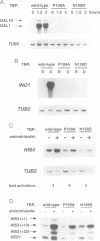
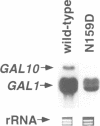
Abstract
The TATA box binding protein (TBP) plays a central and essential role in transcription initiation. At TATA box-containing genes transcribed by RNA polymerase II, TBP binds to the promoter and initiates the assembly of a multiprotein preinitiation complex. Several studies have suggested that binding of TBP to the TATA box is an important regulatory step in transcription initiation in vitro. To determine whether TBP is a target of regulatory factors in vivo, we performed a genetic screen in yeast for TBP mutants defective in activated transcription. One class of TBP mutants identified in this screen comprises inositol auxotrophs that are also defective in using galactose as a carbon source. These phenotypes are due to promoter-specific defects in transcription initiation that are governed by the upstream activating sequence (UAS) and apparently not by the sequence of the TATA element. The finding that these TBP mutants are severely impaired in DNA binding in vitro suggests that transcription initiation at certain genes is regulated at the level of TATA box binding by TBP in vivo.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt K. T., Styles C., Fink G. R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987 Aug 21;237(4817):874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- Bajwa W., Torchia T. E., Hopper J. E. Yeast regulatory gene GAL3: carbon regulation; UASGal elements in common with GAL1, GAL2, GAL7, GAL10, GAL80, and MEL1; encoded protein strikingly similar to yeast and Escherichia coli galactokinases. Mol Cell Biol. 1988 Aug;8(8):3439–3447. doi: 10.1128/mcb.8.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Boyer T. G., Berk A. J. Functional interaction of adenovirus E1A with holo-TFIID. Genes Dev. 1993 Sep;7(9):1810–1823. doi: 10.1101/gad.7.9.1810. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Guarente L., Sharp P. A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989 Feb 24;56(4):549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- Carlson M., Laurent B. C. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994 Jun;6(3):396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Chasman D. I., Kornberg R. D. GAL4 protein: purification, association with GAL80 protein, and conserved domain structure. Mol Cell Biol. 1990 Jun;10(6):2916–2923. doi: 10.1128/mcb.10.6.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Struhl K. Saturation mutagenesis of a yeast his3 "TATA element": genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy B., Green M. R. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature. 1993 Dec 9;366(6455):531–536. doi: 10.1038/366531a0. [DOI] [PubMed] [Google Scholar]
- Conaway J. W., Conaway R. C. Initiation of eukaryotic messenger RNA synthesis. J Biol Chem. 1991 Sep 25;266(27):17721–17724. [PubMed] [Google Scholar]
- Côté J., Quinn J., Workman J. L., Peterson C. L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994 Jul 1;265(5168):53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Devlin C., Tice-Baldwin K., Shore D., Arndt K. T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991 Jul;11(7):3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrin L. K., Mann R. K., Kayne P. S., Grunstein M. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell. 1991 Jun 14;65(6):1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991 Aug 9;66(3):563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Arndt K. M., Ricupero S. L., Rooney J. W., Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992 Jul;6(7):1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Dollard C., Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989 Sep 22;58(6):1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- Flanagan P. M., Kelleher R. J., 3rd, Tschochner H., Sayre M. H., Kornberg R. D. Simple derivation of TFIID-dependent RNA polymerase II transcription systems from Schizosaccharomyces pombe and other organisms, and factors required for transcriptional activation. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7659–7663. doi: 10.1073/pnas.89.16.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., Hoey T., Thut C. J., Admon A., Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993 Nov 5;75(3):519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury P. A., Struhl K. Functional distinctions between yeast TATA elements. Mol Cell Biol. 1989 Dec;9(12):5298–5304. doi: 10.1128/mcb.9.12.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T., Weinzierl R. O., Gill G., Chen J. L., Dynlacht B. D., Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993 Jan 29;72(2):247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Carey M. F., Kakidani H., Roeder R. G. Mechanism of action of a yeast activator: direct effect of GAL4 derivatives on mammalian TFIID-promoter interactions. Cell. 1988 Aug 26;54(5):665–669. doi: 10.1016/s0092-8674(88)80011-4. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Hai T., Lin Y. S., Green M. R., Roeder R. G. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell. 1988 Sep 23;54(7):1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- Horikoshi N., Maguire K., Kralli A., Maldonado E., Reinberg D., Weinmann R. Direct interaction between adenovirus E1A protein and the TATA box binding transcription factor IID. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5124–5128. doi: 10.1073/pnas.88.12.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshizaki D. K., Hill J. E., Henry S. A. The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J Biol Chem. 1990 Mar 15;265(8):4736–4745. [PubMed] [Google Scholar]
- Imbalzano A. N., Zaret K. S., Kingston R. E. Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J Biol Chem. 1994 Mar 18;269(11):8280–8286. [PubMed] [Google Scholar]
- Ingles C. J., Shales M., Cress W. D., Triezenberg S. J., Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991 Jun 13;351(6327):588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- Kim J. L., Nikolov D. B., Burley S. K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993 Oct 7;365(6446):520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kim T. K., Hashimoto S., Kelleher R. J., 3rd, Flanagan P. M., Kornberg R. D., Horikoshi M., Roeder R. G. Effects of activation-defective TBP mutations on transcription initiation in yeast. Nature. 1994 May 19;369(6477):252–255. doi: 10.1038/369252a0. [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994 May 20;77(4):599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger J. H., Hahn S., Sigler P. B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993 Oct 7;365(6446):512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Klein C., Struhl K. Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science. 1994 Oct 14;266(5183):280–282. doi: 10.1126/science.7939664. [DOI] [PubMed] [Google Scholar]
- Koleske A. J., Buratowski S., Nonet M., Young R. A. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell. 1992 May 29;69(5):883–894. doi: 10.1016/0092-8674(92)90298-q. [DOI] [PubMed] [Google Scholar]
- Koleske A. J., Young R. A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994 Mar 31;368(6470):466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- Lee W. S., Kao C. C., Bryant G. O., Liu X., Berk A. J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991 Oct 18;67(2):365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- Leuther K. K., Johnston S. A. Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science. 1992 May 29;256(5061):1333–1335. doi: 10.1126/science.1598579. [DOI] [PubMed] [Google Scholar]
- Lieberman P. M., Berk A. J. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev. 1991 Dec;5(12B):2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Green M. R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991 Mar 8;64(5):971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Ha I., Maldonado E., Reinberg D., Green M. R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991 Oct 10;353(6344):569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- Mahadevan S., Struhl K. Tc, an unusual promoter element required for constitutive transcription of the yeast HIS3 gene. Mol Cell Biol. 1990 Sep;10(9):4447–4455. doi: 10.1128/mcb.10.9.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloff D. M., McGraw P., Henry S. A. The INO2 gene of Saccharomyces cerevisiae encodes a helix-loop-helix protein that is required for activation of phospholipid synthesis. Nucleic Acids Res. 1992 Jun 25;20(12):3253–3253. doi: 10.1093/nar/20.12.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthun M. R., Jaehning J. A. A transcriptionally active form of GAL4 is phosphorylated and associated with GAL80. Mol Cell Biol. 1992 Nov;12(11):4981–4987. doi: 10.1128/mcb.12.11.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Kruger W., Herskowitz I. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell. 1991 Mar 22;64(6):1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- Poon D., Weil P. A. Immunopurification of yeast TATA-binding protein and associated factors. Presence of transcription factor IIIB transcriptional activity. J Biol Chem. 1993 Jul 25;268(21):15325–15328. [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Diverse transcriptional functions of the multisubunit eukaryotic TFIID complex. J Biol Chem. 1992 Jan 15;267(2):679–682. [PubMed] [Google Scholar]
- Reddy P., Hahn S. Dominant negative mutations in yeast TFIID define a bipartite DNA-binding region. Cell. 1991 Apr 19;65(2):349–357. doi: 10.1016/0092-8674(91)90168-x. [DOI] [PubMed] [Google Scholar]
- Roberts S. G., Ha I., Maldonado E., Reinberg D., Green M. R. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993 Jun 24;363(6431):741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- Roeder R. G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991 Nov;16(11):402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- Rose M., Botstein D. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Sayre M. H., Tschochner H., Kornberg R. D. Reconstitution of transcription with five purified initiation factors and RNA polymerase II from Saccharomyces cerevisiae. J Biol Chem. 1992 Nov 15;267(32):23376–23382. [PubMed] [Google Scholar]
- Scafe C., Chao D., Lopes J., Hirsch J. P., Henry S., Young R. A. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990 Oct 4;347(6292):491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. Photofootprinting in vivo detects transcription-dependent changes in yeast TATA boxes. Nature. 1987 Jan 8;325(7000):173–177. doi: 10.1038/325173a0. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer K. F., Ingles C. J., Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990 Jun 28;345(6278):783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundseth R., Hansen U. Activation of RNA polymerase II transcription by the specific DNA-binding protein LSF. Increased rate of binding of the basal promoter factor TFIIB. J Biol Chem. 1992 Apr 15;267(11):7845–7855. [PubMed] [Google Scholar]
- Swanson M. S., Malone E. A., Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991 Jun;11(6):3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese N., Pugh B. F., Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991 Dec;5(12A):2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- Tschopp J. F., Emr S. D., Field C., Schekman R. GAL2 codes for a membrane-bound subunit of the galactose permease in Saccharomyces cerevisiae. J Bacteriol. 1986 Apr;166(1):313–318. doi: 10.1128/jb.166.1.313-318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usheva A., Maldonado E., Goldring A., Lu H., Houbavi C., Reinberg D., Aloni Y. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992 May 29;69(5):871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- Verrijzer C. P., Yokomori K., Chen J. L., Tjian R. Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science. 1994 May 13;264(5161):933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- Winston F., Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992 Nov;8(11):387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S. L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995 Jan;11(1):53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Horikoshi M., Wang J., Hasegawa S., Weil P. A., Roeder R. G. A bipartite DNA binding domain composed of direct repeats in the TATA box binding factor TFIID. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2844–2848. doi: 10.1073/pnas.89.7.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L., Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Lieberman P. M., Boyer T. G., Berk A. J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992 Oct;6(10):1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]
- Zhou Y. H., Zhang X. P., Ebright R. H. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 1991 Nov 11;19(21):6052–6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]