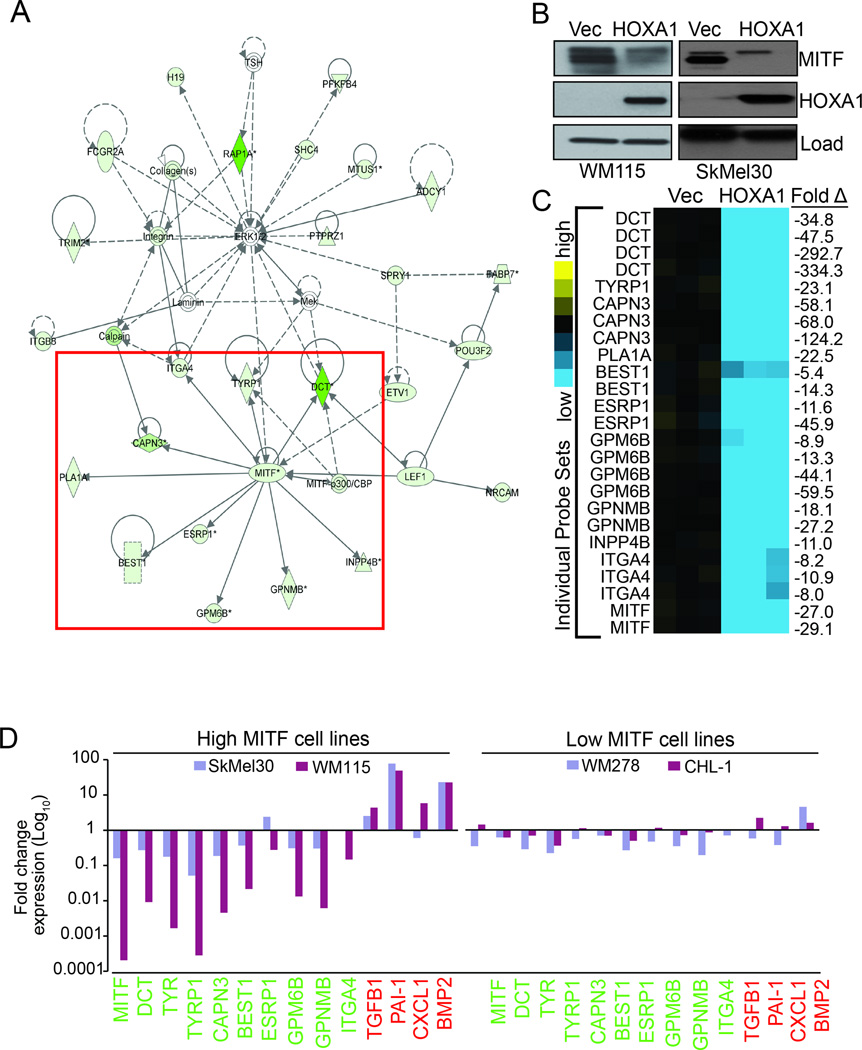
In contrast to the HOXA1-induced up-regulated genes, functional clustering analysis of genes down-regulated by HOXA1 greater than 10-fold, revealed groups related to melanocyte biology, particularly “pigmentation during development” (p=1.9E-05) and “melanocyte differentiation” (p=8.0E-05) (Table S2). In support of this finding, knowledge-based pathway analysis using IPA revealed a top-scoring down-regulated network whose node centered on microphthalmia-associated transcription factor (MITF; average 28-fold down-regulation in HOXA1-expressing cells), a transcription factor critically important for activating expression of genes required for melanocyte differentiation from their neural crest precursors (Fig. 4A, box). Immunoblotting analysis of WM115 protein lysates confirmed decreased MITF protein expression in cells expressing HOXA1 versus vector control (Fig. 4B).
Figure 4. HOXA1 expression down-regulates genes important for melanocyte differentiation and pigmentation.
(A) Molecule network generated using Ingenuity Pathways Analysis. The network is displayed graphically as nodes (genes) and edges (the biological relationships between nodes). Solid lines represent direct interactions and dashed lines represent indirect interactions. Green colors denote genes that were under-expressed >10-fold in WM115 expressing HOXA1 versus control. Red box surrounds MITF node and known MITF target genes differentially expressed >10-fold in WM115-HOXA1 cells. (B) Whole cell lysates from WM115 and SkMel30 cells stably expressing vector control (Vec) or HOXA1 were processed for immunoblot analysis using the indicated antibodies. Load = tubulin (WM115 panel) and GAPDH (SkMel30 panel). (C) Heat map representing Affymetrix probe expression for genes boxed in (A). Expression values at right indicate fold change in gene expression (HOXA1 versus vector control). (D) Expression validation of select genes by RT-qPCR analysis of cDNA prepared from WM115 and SkMel30 (high MITF) and WM278 and CHL-1 (low MITF) cells expressing vector control or HOXA1. All values are normalized based on Actin B expression and plotted as fold change compared to vector control (set as 1.0 for each gene). Values indicate fold change for genes found up-regulated (red) and down-regulated (green) in WM115 transcriptome analysis.
In addition to MITF, we identified components of the MITF signaling network including multiple MITF target genes that include DCT (−177.3×), TYRP1 (−23.1×), and GPNMB (−22.7×) among others based on their probe set expression averages (Fig. 4C), and additional MITF targets that were present in our transcriptome analysis but not included in the IPA-generated network including EDNRB (−90.8×), TYR (−6×) and TBX2 (−5.1×) (Table S1). Quantitative PCR analysis of mRNA isolated from WM115 cells expressing HOXA1 or vector control validated differential expression of select MITF target genes and others found over-expressed in our transcriptome analysis (Fig. 4D), providing additional evidence to support the regulation of these candidates by HOXA1. Importantly, we detected the same trend in up-and down-regulated expression of this target gene set in two WM115 tumors (396 and 397) resulting from our in vivo explant studies (Fig. 2C) that employed the doxycycline-inducible HOXA1 system (Fig. S3). We extended this analysis to three additional melanoma cell lines with documented MITF expression (Fig. S4). Expression of HOXA1 in SkMel30 cells (high MITF) resulted in a trend of panel gene expression similar to observed with WM115 cells and tumors, whereas HOXA1 was less effective at evoking this response in low MITF-expressing cell lines WM278 and CHL-1 (Fig. 4D).
Consistent with this latter finding was our observation that HOXA1 failed to increase cell invasion when expressed in the WM278 and CHL-1 cell lines (Fig. S5) and exhibited weak pro-invasion activity when expressed in the low MITF-expressing WM3211 cell line compared to the high MITF expressing cell lines, HMEL468 and WM115 (Fig. 1D). Moreover, expressing HOXA1 in SkMel30 cells (high MITF) significantly reduced MITF at the protein level similar to observed in WM115 cells (Fig. 4B). Use of a previously described (18) signature scoring metric (“t-score”) to compare the WM115 transcriptome datasets with transcriptome data derived from SkMel30 cells expressing vector control or HOXA1 (Table S1) revealed a high degree of similarity between the signatures (Fig. S6). In contrast, T-score analysis indicated a low degree similarity when comparing the WM115- and WM3211 (low MITF)-derived signatures (Fig. S6). Together, these data suggest that HOXA1 expression drives deregulation of melanocytic development genes, and HOXA1’s phenotypic effects are greatest when expressed in parental cells with high levels of MITF.
HOXA1 promotes an invasive gene signature and prognosticates clinical outcome
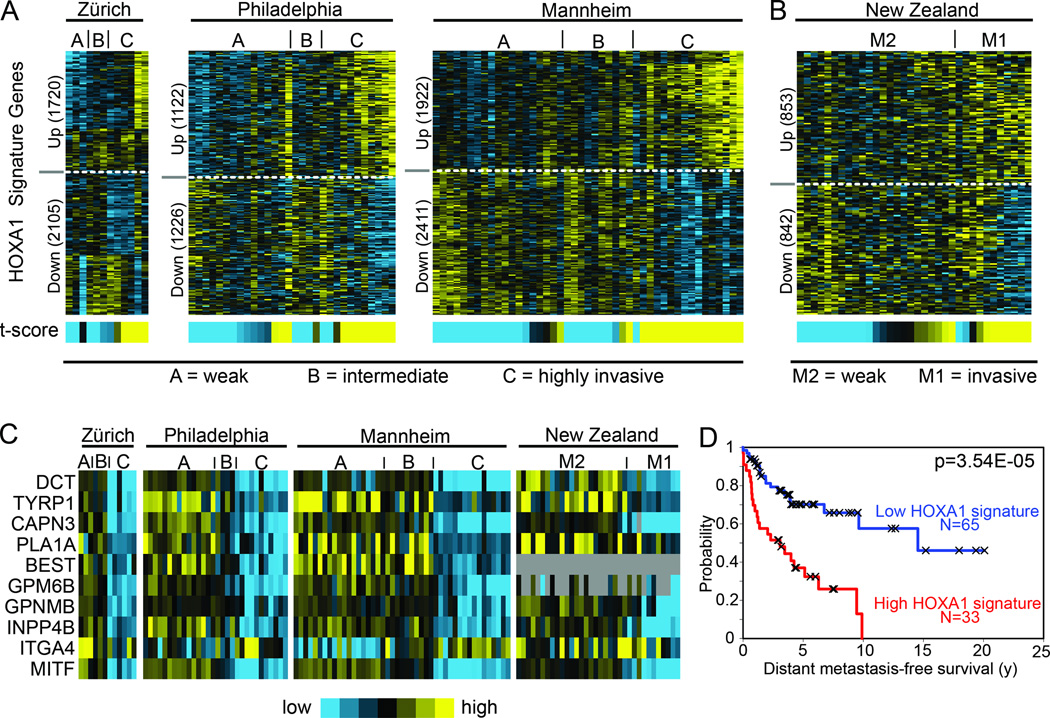
HOXA1’s potent effects on cell invasion led us to explore published transcriptome profiling studies that have produced melanoma invasion gene signatures. One such study by Hoek and colleagues (19) reported an expression clustering analysis that differentiated 86 melanoma specimens from three sample cohorts (Zürich, Philadelphia and Mannheim datasets; GEO accession GSE4845) into three distinct cluster groups (A–C) based on related transcriptome profiles. Comparing our HOXA1-induced expression signature with those of group A (weakly invasive), group B (intermediate) and group C (highly invasive) using the t-score metric indicated a significant degree of similarity with group C in each of the Zürich, Philadelphia and Mannheim cohorts (Fig. 5A; t-test for t-score values: A and B vs. C, p=4.8E-08). Based on the authors’ functional annotation of the genes differentiating groups A–C (19), the genes in the invasive group C differ from those of weakly invasive group A based on the presence of 51 down-regulated genes that are enriched for neural crest and melanoctyic differentiation factors and 54 up-regulated genes enriched for extracellular modifying factors and genes regulated by TGFβ signaling. Of these 105 genes, the HOXA1-induced gene signature contained 31 of 51 down-regulated genes that included MITF, TYRP1, TYR, and DCT among others. In addition, our HOXA1-induced gene signature contained 29 of 54 genes significantly up-regulated in the group C transcriptome.
Figure 5. HOXA1 expression signature correlates with invasiveness and prognosticates patient clinical outcome.
(A–B) Heat map representing expression of HOXA1 signature genes in transcriptome profiles of melanoma specimens collected by (A) Hoek et al (19) (N: Zürich, 15; Philadelphia, 55; Mannheim, 45) and (B) Jeffs et al. (20) (N: 34). Specimens in (A) are annotated based on presence or absence of invasive gene signature defined in (19): A = weakly invasive, B = intermediate, C = highly invasive, whereas specimens in (B) are annotated based on presence or absence of motifs defined in (20): M1 = invasive, M2 = weakly invasive. Signature similarity score (t-score) is represented by heat map bar (blue, low; yellow, high). (C) Heat map representing expression of MITF and select MITF target genes (see Fig.4C–D) in the transcriptome reported by Hoek and Jeffs (19) (20). (D) Differences in patient outcome based on HOXA1 gene signature manifestation (comparing samples in the top 25% of signature scores with those in the bottom 75%) using a cohort of 98 primary melanomas reported by Winnepenninckx et al. (22); p-value calculated by log rank test.
In addition to studies by Hoek et al., we similarly examined expression data recently reported by Jeffs and colleagues (20) who profiled a large cohort of cell lines derived from primary melanomas (New Zealand dataset; GEO accession GSE16404). Like the previous study, Jeffs et al. employed unsupervised clustering analysis that defined two motifs of cell lines based on expression of a core set of genes that included MITF and others involved in such processes as neural crest development, melanocytic differentiation and extracellular matrix remodeling. Comparing the HOXA1 signature with the Jeffs et al. dataset indicated a degree of similarity with expression Motif 1, which is characterized by low MITF and high invasion activity, versus expression Motif 2 that contains higher MITF levels, down-regulation of extracellular remodeling factors and are weakly invasive (Fig. 5B; t-test for t-score values: Motif 1 vs. Motif 2, p=0.0003). Similar to invasive group C specimens defined by Hoek et al. (19), cell lines characterized by the invasive Motif 1 were enriched for MITF target genes which are also down-regulated by HOXA1 expression >10-fold (Fig. 5C) and residing in the MITF network illustrated in Figure 4A. Together, these data suggest that enforced expression of HOXA1 drives cells to adopt an invasion profile through a process reminiscent of de-differentiation into a neural crest-like cell type with higher metastatic potential.
These findings led us to explore the prognostic implications of the HOXA1 gene signature in melanoma. Given the shortage of outcome-associated expression datasets for melanoma, which is primarily due to limited availability of primary human melanoma tissue preserved in a form suitable for expression profiling (21), we focused on a dataset (22) containing a cohort of 98 primary tumor specimens (combined study and validation sets: Stage I, II, III, and IV tumors) with accompanying patient outcome data. We scored these profiles for similarity to the HOXA1 gene signature and found that patients with profiles showing more similarity to the HOXA1-inducible patterns had a shorter time to distant metastasis events (univariate Cox p<0.01, t-score as continuous variable; Log-rank test p=3.5E-05, comparing top 25% of high scoring samples versus others, Fig. 5D). Multivariate Cox analysis incorporating established clinical variables (stage, age, and Breslow thickness) indicate that the HOXA1 gene signature provides additional prognostic power independent of these main clinical variables (Table S3). Together, these data indicate that the HOXA1 gene signature prognosticates patient risk based on expression in primary tumors.
DISCUSSION
The purpose of this study was to functionally validate and elucidate the mechanism-of-action for HOXA1, the top-scoring pro-invasion gene identified from our recently described oncogeneomics-guided screen for melanoma metastasis drivers (10). HOXA1 is a conserved member of the homeobox transcription factor family, which is comprised of 39 genes in humans organized into four different clusters (A–D) that coordinately regulate cell fate, early developmental patterns and organogenesis (23). Complex transcription networks have evolved to spatially and temporally regulate HOX gene expression during development, and a growing body of evidence suggests that disrupting this tight regulation can impact oncogenic and tumor suppressive mechanisms in a context specific manner (23). Indeed, HOX genes have been found to directly impact tumorigenesis via diverse mechanisms in cancers that include lung, breast, and hematological malignancies among others, and HOXA1 up-regulation has specifically been reported in cancers including breast cancer, leukemia, squamous cell carcinoma and melanoma, including in melanomas with distant metastasis (24).
HOXA1’s role as a transcription factor coupled with our current findings that described HOXA1-mediated cancer activities require its functional DNA binding domain led us to examine gene expression changes elicited by HOXA1 in melanoma cells. From those studies an invasive expression profile emerged that includes increased expression of numerous cytokines and their mediators, including those involved with TGFβ/BMP signaling. We demonstrate that HOXA1 expression enhances activation of the TGFβ pathway, and we further show that the SMAD signaling axis required for TGFβ-mediated processes is required for HOXA1 to elicit its full effects on in vitro cell invasion. While it remains unclear how HOXA1 mediates TGFβ signaling and whether HOXA1 also functions through other cytokine signaling pathways identified by our transcription analysis, it is intriguing to speculate that this pathway may be in some way connected to an altered cell differentiation program given the role played by HOX genes in cell fate determination. This notion is supported by our transcriptome analysis that revealed marked de-regulation of genes involved in melanocytic differentiation that included MITF, which is a melanocytic lineage-specific transcription factor that serves as a central regulator in melanocyte determination, as well as key MITF downstream targets. Other studies have provided a mechanism for oncogenesis by which some HOX genes re-express in tumors in a manner temporally different from expression in their parental, normal tissue to disrupt cell differentiation pathways (23). It is interesting to note that mouse studies have shown that HOXA1 expression is tightly regulated between days 7 and 9 of embryonic development where it is expressed in neural crest precursors essential for the generation of mesenchymal derivatives that include melanocytes among other cell lineages (25). Future work will determine whether late, inappropriate mis-expression of HOXA1 promotes melanoma tumor growth and metastasis by a mechanism that may involve de-differentiating cells into a more mobile, invasive neural crest-like precursor state.
While additional studies are required to determine whether HOXA1 directly regulates expression of melanocytic differentiation genes like MITF, it is noteworthy that decreased expression of MITF has previously been associated with increased melanoma invasiveness (26) whereas MITF over-expression has been demonstrated to suppress melanoma metastasis (27). Hoek and colleagues have similarly documented a pattern of decreased MITF expression in invasive cell lines, an observation that led to the “phenotype switching” hypothesis proposed to account for melanoma metastasis (28). In contrast to the stochastic view that pro-metastatic genomic alterations occur in a step-wise manner during tumor evolution, the phenotype switching hypothesis posits that genes required for metastasis are epigenetically modulated to change individual cells from a proliferative to a more invasive cell state (29). It is tempting to speculate that HOXA1 directly influences phenotype switching behavior given similarities between the HOXA1-induced gene signature and those reported by Hoek et al. It is unclear how HOXA1 coordinately down-regulates the melanocytic differentiation pathway and activates signaling through the TGFβ axis, though prior work has suggested that TGFβ represses MITF expression (30). Moreover, other work (31) reported that mobile, metastatic melanoma cells express low levels of MITF and exhibited increased TGFβ activation compared to non-motile cells that expressed high levels of MITF. Given HOXA1’s profound effect on TGFβ signaling and suppression of MITF and its targets, it is possible that HOXA1 could be a regulator of phenotype switching and suggests a model by which decreased expression of MITF and increased signaling through the TGFβ axis coordinately drive melanoma growth and metastasis.
Our finding that the HOXA1 gene signature predicts clinical outcome based on expression profiles of primary tumors suggests that it might be useful to stratify metastatic risk for patients with melanoma, though more work will be required to validate its prognostic utility. Such a molecular prognostic test, if successfully implemented in the clinic, would significantly complement current prognostication standards by identifying high-risk subpopulations from generally low-risk, early-staged patients thereby selecting those patients for aggressive treatment and follow-up regimens. Moreover, the fact that HOXA1 functionally drives metastatic phenotypes and tumor growth in addition to providing a prognostic gene signature also suggests that genes within this signature may represent individual gene targets or pathways, such as the TGFβ pathway (32), suitable for therapeutic intervention.
MATERIALS AND METHODS
Cell culture
All cell lines were propagated at 37°C and 5% CO2 in humidified atmosphere in RPMI 1640 Medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS). HMEL468 melanocytes were a subclone of PMEL/hTERT/CDK4(R24C)/p53DD/BRAFV600E cells, as described (13). The WM115, WM278 and WM3211 cell lines were obtained from the Wistar Institute. The CHL-1 and SkMel30 cell lines were obtained from the ATCC.
Plasmids and RNAi
Full-length cDNA encoding HOXA1 (NM_005522.4) was obtained from the human ORFeome collection and transferred to the following viral vectors via Gateway recombination and virus production following the manufacture’s recommendations: pLenti63/V5 DEST (Invitrogen), pInducer-20 (33). All over-expression studies were performed with newly-transduced stable cells lines. For SMAD3 knockdown, cells were transduced with virus generated from control and pRhetrosuper-SMAD3 (Addgene # 15726; (34)).
Invasion assays
Matrigel coated chambers (BD Biosciences, Franklin Lakes, NJ; 354480) were utilized to assess invasiveness following the manufacture’s suggestions and as described (10). Chambers were seeded in triplicate or quadruplicate and placed in 10% serum-containing media which served as a chemo-attractant as well as in cell culture plates in duplicate as input controls. Following 20 hrs incubation, chambers were fixed in 10% formalin, stained with crystal violet for manual counting. Data were normalized to input cells to control for differences in cell number (loading control).
Soft agar and morphology assays
Soft-agar assays to measure anchorage independent growth were performed as described (10). For 3D Matrigel invasion morphology assays, cells transduced with control or HOXA1 virus were re-suspended in in low density into 12% Matrigel at 800 cells/well.
Animal
All studies using mice were performed in accordance with our IACUC-approved animal protocol at Baylor College of Medicine. For xenograft tumor assays, WM115 cells transduced with control or HOXA1 lentivirus (in pLenti6.3-V5 and pInducer backbones) were stably selected and re-suspended in a 1:1 solution of Hank’s balanced salts (Invitrogen) and Matrigel (BD Bioscience) for subcutaneous implantation into female nude and NOD-SCID (for inducible studies) animals (Harlan, Indianapolis, IN) at 1.0×106 cells/site on both flanks. For the doxycycline induction experiment using pInducer-20-HOXA1, animals injected with transduced cells were separated into two cohorts and maintained with or without chow containing doxycycline (2g/kg) for the duration of the experiment. For lung seeding assays, M3HRAS cells transduced with control of HOXA1 virus were re-suspended in Hank’s balanced salts (1×105 cells in 200 µl) for injection into the tail vein female SCID (Harlan) mice followed by quantitation (lung/animal weight index) of lung tumor burden.
Genomic and pathway analysis
The HOXA1-induced transcription analysis was conducted using RNAs extracted from WM115 cells transduced with either control or HOXA1, followed by hybridization of labeled cDNA onto Affymetrix (Santa Clara, CA) GeneChips (Human Genome U133Plus2.0) by the Baylor College of Medicine Genome Profiling Core Facility (GEO# GSE37136). Data processing was carried out as described previously (18). Two-sided homoscedastic t-tests (using log-transformed data) and fold changes were used to determine differentially expressed genes (for p<0.01 genes, FDR estimated at 5%, using Storey method (35). Functional Annotation Clustering (http://david.abcc.ncifcrf.gov/) and the Ingenuity Pathways Analysis program were used to further analyze the cellular functions and pathways that were significantly regulated in the dataset. In order to define the degree of HOXA1 gene signature manifestation within profiles from an external dataset, we used the previously described “t-score” metric (18). Briefly, the t-score was defined for each external profile as the two-sided t-statistic comparing, within the profile, the average of the HOXA1-induced genes with the average of the HOXA1-repressed genes (genes within external datasets were first centered to standard deviations from the median; where multiple gene probes referred to the same gene, the probe with the highest variation was used).
TGFβ assay
The TGFβ reporter assay was conducted using the p3TPLux reporter (Addgene #11767; (17)). WM115-HOXA1 and control cells were seeded at 2×105 cells per well in triplicate in 6 well plates 24 hours before transfection with the p3TPLux reporter (1ug per well) and control reporter (Renilla, 20 ng per well). Following 24hrs of incubation, cells were treated for 24 hours with TGFβ (20 ng/ml, R&D Systems, Minneapolis, MN) and were subjected to luciferase analysis (Promega, Madison, WI) following manufacture’s protocol to assess reporter activation as indicated by the firefly/Renilla ratio. P-values were calculated using two-tailed t-test.
Real-time qPCR
For analyses of gene expression, total RNA was isolated from cultured cells either expressing HOXA1 or vector control. Coding regions were amplified by quantitative real-time PCR on a real-time PCR system, and the comparative cycle threshold method was used to quantify mRNA copy number. RNA expression levels in were normalized to human Actin, and all validated PCR primer sets were purchased from SABiosciences (Qiagen, Valencia, CA). TGFβ/BMP Signaling Pathway PCR Arrays (Qiagen) and analysis were performed according to the manufacture’s recommendations.
Immunoblot
Cells were washed in twice in PBS and lysed using RIPA buffer containing 1 mM PMSF, 1× Protease Inhibitor Cocktail (Sigma, St. Louis, MO) and 1× Phosphatase inhibitor (Calbiochem, Billerica, MA) for separation on 4–12% Bis-Tris gels (Bio-Rad, Hercules, CA). The following antibodies were used for immunoblotting following the manufacturer’s recommendations: MITF (Thermo); GAPDH (Santa Cruz, Santa Cruz, CA); HOXA1 (Sigma); Tubulin (Sigma).
Supplementary Material
Probe sets differentially regulated > 3-fold in WM115 cells expressing HOXA1 (P<0.01) and SkMel30 expression values for the same probe sets.
Real time imaging analysis of WM115 cells expressing GFP.
Real time imaging analysis of WM115 cells expressing HOXA1.
David Functional Annotation Clustering of genes differentially regulated in WM115 cells expressing HOXA1.
Cox proportional hazards analysis of the HOXA1 gene signature.
WM115 cells expressing vector control (Vec) or HOXA1 were assayed for cell proliferation over a short time course extending beyond the 20 hr duration of in vitro invasion assays. Cells were plated in triplicate at 7000 cells per well and fixed and stained at the stated time points. Following crystal violet staining, cells were hand counted.
Molecule network generated using Ingenuity Pathways Analysis. The network is displayed graphically as nodes (genes) and edges (the biological relationships between nodes). Solid lines represent direct interactions and dashed lines represent indirect interactions. Red colors denote genes that were over-expressed >10-fold in the WM115-HOXA1 transcriptome analysis.
Expression validation of select genes by RT-qPCR analysis of cDNA prepared from two WM115 tumors (396, 397) driven by doxycycline (Dox)-induced expression (see Fig. 2C). All values are normalized based on Actin B expression and plotted as fold change compared to off Dox spontaneous tumor control (set as 1.0 for each gene). Values indicate fold change for genes found up-regulated (red) and down-regulated (green) in WM115 transcriptome analysis.
Quantitative RT-PCR analysis of MITF mRNA expression in the indicated melanoma cell lines. Expression data normalized to expression in normal human melanocytes. Error bars indicated +/− s.d.
CHL-1 and WM278 cells over-expressing vector control (Vec) or HOXA1 were assayed for in vitro invasion activity using transwell invasion chambers. Images show representative invasion readouts. P-values calculated by t-test.
Heat maps representing expression of HOXA1 signature genes significantly up- and down-regulated (>3-fold) in WM115 cells (2718 gene probes, left panel) mapped onto datasets derived from SkMel30 cells (middle panel) and WM3211 cells (right panel) expressing vector control (Vec) or HOXA1. Signature similarity score (t-score) for comparison between WM115 vs. SkMel30 and WM115 vs. WM3211 is represented by histogram in bottom panel.
ACKNOWLEDGEMENTS
K.L.S. was supported by the American Cancer Society, and this work was supported by departmental seed funds to K.L.S. from Baylor College of Medicine. This work was also supported by grants from the NIH (RO1 CA93947, U01 CA84313) to L.C.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. Epub 2011/02/08. [DOI] [PubMed] [Google Scholar]
- 2.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. Epub 2011/10/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418(6900):823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 4.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. Epub 2002/12/20. [DOI] [PubMed] [Google Scholar]
- 5.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464(7291):999–1005. doi: 10.1038/nature08989. Epub 2010/04/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breslow A. Thickness, cross-sectional areas and depth of invasion in the prognosis of cutaneous melanoma. Ann Surg. 1970;172(5):902–908. doi: 10.1097/00000658-197011000-00017. Epub 1970/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuong W, Cheng LS, Armstrong AW. Melanoma: epidemiology, diagnosis, treatment, and outcomes. Dermatologic clinics. 2012;30(1):113–124. ix. doi: 10.1016/j.det.2011.08.006. Epub 2011/11/29. [DOI] [PubMed] [Google Scholar]
- 8.Gimotty PA, Elder DE, Fraker DL, Botbyl J, Sellers K, Elenitsas R, et al. Identification of high-risk patients among those diagnosed with thin cutaneous melanomas. J Clin Oncol. 2007;25(9):1129–1134. doi: 10.1200/JCO.2006.08.1463. Epub 2007/03/21. [DOI] [PubMed] [Google Scholar]
- 9.Criscione VD, Weinstock MA. Melanoma thickness trends in the United States, 1988–2006. J Invest Dermatol. 2010;130(3):793–797. doi: 10.1038/jid.2009.328. Epub 2009/10/16. [DOI] [PubMed] [Google Scholar]
- 10.Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell. 2011;20(1):92–103. doi: 10.1016/j.ccr.2011.05.025. Epub 2011/07/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400(6743):468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 12.Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125(7):1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 14.Remacle S, Shaw-Jackson C, Matis C, Lampe X, Picard J, Rezsohazy R. Changing homeodomain residues 2 and 3 of Hoxa1 alters its activity in a cell-type and enhancer dependent manner. Nucleic Acids Res. 2002;30(12):2663–2668. doi: 10.1093/nar/gkf372. Epub 2002/06/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svingen T, Tonissen KF. Hox transcription factors and their elusive mammalian gene targets. Heredity. 2006;97(2):88–96. doi: 10.1038/sj.hdy.6800847. [DOI] [PubMed] [Google Scholar]
- 16.Kaminska B, Wesolowska A, Danilkiewicz M. TGF beta signalling and its role in tumour pathogenesis. Acta Biochim Pol. 2005;52(2):329–337. [PubMed] [Google Scholar]
- 17.Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, et al. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71(6):1003–1114. doi: 10.1016/0092-8674(92)90395-s. Epub 1992/12/11. [DOI] [PubMed] [Google Scholar]
- 18.Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG, et al. Insulin-Like Growth Factor-I Activates Gene Transcription Programs Strongly Associated With Poor Breast Cancer Prognosis. J Clin Oncol. 2008;26(25):4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19(4):290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 20.Jeffs AR, Glover AC, Slobbe LJ, Wang L, He S, Hazlett JA, et al. A Gene Expression Signature of Invasive Potential in Metastatic Melanoma Cells. PLoS ONE. 2009;4(12):e8461. doi: 10.1371/journal.pone.0008461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tímár J, Győrffy B, Rásó E. Gene signature of the metastatic potential of cutaneous melanoma: too much for too little? Clin Exp Metastasis. 2010;27(6):371–387. doi: 10.1007/s10585-010-9307-2. [DOI] [PubMed] [Google Scholar]
- 22.Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98(7):472–482. doi: 10.1093/jnci/djj103. Epub 2006/04/06. [DOI] [PubMed] [Google Scholar]
- 23.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. Epub 2010/04/02. [DOI] [PubMed] [Google Scholar]
- 24.Maeda K, Hamada J, Takahashi Y, Tada M, Yamamoto Y, Sugihara T, et al. Altered expressions of HOX genes in human cutaneous malignant melanoma. Int J Cancer. 2005;114(3):436–441. doi: 10.1002/ijc.20706. Epub 2004/11/20. [DOI] [PubMed] [Google Scholar]
- 25.Gavalas A, Trainor P, Ariza-McNaughton L, Krumlauf R. Synergy between Hoxa1 and Hoxb1: the relationship between arch patterning and the generation of cranial neural crest. Development. 2001;128(15):3017–3027. doi: 10.1242/dev.128.15.3017. [DOI] [PubMed] [Google Scholar]
- 26.Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes & Development. 2006;20(24):3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lekmine F, Chang CK, Sethakorn N, Das Gupta TK, Salti GI. Role of microphthalmia transcription factor (Mitf) in melanoma differentiation. Biochem Biophys Res Commun. 2007;354(3):830–835. doi: 10.1016/j.bbrc.2007.01.075. [DOI] [PubMed] [Google Scholar]
- 28.Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell & Melanoma Research. 2010;23(6):746–759. doi: 10.1111/j.1755-148X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- 29.Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, et al. In vivo Switching of Human Melanoma Cells between Proliferative and Invasive States. Cancer Res. 2008;68(3):650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura EK, Suzuki M, Igras V, Du J, Lonning S, Miyachi Y, et al. Key Roles for Transforming Growth Factor β in Melanocyte Stem Cell Maintenance. Stem Cell. 2010;6(2):130–140. doi: 10.1016/j.stem.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinner S, Jordan P, Sharrock K, Bazley L, Collinson L, Marais R, et al. Intravital Imaging Reveals Transient Changes in Pigment Production and Brn2 Expression during Metastatic Melanoma Dissemination. Cancer Res. 2009;69(20):7969–7977. doi: 10.1158/0008-5472.CAN-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammad KS, Javelaud D, Fournier PGJ, Niewolna M, Mckenna CR, Peng XH, et al. TGF-β-RI Kinase Inhibitor SD-208 Reduces the Development and Progression of Melanoma Bone Metastases. Cancer Res. 2011;71(1):175–184. doi: 10.1158/0008-5472.CAN-10-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A. 2011;108(9):3665–3670. doi: 10.1073/pnas.1019736108. Epub 2011/02/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MAS, Massagué J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125(5):929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 35.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Probe sets differentially regulated > 3-fold in WM115 cells expressing HOXA1 (P<0.01) and SkMel30 expression values for the same probe sets.
Real time imaging analysis of WM115 cells expressing GFP.
Real time imaging analysis of WM115 cells expressing HOXA1.
David Functional Annotation Clustering of genes differentially regulated in WM115 cells expressing HOXA1.
Cox proportional hazards analysis of the HOXA1 gene signature.
WM115 cells expressing vector control (Vec) or HOXA1 were assayed for cell proliferation over a short time course extending beyond the 20 hr duration of in vitro invasion assays. Cells were plated in triplicate at 7000 cells per well and fixed and stained at the stated time points. Following crystal violet staining, cells were hand counted.
Molecule network generated using Ingenuity Pathways Analysis. The network is displayed graphically as nodes (genes) and edges (the biological relationships between nodes). Solid lines represent direct interactions and dashed lines represent indirect interactions. Red colors denote genes that were over-expressed >10-fold in the WM115-HOXA1 transcriptome analysis.
Expression validation of select genes by RT-qPCR analysis of cDNA prepared from two WM115 tumors (396, 397) driven by doxycycline (Dox)-induced expression (see Fig. 2C). All values are normalized based on Actin B expression and plotted as fold change compared to off Dox spontaneous tumor control (set as 1.0 for each gene). Values indicate fold change for genes found up-regulated (red) and down-regulated (green) in WM115 transcriptome analysis.
Quantitative RT-PCR analysis of MITF mRNA expression in the indicated melanoma cell lines. Expression data normalized to expression in normal human melanocytes. Error bars indicated +/− s.d.
CHL-1 and WM278 cells over-expressing vector control (Vec) or HOXA1 were assayed for in vitro invasion activity using transwell invasion chambers. Images show representative invasion readouts. P-values calculated by t-test.
Heat maps representing expression of HOXA1 signature genes significantly up- and down-regulated (>3-fold) in WM115 cells (2718 gene probes, left panel) mapped onto datasets derived from SkMel30 cells (middle panel) and WM3211 cells (right panel) expressing vector control (Vec) or HOXA1. Signature similarity score (t-score) for comparison between WM115 vs. SkMel30 and WM115 vs. WM3211 is represented by histogram in bottom panel.