Abstract
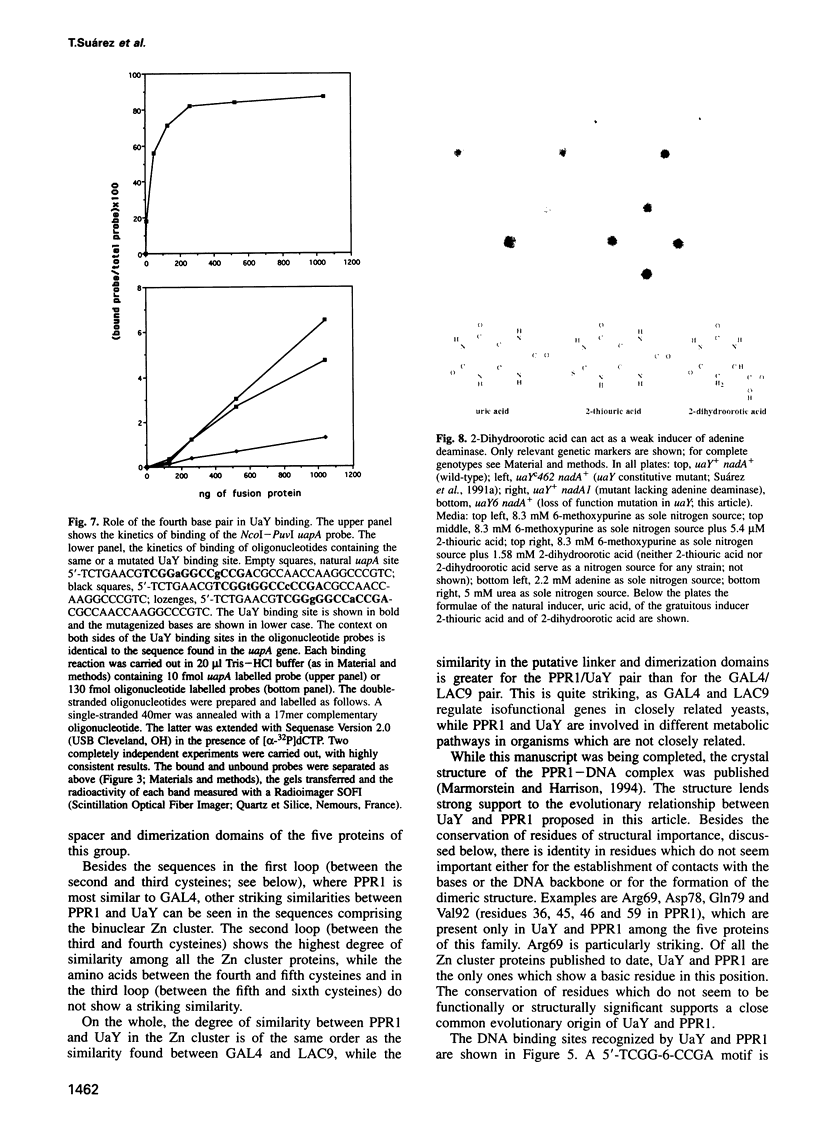
The uaY gene codes for a transcriptional activator mediating the induction of a number of unlinked genes involved in purine utilization in Aspergillus nidulans. Here we present the complete genomic and cDNA nucleotide sequence of this gene. The gene contains two introns. The derived polypeptide of 1060 residues contains a typical zinc binuclear cluster domain and shows a number of similarities with the PPR1 regulatory gene of Saccharomyces cerevisiae. These similarities are most striking in the putative linker and dimerization regions following the zinc cluster. Gel-shift and DNase I footprinting experiments have been carried out for three genes subject to UaY-mediated induction. The binding sequence is 5'-TCGG-6X-CCGA, which is identical to the proposed PPR1 binding sites. Nevertheless, the identity of the base immediately 3' of the 5'-TCGG sequence clearly affects the affinity of the site. The site upstream of the uapA gene has been shown to be active in vivo. Binding to this site has been analysed by a number of interference techniques. There is an interesting chemical similarity between the co-inducer of the purine utilization pathway (uric acid) and that of the genes of the pyrimidine biosynthetic pathway (dihydroorotic acid) and we show that dihydroorotic acid can act as a poor inducer of at least one activity under UaY control. These striking similarities, together with the unique pattern of regulation of pyrimidine biosynthesis in S. cerevisiae, suggest that PPR1 evolved through recruitment into the pyrimidine biosynthetic pathway of an ancestral gene related to uaY.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrianopoulos A., Hynes M. J. Sequence and functional analysis of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol Cell Biol. 1990 Jun;10(6):3194–3203. doi: 10.1128/mcb.10.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramayo R., Timberlake W. E. Sequence and molecular structure of the Aspergillus nidulans yA (laccase I) gene. Nucleic Acids Res. 1990 Jun 11;18(11):3415–3415. doi: 10.1093/nar/18.11.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arst H. N., Jr, Cove D. J. Nitrogen metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1973 Nov 2;126(2):111–141. doi: 10.1007/BF00330988. [DOI] [PubMed] [Google Scholar]
- Arst H. N., Jr, Scazzocchio C. Initiator constitutive mutation with an 'up-promoter' effect in Aspergillus nidulans. Nature. 1975 Mar 6;254(5495):31–34. doi: 10.1038/254031a0. [DOI] [PubMed] [Google Scholar]
- Bai Y. L., Kohlhaw G. B. Manipulation of the 'zinc cluster' region of transcriptional activator LEU3 by site-directed mutagenesis. Nucleic Acids Res. 1991 Nov 11;19(21):5991–5997. doi: 10.1093/nar/19.21.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballance D. J. Sequences important for gene expression in filamentous fungi. Yeast. 1986 Dec;2(4):229–236. doi: 10.1002/yea.320020404. [DOI] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. F. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckholz R. G., Cooper T. G. Oxalurate induction of multiple URA3 transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 1983 Nov;3(11):1889–1897. doi: 10.1128/mcb.3.11.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Strauss J., Scazzocchio C., Lang B. F. nirA, the pathway-specific regulatory gene of nitrate assimilation in Aspergillus nidulans, encodes a putative GAL4-type zinc finger protein and contains four introns in highly conserved regions. Mol Cell Biol. 1991 Nov;11(11):5746–5755. doi: 10.1128/mcb.11.11.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M., Kakidani H., Leatherwood J., Mostashari F., Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989 Oct 5;209(3):423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- Cooper T. G., Chisholm V. T., Cho H. J., Yoo H. S. Allantoin transport in Saccharomyces cerevisiae is regulated by two induction systems. J Bacteriol. 1987 Oct;169(10):4660–4667. doi: 10.1128/jb.169.10.4660-4667.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubero B., Scazzocchio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994 Jan 15;13(2):407–415. doi: 10.1002/j.1460-2075.1994.tb06275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington A. J., Scazzocchio C. Evidence for an alternative pathway of xanthine oxidation in Aspergillus nidulans. Biochim Biophys Acta. 1968 Sep 24;166(2):569–571. doi: 10.1016/0005-2787(68)90244-x. [DOI] [PubMed] [Google Scholar]
- Darlington A. J., Scazzocchio C., Pateman J. A. Biochemical and genetical studies of purine breakdown in Aspergillus. Nature. 1965 May 8;206(984):599–600. doi: 10.1038/206599a0. [DOI] [PubMed] [Google Scholar]
- Darlington A. J., Scazzocchio C. Use of analogues and the substrate-sensitivity of mutants in analysis of purine uptake and breakdown in Aspergillus nidulans. J Bacteriol. 1967 Mar;93(3):937–940. doi: 10.1128/jb.93.3.937-940.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallinas G., Scazzocchio C. A gene coding for the uric acid-xanthine permease of Aspergillus nidulans: inactivational cloning, characterization, and sequence of a cis-acting mutation. Genetics. 1989 Jun;122(2):341–350. doi: 10.1093/genetics/122.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Laskey R. A. Nuclear targeting sequences--a consensus? Trends Biochem Sci. 1991 Dec;16(12):478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Dowzer C. E., Kelly J. M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991 Nov;11(11):5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyereisen R., Koener J. F., Farnsworth D. E., Nebert D. W. Isolation and sequence of cDNA encoding a cytochrome P-450 from an insecticide-resistant strain of the house fly, Musca domestica. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1465–1469. doi: 10.1073/pnas.86.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidel S., Doonan J. H., Morris N. R. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a gamma-actin. Gene. 1988 Oct 30;70(2):283–293. doi: 10.1016/0378-1119(88)90200-4. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Glatigny A., Scazzocchio C. Cloning and molecular characterization of hxA, the gene coding for the xanthine dehydrogenase (purine hydroxylase I) of Aspergillus nidulans. J Biol Chem. 1995 Feb 24;270(8):3534–3550. doi: 10.1074/jbc.270.8.3534. [DOI] [PubMed] [Google Scholar]
- Gorfinkiel L., Diallinas G., Scazzocchio C. Sequence and regulation of the uapA gene encoding a uric acid-xanthine permease in the fungus Aspergillus nidulans. J Biol Chem. 1993 Nov 5;268(31):23376–23381. [PubMed] [Google Scholar]
- Halvorsen Y. D., Nandabalan K., Dickson R. C. Identification of base and backbone contacts used for DNA sequence recognition and high-affinity binding by LAC9, a transcription activator containing a C6 zinc finger. Mol Cell Biol. 1991 Apr;11(4):1777–1784. doi: 10.1128/mcb.11.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kammerer B., Guyonvarch A., Hubert J. C. Yeast regulatory gene PPR1. I. Nucleotide sequence, restriction map and codon usage. J Mol Biol. 1984 Dec 5;180(2):239–250. doi: 10.1016/s0022-2836(84)80002-9. [DOI] [PubMed] [Google Scholar]
- Kudla B., Caddick M. X., Langdon T., Martinez-Rossi N. M., Bennett C. F., Sibley S., Davies R. W., Arst H. N., Jr The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990 May;9(5):1355–1364. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulmburg P., Prangé T., Mathieu M., Sequeval D., Scazzocchio C., Felenbok B. Correct intron splicing generates a new type of a putative zinc-binding domain in a transcriptional activator of Aspergillus nidulans. FEBS Lett. 1991 Mar 11;280(1):11–16. doi: 10.1016/0014-5793(91)80193-7. [DOI] [PubMed] [Google Scholar]
- Lacroute F. Regulation of pyrimidine biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1968 Mar;95(3):824–832. doi: 10.1128/jb.95.3.824-832.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockington R. A., Sealy-Lewis H. M., Scazzocchio C., Davies R. W. Cloning and characterization of the ethanol utilization regulon in Aspergillus nidulans. Gene. 1985;33(2):137–149. doi: 10.1016/0378-1119(85)90088-5. [DOI] [PubMed] [Google Scholar]
- Marczak J. E., Brandriss M. C. Analysis of constitutive and noninducible mutations of the PUT3 transcriptional activator. Mol Cell Biol. 1991 May;11(5):2609–2619. doi: 10.1128/mcb.11.5.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczak J. E., Brandriss M. C. Isolation of constitutive mutations affecting the proline utilization pathway in Saccharomyces cerevisiae and molecular analysis of the PUT3 transcriptional activator. Mol Cell Biol. 1989 Nov;9(11):4696–4705. doi: 10.1128/mcb.9.11.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R., Carey M., Ptashne M., Harrison S. C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Marmorstein R., Harrison S. C. Crystal structure of a PPR1-DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster. Genes Dev. 1994 Oct 15;8(20):2504–2512. doi: 10.1101/gad.8.20.2504. [DOI] [PubMed] [Google Scholar]
- Marshall M. A., Timberlake W. E. Aspergillus nidulans wetA activates spore-specific gene expression. Mol Cell Biol. 1991 Jan;11(1):55–62. doi: 10.1128/mcb.11.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
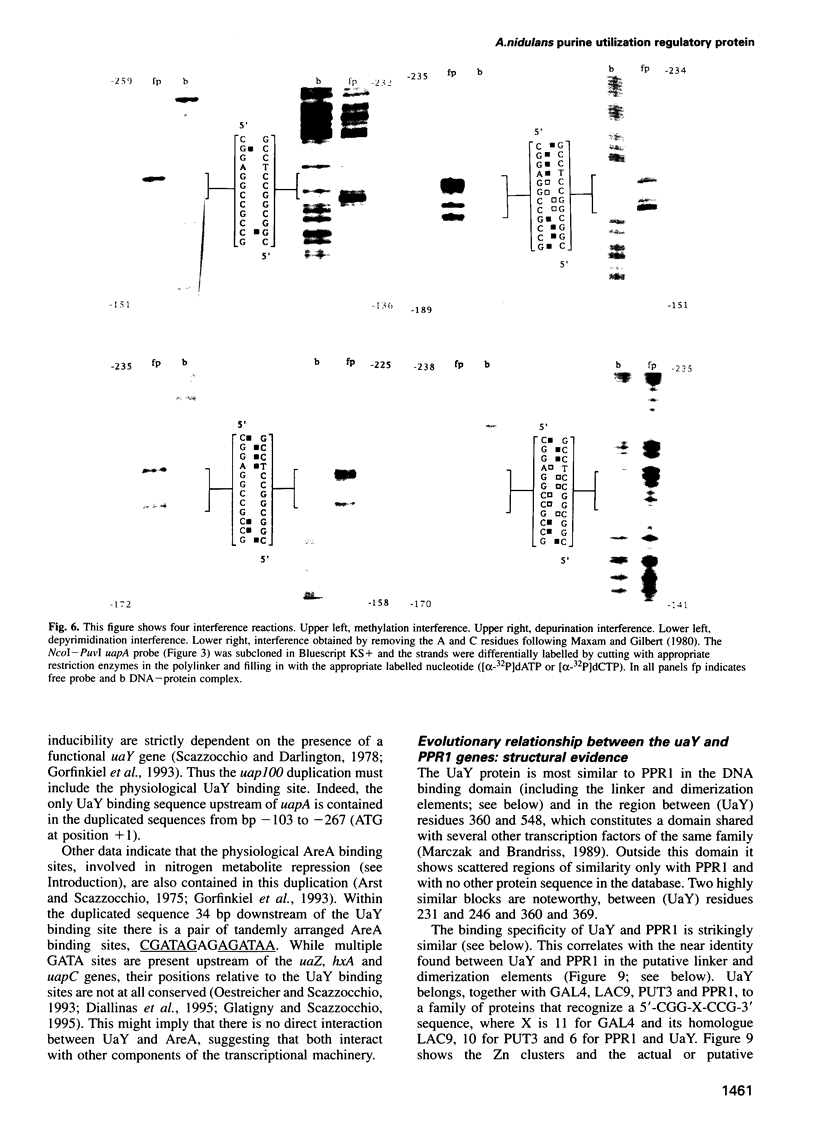
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Myers E. W., Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988 Mar;4(1):11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Nagy M., Lacroute F., Thomas D. Divergent evolution of pyrimidine biosynthesis between anaerobic and aerobic yeasts. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8966–8970. doi: 10.1073/pnas.89.19.8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreicher N., Scazzocchio C. Sequence, regulation, and mutational analysis of the gene encoding urate oxidase in Aspergillus nidulans. J Biol Chem. 1993 Nov 5;268(31):23382–23389. [PubMed] [Google Scholar]
- Oestreicher N., Sealy-Lewis H. M., Scazzocchio C. Characterisation, cloning and integrative properties of the gene encoding urate oxidase in Aspergillus nidulans. Gene. 1993 Oct 15;132(2):185–192. doi: 10.1016/0378-1119(93)90194-8. [DOI] [PubMed] [Google Scholar]
- Osmani S. A., Pu R. T., Morris N. R. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell. 1988 Apr 22;53(2):237–244. doi: 10.1016/0092-8674(88)90385-6. [DOI] [PubMed] [Google Scholar]
- Philippides D., Scazzocchio C. Positive regulation in a eukaryote, a study of the uaY gene of Aspergillus nidulans. II. Identification of the effector binding protein. Mol Gen Genet. 1981;181(1):107–115. doi: 10.1007/BF00339013. [DOI] [PubMed] [Google Scholar]
- Polacco J. C., Gross S. R. The product of the leu-3 cistron as a regulatory element for the production of the leucine biosynthetic enzymes of Neurospora. Genetics. 1973 Jul;74(3):443–459. doi: 10.1093/genetics/74.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R. J., Ptashne M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science. 1993 Aug 13;261(5123):909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- Robbins J., Dilworth S. M., Laskey R. A., Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991 Feb 8;64(3):615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Roy A., Exinger F., Losson R. cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol Cell Biol. 1990 Oct;10(10):5257–5270. doi: 10.1128/mcb.10.10.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C., Arst H. N., Jr The nature of an initiator constitutive mutation in Aspergillus nidulans. Nature. 1978 Jul 13;274(5667):177–179. doi: 10.1038/274177a0. [DOI] [PubMed] [Google Scholar]
- Scazzocchio C., Darlington A. J. The induction and repression of the enzymes of purine breakdown in Aspergillus nidulans. Biochim Biophys Acta. 1968 Sep 24;166(2):557–568. doi: 10.1016/0005-2787(68)90243-8. [DOI] [PubMed] [Google Scholar]
- Scazzocchio C., Holl F. B., Foguelman A. I. The genetic control of molybdoflavoproteins in Aspergillus nidulans. Allopurinol-resistant mutants constitutive for xanthine-dehydrogenase. Eur J Biochem. 1973 Jul 16;36(2):428–445. doi: 10.1111/j.1432-1033.1973.tb02928.x. [DOI] [PubMed] [Google Scholar]
- Scazzocchio C., Sdrin N., Ong G. Positive regulation in a eukaryote, a study of the uaY gene of Aspergillus nidulans: I. Characterization of alleles, dominance and complementation studies, and a fine structure map of the uaY--oxpA cluster. Genetics. 1982 Feb;100(2):185–208. doi: 10.1093/genetics/100.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C. The genetic control of molybdoflavoproteins in Aspergillus nidulans. II. Use of NADH dehydrogenase activity associated with xanthine dehydrogenase to investigate substrate and product inductions. Mol Gen Genet. 1973 Sep 5;125(2):147–155. doi: 10.1007/BF00268868. [DOI] [PubMed] [Google Scholar]
- Sealy-Lewis H. M., Scazzocchio C., Lee S. A mutation defective in the xanthine alternative pathway of Aspergillus nidulans: its use to investigate the specificity of uaY mediated induction. Mol Gen Genet. 1978 Sep 8;164(3):303–308. doi: 10.1007/BF00333161. [DOI] [PubMed] [Google Scholar]
- Sharma K. K., Arst H. N., Jr The product of the regulatory gene of the proline catabolism gene cluster of Aspergillus nidulans is a positive-acting protein. Curr Genet. 1985;9(4):299–304. doi: 10.1007/BF00419959. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Suárez T., Oestreicher N., Kelly J., Ong G., Sankarsingh T., Scazzocchio C. The uaY positive control gene of Aspergillus nidulans: fine structure, isolation of constitutive mutants and reversion patterns. Mol Gen Genet. 1991 Dec;230(3):359–368. doi: 10.1007/BF00280292. [DOI] [PubMed] [Google Scholar]
- Suárez T., Oestreicher N., Peñalva M. A., Scazzocchio C. Molecular cloning of the uaY regulatory gene of Aspergillus nidulans reveals a favoured region for DNA insertions. Mol Gen Genet. 1991 Dec;230(3):369–375. doi: 10.1007/BF00280293. [DOI] [PubMed] [Google Scholar]
- Vogels G. D., Van der Drift C. Degradation of purines and pyrimidines by microorganisms. Bacteriol Rev. 1976 Jun;40(2):403–468. doi: 10.1128/br.40.2.403-468.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]