Abstract
Purpose
To evaluate the feasibility of a retrospective binning technique for 4-dimensional magnetic resonance imaging (4D-MRI) using body area (BA) as a respiratory surrogate.
Methods and Materials
Seven patients with hepatocellular carcinoma (4 of 7) or liver metastases (3 of 7) were enrolled in an institutional review board-approved prospective study. All patients were simulated with both computed tomography (CT) and MRI to acquire 3-dimensinal and 4D images for treatment planning. Multiple-slice multiple-phase cine-MR images were acquired in the axial plane for 4D-MRI reconstruction. Image acquisition time per slice was set to 10-15 seconds. Single-slice 2-dimensinal cine-MR images were also acquired across the center of the tumor in orthogonal planes. Tumor motion trajectories from 4D-MRI, cine-MRI, and 4D-CT were analyzed in the superior–inferior (SI), anterior–posterior (AP), and medial–lateral (ML) directions, respectively. Their correlation coefficients (CC) and differences in tumor motion amplitude were determined. Tumor-to-liver contrast-to-noise ratio (CNR) was measured and compared between 4D-CT, 4D-MRI, and conventional T2-weighted fast spin echo MRI.
Results
The means (±standard deviations) of CC comparing 4D-MRI with cine-MRI were 0.97 ± 0.03, 0.97 ± 0.02, and 0.99 ± 0.04 in SI, AP, and ML directions, respectively. The mean differences were 0.61 ± 0.17 mm, 0.32 ± 0.17 mm, and 0.14 ± 0.06 mm in SI, AP, and ML directions, respectively. The means of CC comparing 4D-MRI and 4D-CT were 0.95 ± 0.02, 0.94 ± 0.02, and 0.96 ± 0.02 in SI, AP, and ML directions, respectively. The mean differences were 0.74 ± 0.02 mm, 0.33 ± 0.13 mm, and 0.18 ± 0.07 mm in SI, AP, and ML directions, respectively. The mean tumor-to-tissue CNRs were 2.94 ± 1.51, 19.44 ± 14.63, and 39.47 ± 20.81 in 4D-CT, 4D-MRI, and T2-weighted MRI, respectively.
Conclusions
The preliminary evaluation of our 4D-MRI technique results in oncologic patients demonstrates its potential usefulness to accurately measure tumor respiratory motion with improved tumor CNR compared with 4D-CT.
Introduction
Respiration-induced organ motion poses significance challenges to treatment with radiation. Suboptimal management of organ motion can lead to degradation of the delivered dose distribution to the irradiated volume, with underdosage of target tissues and overdosage of surrounding normal tissues (1). Four-dimensional computed tomography (4D-CT) has been widely used in clinical radiation therapy to acquire patient-specific respiratory motion data (2-7). However, 4D-CT involves a substantial image radiation dose to nontarget tissues, owing to multiple volumetric data acquisitions at the same body location, and has relatively poor soft-tissue contrast, potentially resulting in inaccurate tumor delineation (8-10). As an alternative, the emerging 4D magnetic resonance imaging (4D-MRI) technique is able to image respiratory associated organ motion with improved soft-tissue contrast and zero nontarget ionizing radiation.
Several 4D-MRI techniques have been recently reported (11-14). Hu et al (11) introduced a prospective amplitude-based 4D-MRI technique using a respiratory triggering system. Although this technique improved the tumor-to-tissue contrast-to-noise ratio (CNR) by acquiring T2-weighted 4D-MRI image datasets, the acquisition efficiency was affected by inconsistent breathing patterns between the preparation and data acquisition stages. Tryggestad et al (12) demonstrated a retrospective 4D-MRI technique for dynamic MRI, which used a pneumatic device strapped around the subjects’ upper abdomen to acquire the respiratory signals. Two-pass approaches were used in retrospective sorting to acquire “de-blurred 4D-MRI”; however, unavoidable artifacts in the first-pass “average 4D-MRI” limited the robustness of second-pass “de-blurred 4D-MRI.”
We have previously demonstrated a retrospective 4D-MRI technique using body area (BA) as an internal respiratory surrogate. The technical feasibility of this approach has been tested and validated on a motion phantom, a digital phantom, and healthy human subjects (13).
The purpose of this study was to assess the effectiveness of our 4D-MRI technique in real cancer patients. We evaluated the accuracy of tumor motion measurement using 2-dimensional (2D) single-slice cine MR in 3 orthogonal directions (14) and 4D-CT and the improvement in tumor-to-tissue CNR of the 4D-MRI technique.
Methods and Materials
Patients and imaging study
Seven patients (3 male, 4 female, mean age 65.0 years) who had hepatocellular carcinoma (n = 4) or liver metastases (n = 3) were enrolled in this institutional review board-approved prospective study. Note that patient 7 had 2 tumors. Table 1 summarizes the patients’ clinical characteristics. All patients underwent CT and MRI simulations on the same day for treatment planning. The CT scans were performed on a 4-slice CT scanner (Lightspeed; GE Healthcare, Milwaukee, WI) equipped with the Real-time Position Management (RPM) system (Varian Medical Systems, Palo Alto, CA) and Advantage 4D software (GE Healthcare). The CT simulation included a free-breathing helical 3-dimensional (3D)-CT scan, a breath-hold helical 3D-CT scan with contrast, and a cine 4-dimensional (4D)-CT scan. All patients were positioned head first supine in an immobilization device during the scans. The 4D-CT scan was acquired in the cine mode, in which the cine duration per slice was set to the patient’s breathing period plus 1 second, and the cine time (image acquisition time per phase per slice) was set to one-tenth of the breathing period. The following imaging parameters were used: 120 kV, 290 mA, 2.5-mm slice thickness, gantry rotation of 0.5 seconds per cycle, reconstruction matrix of 512 × 512, field of view of 450-500 mm.
Table 1.
Summary of patient characteristics and measurements
| Patient | Age (y) | Sex | Cancer site | Scanner type | 4D-MRI vs cine-MRI & 4D-MRI vs 4D-CT
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC
|
D (mm)
|
CNR
|
|||||||||||
| SI | AP | ML | SI | AP | ML | CT | 4D-MRI | T2-weighted MRI | |||||
| 1 | 48 | M | Liver metastases | 1.5 T | 0.98 & 0.96 | 0.99 & 0.96 | 0.99 & 0.95 | 0.57 & 0.55 | 0.27 & 0.30 | 0.12 & 0.10 | 4.97 | 16.17 | 51.15 |
| 2 | 66 | F | HCC | 1.5 T | 0.93 & 0.95 | 0.95 & 0.95 | 0.99 & 0.96 | 0.65 & 0.70 | 0.30 & 0.32 | 0.05 & 0.08 | 3.33 | 32.36 | 72.70 |
| 3 | 69 | F | HCC | 3.0 T | 0.96 & 0.97 | 0.98 & 0.96 | 0.99 & 0.94 | 1.50 & 1.00 | 0.55 & 0.60 | 0.20 & 0.30 | 0.8 | 5.00 | 13.43 |
| 4 | 70 | M | HCC | 3.0 T | 0.99 & 0.96 | 0.95 & 0.94 | 0.98 & 0.93 | 0.40 & 0.50 | 0.15 & 0.10 | 0.15 & 0.20 | 1.61 | 4.57 | 22.33 |
| 5 | 76 | F | Liver metastases | 1.5 T | 0.99 & 0.95 | 0.98 & 0.95 | 0.99 & 0.94 | 0.80 & 1.05 | 0.35 & 0.30 | 0.20 & 0.22 | 4.33 | 18.33 | 38.81 |
| 6 | 61 | M | HCC | 1.5 T | 0.97 & 0.92 | 0.97 & 0.94 | 0.98 & 0.98 | 0.61 & 0.88 | 0.32 & 0.30 | 0.13 & 0.15 | 3.50 | 14.65 | 25.05 |
| 7a | 65 | F | Liver metastases | 1.5 T | 0.96 & 0.93 | 0.96 & 0.90 | 0.98 & 0.98 | 0.55 & 0.60 | 0.34 & 0.32 | 0.15 & 0.18 | 2.01 | 45.00 | 52.80 |
| 7b | 65 | F | Liver metastases | 1.5 T | 0.97 & 0.95 | 0.96 & 0.92 | 0.98 & 0.96 | 0.57 & 0.60 | 0.35 & 0.36 | 0.20 & 0.22 | 2.50 | 40.20 | 50.56 |
| Mean | 65 | - | - | - | 0.97 & 0.95 | 0.97 & 0.94 | 0.99 & 0.96 | 0.61 & 0.74 | 0.32 & 0.33 | 0.14 & 0.18 | 2.94 | 19.44 | 39.47 |
| SD | 9.7 | - | - | - | 0.03 & 0.02 | 0.02 & 0.02 | 0.01 & 0.02 | 0.17 & 0.20 | 0.17 & 0.13 | 0.06 & 0.07 | 1.51 | 14.63 | 20.81 |
Abbreviations: 4D-MRI = 4-dimensional magnetic resonance imaging; AP = anterior–posterior; CC = coefficient correlation; CNR = contrast-to-noise ratio; CT = computed tomography; D = difference in motion amplitude; F = female; HCC = hepatocellular carcinoma; M = male; ML = medial–lateral; MRI = magnetic resonance imaging; SD = standard deviation; SI = superior–inferior.
All MR scans were performed on a 1.5-T (Signa; GE Healthcare) or a 3.0-T MR system (MAGNETOM Trio; Siemens Healthcare, Erlangen, Germany). An immobilization device was used when it was possible to fit the device with the patient inside the MR scanner bore. The MR simulation included a 4D-MRI scan, multiple free-breathing cine MR scans, and a breath-hold T2-weighted MR scan using a respiratory-triggered fast relaxation fast spin echo sequence.
The unsorted 4D-MRI images were acquired in the axial plane using a fast steady-state acquisition imaging technique (labeled as FIESTA by GE and TrueFISP by Siemens), including multiple slices to cover a volume of interest. Scan time per axial slice was set to approximately 2 to 3 times the patient’s breathing period. Magnetic resonance images were interpolated to 256 × 256 before further analysis.
Single-slice 2D cine MR images were acquired across the center of the tumor in 3 orthogonal planes (axial, sagittal, and coronal) for 30 seconds using the same sequence as the 4D-MRI scan. The orthogonal slices were acquired in separate but contiguous acquisitions. The MRI parameters were optimized to achieve fast image acquisition (>3 frames per second) while maintaining adequate spatial resolution: repetition time (TR)/echo time (TE): 3.005 ms/1.128 ms; field of view (FOV): 300-480 × 360-480 mm; flip angle: 50°; slice thickness: 5 mm; bandwidth: 976.562 Hz per pixel; acquisition matrix: 192 × 128.
The imaging parameters for T2-weighted MRI were as follows: TR/TE: 12,857.1 ms/64.196 ms; FOV: 350 × 227 mm; flip angle: 50°; slice thickness: 3 mm; bandwidth: 390.625 Hz per pixel; acquisition matrix: 256 × 192.
4D-MRI using BA as respiratory surrogate
Our 4D-MRI technique was achieved by using a fast 2D MR sequence to acquire axial images continuously throughout the breathing cycle, and then retrospectively sorting the MR images according to respiratory phase. The 4D-MRI technique and the use of BA as respiratory surrogate have been described in detail previously (13) and will only be briefly described here.
To determine the breathing signal, each MR image was first processed to determine the body contour. Individual breathing curves were then generated at each axial location by plotting the BA as a function of image acquisition time. The complete breathing signal was obtained by plotting individual breathing curves continuously, followed by removing the low-frequency component (called base BA) of the signal, which is caused by anatomic changes. To reconstruct the 4D-MRI, respiratory peaks of the complete breathing signal were determined using an automatic search algorithm, followed by a manual correction method to remove erroneous peak detections. Peaks were assigned to phase 50%, and the rest of the phases were then linearly interpolated. In cases in which a phase was missing, the nearest phase was instead used in the 4D-MRI reconstruction. In addition, the first 3 images in the image series at each axial location were excluded for reconstruction, because this is the approximate time required for MRI signals to reach a steady state (ie, consistent signal). MATLAB (The MathWorks, Natick, MA) was used to perform the image processing and data analysis.
Comparison of 4D tumor trajectories
Single-slice cine MR and 4D-CT were used to validate the motion accuracy of 4D-MRI. Notably, there were differences between the cine MR used here and the other one used for 4D-MRI. The single-slice cine MR imaged only 1 slice across the center of the tumor in 3 orthogonal directions (superior–inferior [SI], anterior–posterior [AP], and medial–lateral [ML]), whereas the multiple-slice cine MR used for 4D-MRI was acquired in the transverse plane. Coronal and sagittal 4D-MRI images were reconstructed. For the orthogonal single-slice cine MR, tumor motion trajectories in both the vertical and horizontal directions were extracted from the images using an automatic tracking algorithm based on cross-correlation. Tumor motion tracking using a cross-correlation algorithm is an established method and has been used in previous studies (15, 16). For quantitative comparison with tumor motion trajectories determined from the 4D-MRI, each of the 30-second-long tumor motion trajectories of the single-slice cine MRI was processed to generate average tumor motion trajectories that contain only 1 breathing cycle. Tumor motion trajectories were also extracted from 4D-CT and 4D-MRI using the motion tracking method in all 3 directions (SI, AP, and ML). Tumor trajectories in the SI and AP directions were extracted from sagittal MR images (for both 4D and cine), and tumor trajectories in the ML direction were extracted from coronal MR images (for both 4D and cine). Although coronal MR images also provided SI tumor motion information, we did not use that information because of concern about errors caused by through-plane (ie, AP) tumor motion. For sagittal MR images, the through-plane (ie, ML) tumor motion is less concerning because tumor motion in the ML direction is typically very small. The tracking process was repeated 5 times for each 4D-CT and 4D-MRI, to remove human variation in selecting the base template that is used for tracking. Although our intention was to use the same template in all tracking processes, the base template was manually drawn and could be slightly different between different tracking processes. Using repeated measurements to determine the average tumor motion trajectories can eliminate this human variation.
Tumor motion trajectories determined from cine MR, 4D-CT, and 4D-MRI were then compared. Because the cine MR images were acquired near real-time relative to the tumor motion, they were used as the reference for evaluating the tumor motion measurement of 4D-MRI. Specifically, the correlation coefficient (CC) and the difference in motion amplitude between the motion trajectories derived by the 3 methods were calculated for each patient. The difference in motion amplitude, D, was calculated as the mean difference in amplitude of the 10 respiratory phases among cine MR, 4D-CT, and 4D-MRI.
Tumor-to-liver CNR was measured on and compared among CT, 4D-MRI, and T2-weighted MRI for each patient. To determine CNR, a circular region of interest was placed on the tumor, the liver, and the background of the image to measure the mean intensity (tumor and liver) or standard deviation (background). The CNR was then calculated as the difference between average signal intensity of each tumor and average signal intensity of the surrounding liver parenchyma, divided by the standard deviation of signal intensity within the tumor.
Results
Results of 4D-MRI for a representative patient are shown in Figures 1-4. Figure 1 illustrates 1 subject’s breathing signals and respiratory phases, extracted directly from MR images using the BA method. Figure 2 shows example images of 4D-CT (Fig. 2a), 4D-MRI (Fig. 2b), and cine-MRI (Fig. 2c) of the same subject in the same sagittal plane. Respiratory motion of the liver was clearly seen in all image sets.
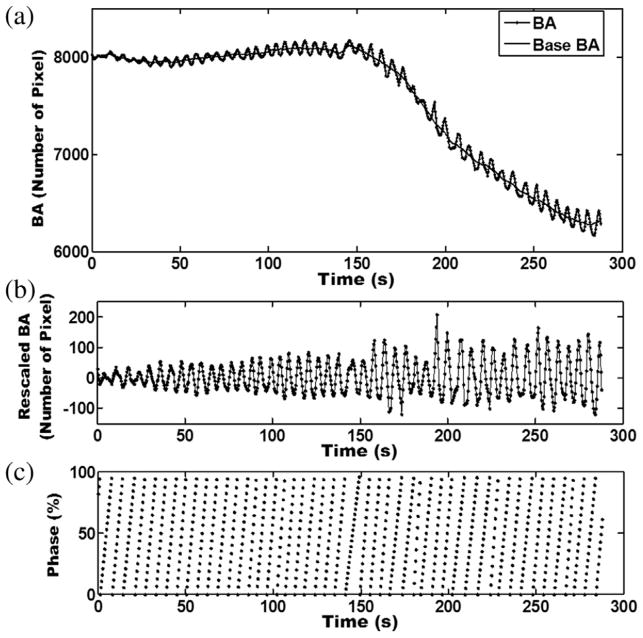
Fig. 1.
Examples of (a) body area (BA) and base BA curves, (b) rescaled BA curve by subtracting BA from base BA, and (c) respiratory phase measured using the BA method. Four-dimensional magnetic resonance images were acquired in axial planes; each slice was imaged continuously throughout the breathing cycle. The BA was determined for each image. For each image slice an individual breathing curve was generated by plotting BA as a function of image acquisition time. The BA curve was generated by plotting all individual breathing curves as a function of acquisition time.
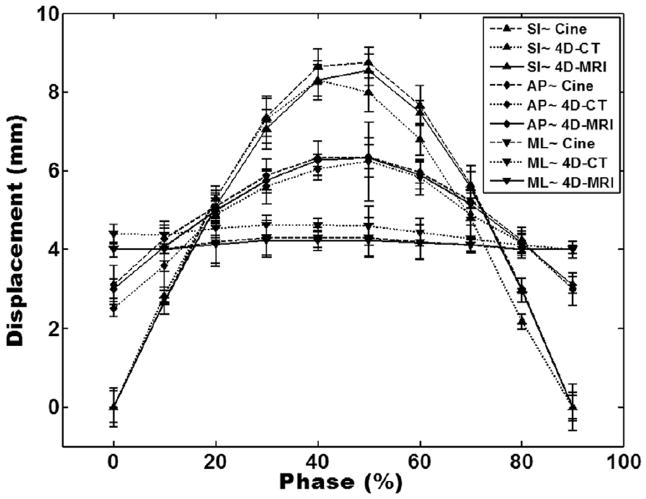
Fig. 4.
Comparison of tumor motion trajectories from cine-MRI (magnetic resonance imaging), 4-dimensional (4D) computed tomography (CT), and 4D-MRI in orthogonal planes (superior–inferior [SI], anterior–posterior [AP], and medial–lateral [ML]). Error bars are standard deviations of multiple measurements in 4D computed tomography and 4D-MRI and are standard deviations of multiple breathing cycles in cine-MRI. Each of the 30-second-long tumor motion trajectories of the single-slice cine MRI was processed to generate average tumor motion trajectories that contain only 1 breathing cycle for motion comparison.
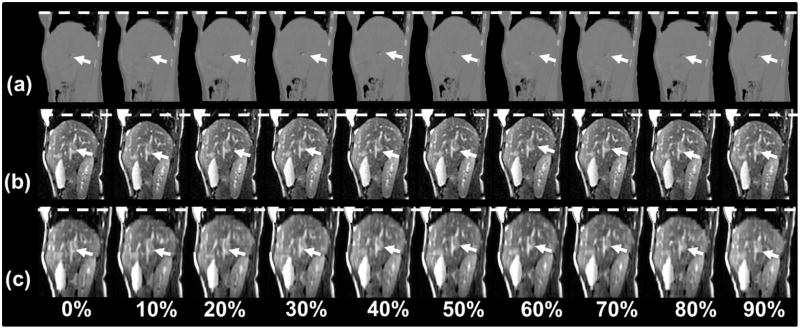
Fig. 2.
Representative sagittal 4-dimensional (4D) computed tomography (a), 4D magnetic resonance imaging (MRI) (b), and cine-MRI (c) for a liver cancer patient. White dashed lines are drawn to facilitate the visualization of the respiratory motion. Arrows indicate the tumor. Although the tumor (indicted by arrows) is nearly invisible in the 4D computed tomography images, it can be readily seen in both the cine-MRI and 4D-MRI images.
Although the tumor (arrows in Fig. 2) was nearly invisible in the 4D-CT images (window/level of 4D-CT images was set to “abdomen” for best possible visibility for soft-tissue contrast), it can be readily seen in both the cine-MRI and 4D-MRI images. In all cases, tumor-to-liver CNRs were lowest for 4D-CT, much higher for 4D-MRI, and still higher for T2-weighted MRI. Mean tumor-to-liver CNRs were 2.94 ± 1.51, 19.44 ± 14.63, and 39.47 ± 20.81 for 4D-CT, 4D-MRI, and T2-weighted MRI, respectively, across the study population. Figure 3 shows the tumor motion trajectories determined from the cine-MRI images in all 3 directions. Note that the tumor motion trajectory in the SI direction was determined from both sagittal and coronal cine MR images. Figure 4 shows the comparison of tumor motion trajectories between cine MR, 4D-CT, and 4D-MRI. Good matching was observed between the cine MR and 4D-MRI: the CC ranged from 0.98 to 0.99, and the mean difference in motion amplitude ranged from 0.12 mm to 0.57 mm, with the largest difference in the SI direction. Good agreement was also found between 4D-CT and 4D-MRI: the CC ranged from 0.93 to 0.98, and the mean difference in motion amplitude ranged from 0.08 mm to 0.60 mm.
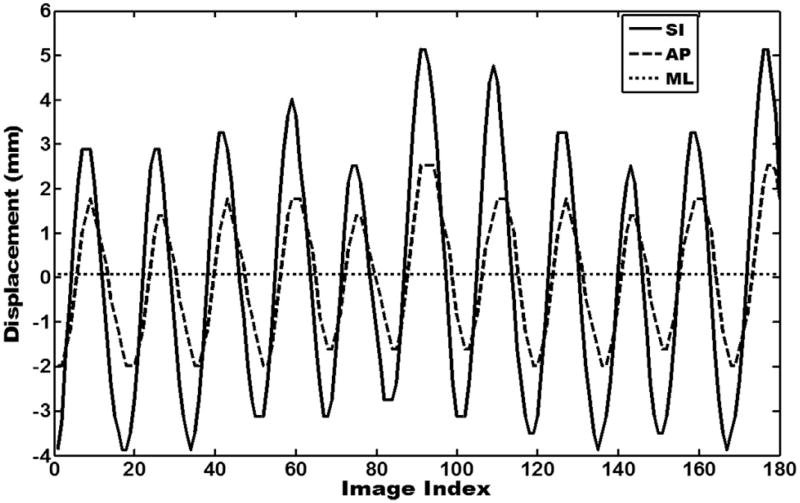
Fig. 3.
Tumor motion trajectories of cine-MR (magnetic resonance) images in orthogonal planes (superior–inferior [SI], anterior–posterior [AP], and medial–lateral [ML]) by tracking the sagittal plane and coronal plane. Tumor trajectories in the superior–inferior and anterior–posterior directions were extracted from sagittal MR images, and tumor trajectories in the medial–lateral direction were extracted from coronal MR images because of concern regarding errors caused by through-plane.
Table 1 summarizes the results of all patients. On average, the tumor motion trajectories matched well between 4D-MRI and cine MR: the means of CC were 0.97 ± 0.03, 0.97 ± 0.02, and 0.99 ± 0.04 in the SI, AP, and ML directions, respectively. The mean differences in tumor motion amplitude were 0.61 ± 0.17 mm, 0.32 ± 0.17 mm, and 0.14 ± 0.06 mm in the SI, AP, and ML directions, respectively. Tumor motion trajectories also matched well between 4D-MRI and 4D-CT: the means of CC were 0.95 ± 0.02, 0.94 ± 0.02, and 0.96 ± 0.02 in the SI, AP, and ML directions, respectively. The mean differences in tumor motion amplitude were 0.74 ± 0.02 mm, 0.33 ± 0.13 mm, and 0.18 ± 0.07 mm in the SI, AP, and ML directions, respectively. The mean tumor-to-liver CNRs were 2.94 ± 1.51, 19.44 ± 14.63, and 39.47 ± 20.81 in CT, 4D-MRI, and T2-weighted MRI, respectively.
Discussion
In this study we reported the initial results of 4D-MRI reconstruction analysis using BA as a surrogate on oncologic patients. Tumor motion trajectories derived from 4D-MRI were comparable to the trajectories derived from cine MR and 4D-CT, indicating that our 4D-MRI technique is potentially capable of accurately measuring tumor respiratory motion. However, because 4D-MRI captures an entire volume of interest, rather than just 3 orthogonal planes as in cine-MRI, it may provide a more robust evaluation when respiratory motion is complex. In addition, 4D-MRI dramatically improved the tumor-to-tissue CNR (a surrogate for lesion conspicuity/visibility) in these patients as compared with the 4D-CT. Notably, however, there was a large degree of intersubject variation in the tumor-to-tissue CNR of the 4D-MRI (range, 4.57-45.00; standard deviation 14.63). The tumor-to-tissue CNR may be affected by multiple factors, including histology and stage of the cancer, patient size, as well as characteristics of the MRI system (eg, scanner type, magnetic field strength [17] and the number of receiver coil elements, T2-weighting of the pulse sequence). Both 1.5-T and 3-T MR scanners were used in this study. Our results demonstrated that tumor-to-tissue CNR was improved in 4D-MRI as compared with 4D-CT in both scanners. In addition, the FIESTA/TrueFISP sequence used for 4D-MRI has a complex T2*/T1 weighting, which provides less optimal tumor contrast relative to conventional T2-weighted MR sequences. It therefore may be of interest to develop a T2-weighted 4D-MRI technique to further improve the tumor-to-tissue CNR and thus tumor conspicuity/visibility (11, 12).
Artifacts were observed in some 4D-MRI images, as shown in an example in Figure 5. Compared with cine MR images (Fig. 5a), the 4D-MRI volume images (Fig. 5b) exhibited “gaps” at multiple locations and at multiple respiratory phases (indicated by white arrows). These artifacts were presumably caused by patient respiratory variations, as commonly observed in 4D-CT images (18). The 4D techniques (both CT and MRI) acquire images at each slice location intermittently, rather than continuously during whole breathing cycles as in the 2D MRI method. Thus, the collection of an image at every slice position in every respiratory phase is not guaranteed, resulting in the “gaps” demonstrated in Figure 5. In addition, the FIESTA/TrueFISP sequences are typically affected by dark phase dispersion bands, which are caused by patient-related local field inhomogeneities and made prominent by the relatively long TR used. Furthermore, ghost artifacts may also appear in image acquisitions using these sequences. If these artifacts appear in the same locations, such as tumor and organs, errors may occur in tumor identification or organ edge detection.
Fig. 5.
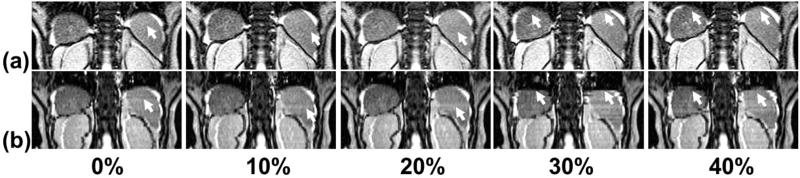
Artifacts (indicated by arrows) in coronal 4-dimensional magnetic resonance imaging (b) as compared with coronal cine-magnetic resonance imaging (a) for patient 2.
Furthermore, inaccurate determination of respiratory phase can also contribute to the artifacts. Our previous study has shown that space-dependent phase shift is a limiting factor of the accuracy of the BA surrogate (19). Space-dependent phase shift affects the BA surrogate given that the respiratory peaks were determined separately for different body locations using the images acquired at the corresponding locations. It is important to understand the causes and characteristics of these artifacts, to minimize their adverse effects in clinical application of 4D-MRI. It is worth noting that some external surrogates, such as respiratory belts, are also practical and inexpensive for 4D-MRI imaging. Although a full evaluation of its pros and cons to their advantages and disadvantages is yet to be performed, we have observed that the placement of the belt around the body frequently causes anatomic changes, introducing uncertainties in fusion with CT and subsequently dose calculation.
This pilot study included a limited number of patients and assessed only patients with intrahepatic cancers. A larger pool of patients is needed in future studies. It should be noted that knowledge and experience gained from 4D-CT, although useful, cannot be simply directly applied to 4D-MRI, owing to the differences in image acquisition (per couch position in 4D-CT, per slice in 4D-MRI), characteristics of image artifacts, and image reconstruction. In addition, errors in tumor motion tracking using the cross-correlation algorithm may occur during tracking due to various reasons, such as choice of template image and interpolative/resampling process. Because validating tumor motion in 4D images with breathing irregularity has been challenging owing to the lack of a standard, we believe that cine MRI, to some extent, is a good reference for evaluating 4D-MRI tumor motion. Cine MRI is near real-time imaging (3-10 frames per second) with high spatial resolution (1-2 mm). It contains tumor motion information with a long time period (30 seconds), meaning it captures some irregularities of the breathing. The average motion curves derived from cine MRI most likely represent the typical breathing pattern of the patients. However, at present, there is no standard method for evaluating tumor motion with irregularities (the same problem exists for 4D-CT imaging). Cine MRI remains one of the best choices for evaluating retrospective 4D images.
Conclusions
Four-dimensional MRI using BA as a respiratory surrogate with axial image acquisition has been successfully demonstrated in patients with intrahepatic cancers. It has been shown that this 4D-MRI technique can accurately image tumor respiratory motion, with improved soft-tissue contrast compared with 4D-CT.
Summary.
This study performed preliminary patient validation of the accuracy of tumor motion measurements and the improvement of tumor-to-tissue contrast-to-noise ratio in the proposed 4D-MRI technique using body area as the respiratory surrogate. Seven patients with liver cancer(s) were enrolled in the IRB-approved study. The results demonstrated that this 4D-MRI technique was a promising technique for more accurately imaging tumor respiratory motion, with improved soft-tissue contrast.
Acknowledgments
This work is partly supported by research grants from the National Institutes of Health (1R21CA165384-01A1) and the Golfers Against Cancer Foundation, as well as by an educational fund from the China Scholarship Council.
Conflict of interest: J.Y. received personal fees from the China Scholarship Council during the conduct of the study. J.C. received grants from the National Institutes of Health and the Golfers Against Cancer Foundation during the conduct of the study; and grants from Phillips, outside the submitted work. M.B. received support from Siemens Healthcare, outside the submitted work. F.-F.Y. received grants from the National Institutes of Health during the conduct of the study.
References
- 1.Lambert J, Suchowerska N, Mckenzie DR, et al. Intrafractional motion during proton beam scanning. Phys Med Biol. 2005;50:4853–4862. doi: 10.1088/0031-9155/50/20/008. [DOI] [PubMed] [Google Scholar]
- 2.Keall P. 4-dimensional computed tomography imaging and treatment planning. Semin Radiat Oncol. 2004;14:81–90. doi: 10.1053/j.semradonc.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Low DA, Nystrom M, Kalinin E, et al. A method for the reconstruction of four-dimensional synchronized CT scans acquired during free breathing. Med Phys. 2003;30:1254–1263. doi: 10.1118/1.1576230. [DOI] [PubMed] [Google Scholar]
- 4.Mageras GS, Pevsner A, Yorke ED, et al. Measurement of lung tumor motion using respiration-correlated CT. Int J Radiat Oncol Biol Phys. 2004;60:933–941. doi: 10.1016/j.ijrobp.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 5.Pan T. Comparison of helical and cine acquisitions for 4D-CT imaging with multislice CT. Med Phys. 2005;32:627–634. doi: 10.1118/1.1855013. [DOI] [PubMed] [Google Scholar]
- 6.Rietzel E, Pan T, Chen GT. Four-dimensional computed tomography: Image formation and clinical protocol. Med Phys. 2005;32:874–889. doi: 10.1118/1.1869852. [DOI] [PubMed] [Google Scholar]
- 7.Ford EC, Mageras GS, Yorke E, et al. Respiration-correlated spiral CT: A method of measuring respiratory induced anatomic motion for radiation treatment planning. Med Phys. 2003;30:88–97. doi: 10.1118/1.1531177. [DOI] [PubMed] [Google Scholar]
- 8.Keall PJ, Starkschall G, Shukla H, et al. Acquiring 4D thoracic CT scans using a multi-slice helical method. Phys Med Biol. 2004;49:2053–2067. doi: 10.1088/0031-9155/49/10/015. [DOI] [PubMed] [Google Scholar]
- 9.Murphy MJ, Balter J, Balter S, et al. The management of imaging dose during image-guided radiotherapy: Report of the AAPM Task Group 75. Med Phys. 2007;34:4041–4063. doi: 10.1118/1.2775667. [DOI] [PubMed] [Google Scholar]
- 10.Koste JRS, Senan S, Kleynen CE, et al. Renal mobility during uncoached quiet respiration: An analysis of 4DCT scans. Int J Radiat Oncol Biol Phys. 2006;64:799–803. doi: 10.1016/j.ijrobp.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y, Caruthers SD, Low DA, et al. Respiratory amplitude guised 4-dimensional magnetic resonance imaging. Int J Radiat Oncol Biol Phys. 2013;86:198–204. doi: 10.1016/j.ijrobp.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tryggestad E, Flammang A, Han-Oh S, et al. Respiration-based sorting of dynamic MRI to derive representative 4D-MRI for radiotherapy planning. Med Phys. 2013;40:51909. doi: 10.1118/1.4800808. [DOI] [PubMed] [Google Scholar]
- 13.Cai J, Chang Z, Wang ZH, et al. Four-dimensional magnetic resonance imaging (4D-MRI) using image-based respiratory surrogate: A feasibility study. Med Phys. 2011;38:63–84. doi: 10.1118/1.3658737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tryggestad E, Flammang A, Hales R, et al. 4D tumor centroid tracking using orthogonal 2D dynamic MRI: Implications for radiotherapy planning. Med Phys. 2013;40:1–12. doi: 10.1118/1.4818656. [DOI] [PubMed] [Google Scholar]
- 15.Leese JA, Novak CS, Clark BB. An automatic technique for obtaining cloud motion from geosynchronous satellite data using cross correlation. J Appl Meteor. 1971;10:118–132. [Google Scholar]
- 16.Briechle K, Hanebeck UD. Template matching using fast normalized cross correlation (Abstr.) Optical Pattern Recognition XII. 2001;4387:95. [Google Scholar]
- 17.Fabel M, Wintersperger BJ, Dietrich O, et al. MRI of respiratory dynamics with 2D steady-state free-precession and 2D gradient echo sequences at 1.5 and 3 Tesla: An observer preference study. ESR. 2009;19:391–399. doi: 10.1007/s00330-008-1148-x. [DOI] [PubMed] [Google Scholar]
- 18.Pan T, Lee TY, Rietzel E, et al. 4D-CT imaging of a volume influenced by respiratory motion on multi-slice CT. Med Phys. 2004;31:333–340. doi: 10.1118/1.1639993. [DOI] [PubMed] [Google Scholar]
- 19.Cai J, Chang Z, O’ Daniel J, et al. Investigation of sliced body volume (SBV) as respiratory surrogate. J Appl Clin Med Phys. 2013;14:3987–3996. doi: 10.1120/jacmp.v14i1.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]