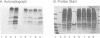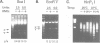Abstract
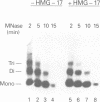
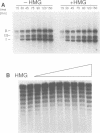
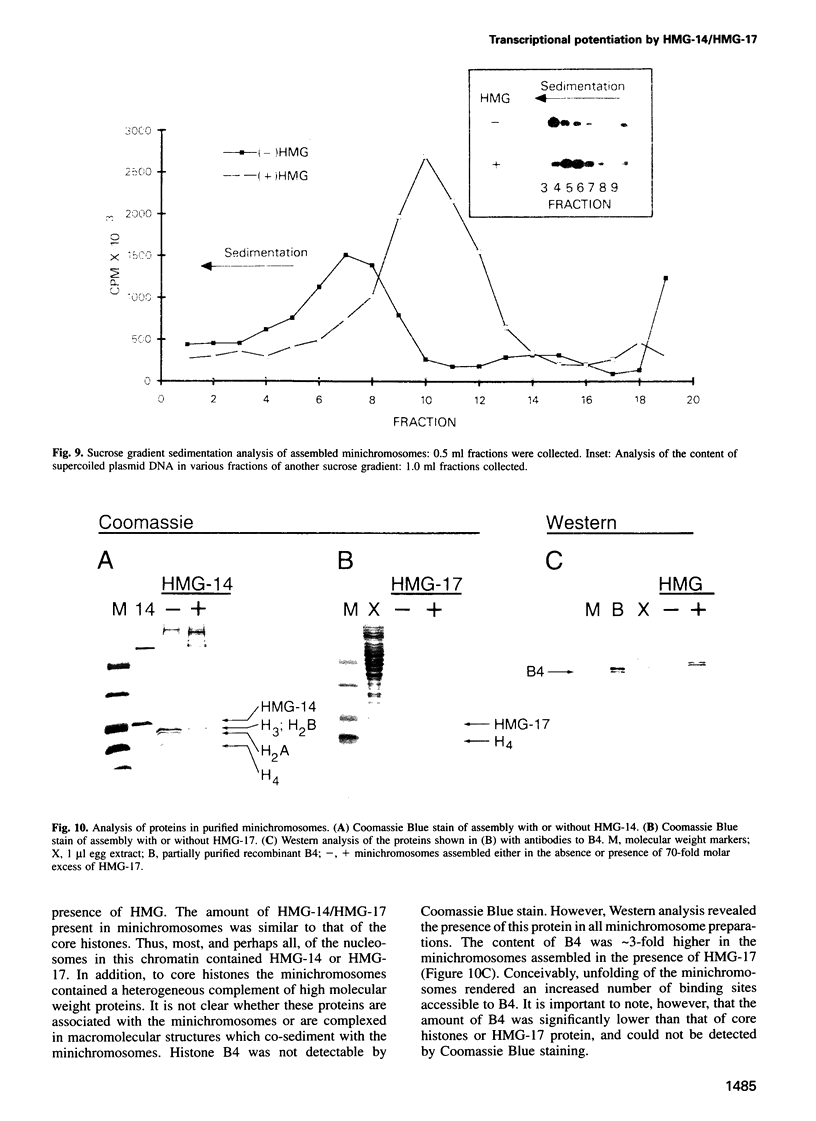
The role of chromosomal proteins HMG-14 and HMG-17 in the generation of transcriptionally active chromatin was studied in a Xenopus laevis egg extract which supports complementary DNA strand synthesis and chromatin assembly. Chromosomal proteins HMG-14/HMG-17 enhanced transcription from a chromatin template carrying a 5S rRNA gene, but not from a DNA template. The transcriptional potential of chromatin was enhanced only when these proteins were incorporated into the template during, but not after, chromatin assembly. HMG-14 and HMG-17 stimulate transcription by increasing the activity, and not the number, of transcribed templates. They unfold the chromatin template without affecting the nucleosomal repeat or decreasing the content of histone B4. We suggest that HMG-14/HMG-17 enhance transcription by inducing an extended conformation in the chromatin fiber, perhaps due to interactions with histone tails in nucleosomes. By disrupting the higher order chromatin structure HMG-14/HMG-17 increase the accessibility of target sequences to components of the transcriptional apparatus.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albright S. C., Wiseman J. M., Lange R. A., Garrard W. T. Subunit structures of different electrophoretic forms of nucleosomes. J Biol Chem. 1980 Apr 25;255(8):3673–3684. [PubMed] [Google Scholar]
- Alfonso P. J., Crippa M. P., Hayes J. J., Bustin M. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J Mol Biol. 1994 Feb 11;236(1):189–198. doi: 10.1006/jmbi.1994.1128. [DOI] [PubMed] [Google Scholar]
- Allan J., Harborne N., Rau D. C., Gould H. Participation of core histone "tails" in the stabilization of the chromatin solenoid. J Cell Biol. 1982 May;93(2):285–297. doi: 10.1083/jcb.93.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G., Clark D. J., Méchali M., Wolffe A. P. Chromatin assembly on replicating DNA in vitro. Nucleic Acids Res. 1990 Oct 11;18(19):5767–5774. doi: 10.1093/nar/18.19.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G., Méchali M., Wolffe A. P. Competition between transcription complex assembly and chromatin assembly on replicating DNA. EMBO J. 1990 Feb;9(2):573–582. doi: 10.1002/j.1460-2075.1990.tb08145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G., Méchali M., Wolffe A. P. Transcription complex disruption caused by a transition in chromatin structure. Mol Cell Biol. 1991 Feb;11(2):655–665. doi: 10.1128/mcb.11.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G., Wolffe A. P. Nuclear assembly, structure, and function: the use of Xenopus in vitro systems. Exp Cell Res. 1993 Mar;205(1):1–15. doi: 10.1006/excr.1993.1051. [DOI] [PubMed] [Google Scholar]
- Almouzni G., Wolffe A. P. Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev. 1993 Oct;7(10):2033–2047. doi: 10.1101/gad.7.10.2033. [DOI] [PubMed] [Google Scholar]
- Barratt M. J., Hazzalin C. A., Zhelev N., Mahadevan L. C. A mitogen- and anisomycin-stimulated kinase phosphorylates HMG-14 in its basic amino-terminal domain in vivo and on isolated mononucleosomes. EMBO J. 1994 Oct 3;13(19):4524–4535. doi: 10.1002/j.1460-2075.1994.tb06774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Brotherton T. W., Ginder G. D. Preferential in vitro binding of high mobility group proteins 14 and 17 to nucleosomes containing active and DNase I sensitive single-copy genes. Biochemistry. 1986 Jun 3;25(11):3447–3454. doi: 10.1021/bi00359a053. [DOI] [PubMed] [Google Scholar]
- Brotherton T. W., Reneker J., Ginder G. D. Binding of HMG 17 to mononucleosomes of the avian beta-globin gene cluster in erythroid and non-erythroid cells. Nucleic Acids Res. 1990 Apr 25;18(8):2011–2016. doi: 10.1093/nar/18.8.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M., Becerra P. S., Crippa M. P., Lehn D. A., Pash J. M., Shiloach J. Recombinant human chromosomal proteins HMG-14 and HMG-17. Nucleic Acids Res. 1991 Jun 11;19(11):3115–3121. doi: 10.1093/nar/19.11.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M., Lehn D. A., Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990 Jul 30;1049(3):231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- Crippa M. P., Alfonso P. J., Bustin M. Nucleosome core binding region of chromosomal protein HMG-17 acts as an independent functional domain. J Mol Biol. 1992 Nov 20;228(2):442–449. doi: 10.1016/0022-2836(92)90833-6. [DOI] [PubMed] [Google Scholar]
- Crippa M. P., Trieschmann L., Alfonso P. J., Wolffe A. P., Bustin M. Deposition of chromosomal protein HMG-17 during replication affects the nucleosomal ladder and transcriptional potential of nascent chromatin. EMBO J. 1993 Oct;12(10):3855–3864. doi: 10.1002/j.1460-2075.1993.tb06064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov S., Dasso M. C., Wolffe A. P. Remodeling sperm chromatin in Xenopus laevis egg extracts: the role of core histone phosphorylation and linker histone B4 in chromatin assembly. J Cell Biol. 1994 Aug;126(3):591–601. doi: 10.1083/jcb.126.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H. F., Rimsky S., Batson S. C., Bustin M., Hansen U. Stimulation of RNA polymerase II elongation by chromosomal protein HMG-14. Science. 1994 Aug 5;265(5173):796–799. doi: 10.1126/science.8047885. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Reconstitution of short-spaced chromatin from the histone octamer and either HMG-14,17 or histone H1. J Mol Biol. 1993 Apr 5;230(3):824–836. doi: 10.1006/jmbi.1993.1204. [DOI] [PubMed] [Google Scholar]
- Druckmann S., Mendelson E., Landsman D., Bustin M. Immunofractionation of DNA sequences associated with HMG-17 in chromatin. Exp Cell Res. 1986 Oct;166(2):486–496. doi: 10.1016/0014-4827(86)90493-3. [DOI] [PubMed] [Google Scholar]
- Du W., Thanos D., Maniatis T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell. 1993 Sep 10;74(5):887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- Einck L., Bustin M. The intracellular distribution and function of the high mobility group chromosomal proteins. Exp Cell Res. 1985 Feb;156(2):295–310. doi: 10.1016/0014-4827(85)90539-7. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espel E., Bernués J., Guasch M. D., Querol E., Plana M., Itarte E. Phosphorylation of high-mobility-group protein 14 by two specific kinases modifies its interaction with histone oligomers in free solution. Biochim Biophys Acta. 1987 Aug 25;909(3):190–200. doi: 10.1016/0167-4781(87)90077-7. [DOI] [PubMed] [Google Scholar]
- Ferrari S., Harley V. R., Pontiggia A., Goodfellow P. N., Lovell-Badge R., Bianchi M. E. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992 Dec;11(12):4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ramirez M., Dong F., Ausio J. Role of the histone "tails" in the folding of oligonucleosomes depleted of histone H1. J Biol Chem. 1992 Sep 25;267(27):19587–19595. [PubMed] [Google Scholar]
- Godde J. S., Widom J. Chromatin structure of Schizosaccharomyces pombe. A nucleosome repeat length that is shorter than the chromatosomal DNA length. J Mol Biol. 1992 Aug 20;226(4):1009–1025. doi: 10.1016/0022-2836(92)91049-u. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J., Bloomer L. S. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982 Apr;28(4):781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- Graziano V., Ramakrishnan V. Interaction of HMG14 with chromatin. J Mol Biol. 1990 Aug 20;214(4):897–910. doi: 10.1016/0022-2836(90)90344-L. [DOI] [PubMed] [Google Scholar]
- Hansen J. C., Wolffe A. P. A role for histones H2A/H2B in chromatin folding and transcriptional repression. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2339–2343. doi: 10.1073/pnas.91.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. C., Wolffe A. P. Influence of chromatin folding on transcription initiation and elongation by RNA polymerase III. Biochemistry. 1992 Sep 1;31(34):7977–7988. doi: 10.1021/bi00149a032. [DOI] [PubMed] [Google Scholar]
- Kamakaka R. T., Bulger M., Kadonaga J. T. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993 Sep;7(9):1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990 Jan 26;60(2):235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landsman D., Bustin M. Assessment of the transcriptional activation potential of the HMG chromosomal proteins. Mol Cell Biol. 1991 Sep;11(9):4483–4489. doi: 10.1128/mcb.11.9.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991 Oct 11;254(5029):238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik N., Smulson M., Bustin M. Enrichment of acetylated histones in polynucleosomes containing high mobility group protein 17 revealed by immunoaffinity chromatography. J Biol Chem. 1984 Jan 25;259(2):699–702. [PubMed] [Google Scholar]
- Ohsumi K., Katagiri C., Kishimoto T. Chromosome condensation in Xenopus mitotic extracts without histone H1. Science. 1993 Dec 24;262(5142):2033–2035. doi: 10.1126/science.8266099. [DOI] [PubMed] [Google Scholar]
- Pash J. M., Alfonso P. J., Bustin M. Aberrant expression of high mobility group chromosomal protein 14 affects cellular differentiation. J Biol Chem. 1993 Jun 25;268(18):13632–13638. [PubMed] [Google Scholar]
- Paton A. E., Wilkinson-Singley E., Olins D. E. Nonhistone nuclear high mobility group proteins 14 and 17 stabilize nucleosome core particles. J Biol Chem. 1983 Nov 10;258(21):13221–13229. [PubMed] [Google Scholar]
- Paull T. T., Haykinson M. J., Johnson R. C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993 Aug;7(8):1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- Peterson R. C., Doering J. L., Brown D. D. Characterization of two xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980 May;20(1):131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- Postnikov Y. V., Shick V. V., Belyavsky A. V., Khrapko K. R., Brodolin K. L., Nikolskaya T. A., Mirzabekov A. D. Distribution of high mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5 on transcribed and non-transcribed regions of chicken erythrocyte chromatin. Nucleic Acids Res. 1991 Feb 25;19(4):717–725. doi: 10.1093/nar/19.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring D., Cole R. D. Chemical cross-linking of H1 histone to the nucleosomal histones. J Biol Chem. 1979 Nov 25;254(22):11688–11695. [PubMed] [Google Scholar]
- Sandeen G., Wood W. I., Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980 Sep 11;8(17):3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter H., Bode J. The binding sites for large and small high-mobility-group (HMG) proteins. Studies on HMG-nucleosome interactions in vitro. Eur J Biochem. 1982 Oct;127(2):429–436. doi: 10.1111/j.1432-1033.1982.tb06890.x. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Jackson I. J., Brown D. D. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984 Jun;37(2):645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- Smith R. C., Dworkin-Rastl E., Dworkin M. B. Expression of a histone H1-like protein is restricted to early Xenopus development. Genes Dev. 1988 Oct;2(10):1284–1295. doi: 10.1101/gad.2.10.1284. [DOI] [PubMed] [Google Scholar]
- Spaulding S. W., Fucile N. W., Bofinger D. P., Sheflin L. G. Cyclic adenosine 3',5'-monophosphate-dependent phosphorylation of HMG 14 inhibits its interactions with nucleosomes. Mol Endocrinol. 1991 Jan;5(1):42–50. doi: 10.1210/mend-5-1-42. [DOI] [PubMed] [Google Scholar]
- Svaren J., Chalkley R. The structure and assembly of active chromatin. Trends Genet. 1990 Feb;6(2):52–56. doi: 10.1016/0168-9525(90)90074-g. [DOI] [PubMed] [Google Scholar]
- Tahourdin C. S., Neihart N. K., Isenberg I., Bustin M. Immunochemical detection of chromosomal protein HMG-17 in chromatin subunits. Biochemistry. 1981 Feb 17;20(4):910–915. doi: 10.1021/bi00507a040. [DOI] [PubMed] [Google Scholar]
- Thanos D., Maniatis T. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell. 1992 Nov 27;71(5):777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- Tremethick D. J., Drew H. R. High mobility group proteins 14 and 17 can space nucleosomes in vitro. J Biol Chem. 1993 May 25;268(15):11389–11393. [PubMed] [Google Scholar]
- Tremethick D. J. High mobility group proteins 14 and 17 can space nucleosomal particles deficient in histones H2A and H2B creating a template that is transcriptionally active. J Biol Chem. 1994 Nov 11;269(45):28436–28442. [PubMed] [Google Scholar]
- Weisbrod S., Weintraub H. Isolation of a subclass of nuclear proteins responsible for conferring a DNase I-sensitive structure on globin chromatin. Proc Natl Acad Sci U S A. 1979 Feb;76(2):630–634. doi: 10.1073/pnas.76.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P. Dominant and specific repression of Xenopus oocyte 5S RNA genes and satellite I DNA by histone H1. EMBO J. 1989 Feb;8(2):527–537. doi: 10.1002/j.1460-2075.1989.tb03407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P., Jordan E., Brown D. D. A bacteriophage RNA polymerase transcribes through a Xenopus 5S RNA gene transcription complex without disrupting it. Cell. 1986 Feb 14;44(3):381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Morse R. H. The transcription complex of the Xenopus somatic 5 S RNA gene. A functional analysis of protein-DNA interactions outside of the internal control region. J Biol Chem. 1990 Mar 15;265(8):4592–4599. [PubMed] [Google Scholar]
- Wolffe A. P. Replication timing and Xenopus 5S RNA gene transcription in vitro. Dev Biol. 1993 May;157(1):224–231. doi: 10.1006/dbio.1993.1126. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Schild C. Chromatin assembly. Methods Cell Biol. 1991;36:541–559. [PubMed] [Google Scholar]
- Wolffe A. P. Transcription fraction TFIIIC can regulate differential Xenopus 5S RNA gene transcription in vitro. EMBO J. 1988 Apr;7(4):1071–1079. doi: 10.1002/j.1460-2075.1988.tb02915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]