Abstract
Long-term memory formation requires de novo protein synthesis and gene transcription. During contextual long-term memory formation brain-derived neurotrophic factor (BDNF) gene expression changes in conjunction with alterations of DNA methylation in the Bdnf gene. However, little is known about the molecular mechanisms underlying the maintenance and persistence of contextual long-term memory. Here, we examined the transcription of specific Bdnf exons in the hippocampus for long periods after contextual fear conditioning. We found changes in transcription lasting for at least 24 hours after contextual fear conditioning, with some sex-specific effects. In addition, hypomethylation at a CpG site in CpG island 2 located at the end of Bdnf exon III sequence was detected at 0.5 h and maintained for up to 24 h after contextual fear conditioning. The identification of these long-lasting changes in transcription and DNA methylation at the Bdnf gene suggests that BDNF might have a role for storage of contextual long-term memory in the hippocampus.
Keywords: contextual fear memory, DNA methylation, gene expression, hippocampus, Nr4a1, sex differences
Introduction
Long-term memory formation requires de novo protein synthesis and gene transcription (Dudai 2004; Silva & Giese 1994). However, little is known about the molecular mechanisms underlying the maintenance and persistence of memory. Recent studies suggest that cellular development and memory processes have homologous molecular mechanisms (Day & Sweatt 2011). Thus, epigenetic coding, which is important for development, might be critical for memory. A key epigenetic mechanism mediating the dynamic regulation of gene transcription is DNA methylation occurring primarily at CpG dinucleotides in the genome and catalyzed by DNA methyltransferases (DNMTs) (Sweatt 2009; Wu & Zhang 2010).
Recent studies have indicated that DNA methylation regulates processes in the mature nervous system including synaptic plasticity and memory formation in adult rodents (Lubin et al. 2008; Martinowich et al. 2003; Miller & Sweatt 2007; Nelson et al. 2008). In contextual fear conditioning, where a neutral environment is associated with an aversive shock, DNMT inhibitors block memory formation (Lubin et al. 2008; Miller & Sweatt 2007; Monsey et al. 2011). Furthermore, contextual fear conditioning leads to hypermethylation and transcriptional silencing of the memory suppressor gene PP1 and to rapid demethylation and transcriptional activation of the synaptic plasticity gene reelin (Miller & Sweatt 2007). Additionally, after contextual fear conditioning, DNA methylation regulates exon-specific transcription of the Bdnf gene (Lubin et al. 2008).
Brain-derived neurotrophic factor (BDNF) regulates not only the survival and differentiation of neurons during development, but also synaptic plasticity and memory in the adult brain (Cunha et al. 2010; Tyler et al. 2002; Yamada et al. 2002). BDNF plays an important role in hippocampus-dependent memory including contextual fear conditioning and spatial memory formation (Gorski et al. 2003; Lee et al. 2004; Liu et al. 2004). Furthermore, recent studies have shown that there is a novel protein synthesis- and BDNF-dependent phase in the hippocampus for the persistence of long-term memory storage (Bekinschtein et al. 2007, 2008b). This demonstrates that both BDNF and protein synthesis are required not only for the formation of memories soon after training, but also for memory persistence days after training (Bekinschtein et al. 2008a). Additionally, reactivation of long-term memory can induce BDNF transcription in the hippocampus (Kirtley and Thomas, 2010), although such transcription may not be essential for the maintenance of long-term memory (Lee et al., 2004).
The Bdnf gene is highly complex, consisting of nine 5’ noncoding exons each linked to individual promoter regions, and a 3’ coding exon (IX), which codes for the BDNF precursor-protein amino acid sequence (Aid et al. 2007). For example, Bdnf promoter IV regulates Bdnf gene transcription and is correlated with DNA methylation state at CpG sites within promoter IV during memory formation or stress in rats (Lubin et al. 2008; Roth et al. 2009, 2011).
In this study we examined whether exon-specific Bdnf gene transcription is induced for long periods in the hippocampus after contextual fear conditioning and, if so, whether transcription is recapitulated with reactivation of the long-term memory. We also tested whether long-lasting changes in Bdnf mRNA expression are linked to altered DNA methylation. All experiments were performed in both male and female mice, because some molecular mechanisms in memory formation are known to be sex-specific (Mizuno & Giese 2010). We identified significant up-regulation in transcription of the Bdnf gene that persisted for at least 24 hours after contextual fear conditioning. These changes correlated with altered DNA methylation at a number of specific CpG sites in CpG islands associated with the Bdnf gene.
Material and methods
Animals
C57BL/6J mice (10 weeks old) were obtained from Charles River Laboratories. Mice were housed of 4 to 5 mice group per cage under a 12:12 light/dark cycle with foods and water ad libitum. All animal procedures were conducted under the United Kingdom Animals (Scientific Procedures) Act, 1986.
Contextual fear conditioning
All animals used for experiments were handled 5 min/day for three days before conditioning. All experiments were performed during light cycle. Each mouse was placed into the conditioning chamber (Med Associates Inc., St. Albans, VT, USA) in a soundproof box. After a 148 s introductory period, the mouse received a 2 s foot shock (0.75 mA), followed by two foot shocks with a 30 s interval between each. After an additional 30 s, the mouse was returned to the home cage. For contextual re-exposure, the mice were brought back to the conditioning chamber for 10 min.
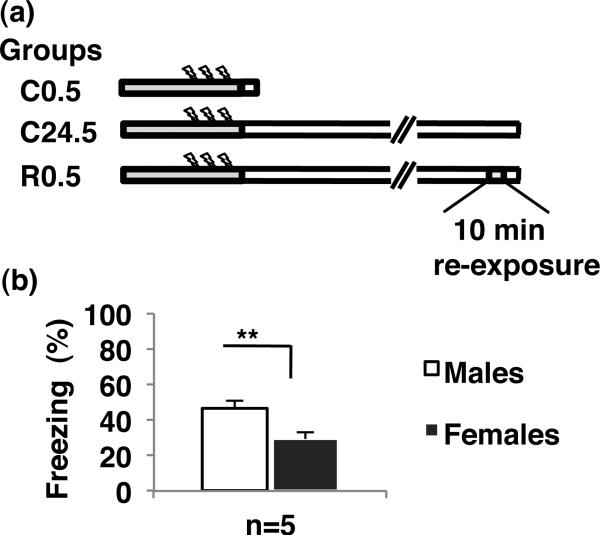
The mice were divided into four groups (males n=5, females n=5 for each group): (1) naïve; (2) C0.5, consolidation group, killed 0.5 h after training; (3) C24.5, consolidation group, killed 24.5 h after training; and (4) R0.5, memory reactivation group (reconsolidation), re-exposure to the training context for 10 min 24 h after training and killed 0.5 h after re-exposure. 10 min re-exposure to the context has been reported to cause the reactivation of memory, but not extinction after 3 shocks conditioning (Susuki et al. 2004). A reconsolidation group was included as previous work has highlighted transcriptional differences between consolidation and reconsolidation (von Hertzen & Giese, 2005).
The behavior of the mice (R0.5 group) during re-exposure was videotaped and freezing was scored every 5 s for 2 s during the initial 5 min if no movements other than respiratory movements were detected.
Quantitative real-time PCR
Hippocampi were fresh-frozen on dry ice and stored at -80°C. Total RNA and genomic DNA were simultaneously purified using the AllPrep DNA/RNA/Protein mini kit (Qiagen). Total RNA (2 μg) was reverse transcribed using superscript II reverse transcriptase (Invitrogen). The obtained cDNA was diluted 1: 10 and stored at -20°C. The cDNA for each sample was checked for genomic DNA contamination using a PCR that distinguishes between genomic DNA and cDNA for the hypoxanthine phosphoribosyltransferase (HPRT) gene. The primer sequences used were HPRT forward 5’-GCTGGTGAAAAGGACCTCT-3’ and HPRT reverse 5’-CACAGGACTAGAACACCTGC-3’, and PCR amplification conditions were 93°C for 2 min, 35 amplification cycles (93°C for 30 s, 58°C for 45 s, 72°C for 1 min), and 72°C for 10 min. The specific Bdnf primers used for quantitative real-time PCR (qRCR) are listed in Supplementary Table 1.
qPCR was performed in triplicate on the DNA Engine (Bio-Rad) using SYBR Green (PrimerDesign Ltd., Hants, UK) and analyzed using Opticon Monitor analysis software 3.1 (Bio-Rad). Reactions were performed in 96-well ABgene PCR Plates (Thermo Scientific, Hampshire, UK) capped with cap strips. The cycle conditions were 95°C for 10 min, followed by 50 amplification cycles (95°C for 15s, 60°C for 1 min). Validation experiments were performed to demonstrate that the efficiency of target and reference amplicons was approximately equal throughout a range of cDNA dilutions after the optimization of primer concentrations. For each sample, the mean threshold cycle (Ct) was determined for target and reference genes. If the absolute value of the slope of log input amount against differences between target and reference gene (dCt) was less than 0.1, then these primer combinations were used for qPCR. The comparative Ct method was used to normalize target mRNA amount to reference (HPRT or glyceraldehyde 3-phosphate dehydrogenase; GAPDH) mRNA, and compare the test animals to the naïve group.
DNA methylation analysis
Genomic DNA (500 ng) was treated with sodium bisulfite using the EZ-96 DNA Methylation Kit (Zymo Research, CA, USA) following the manufacturers' standard protocol. DNA methylation assays were designed using the online Sequenom EpiDesigner software (www.epidesigner.org). The oligo sequences and the location of the amplicons across which DNA methylation was assessed in this study are given in Supplementary Table 2. Bisulfite-PCR amplification was conducted using Hot Star Taq DNA polymerase (Qiagen, UK) and cycling conditions of 45 cycles with an annealing temperature of 56°C for all amplicons. Subsequent to bisulfite-PCR amplification, DNA methylation analysis was conducted using the Sequenom EpiTYPER system (Sequenom Inc, CA, USA). All bisulfite-PCR reactions were performed in duplicate. Positive controls, including both artificially methylated and artificially unmethylated DNA samples were included in all experimental procedures to ensure unambiguous PCR amplification of bisulfite-treated samples. Data generated from the EpiTYPER software were treated with stringent quality control analysis where CpG units with low calling rates and individuals with a high number of missing CpG units were removed.
Data analysis
Behavior data were analyzed with t-test and qPCR data were analyzed with one-way analysis of variance (ANOVA), Krushkal-Wallis one-way ANOVA on ranks, or two way ANOVA, followed by Student-Newman-Keuls post hoc tests, if significant differences were found. Data for DNA methylation were analyzed with standard planned t-tests.
Results
Contextual fear conditioning
We trained mice in contextual fear conditioning in three different conditions and investigated long-term changes in Bdnf gene expression and DNA methylation in the hippocampus (Fig. 1a). The contextual fear conditioning protocol induced freezing 24 h after training in both sexes (males 46.3 ± 4.4%, females 29.4 ± 3.7%), with males freezing significantly more than females (t8 = 2.97; p=0.018) (Fig. 1b).
Figure 1.
Experimental design and contextual memory after conditioning. (a) Groups to investigate changes in hippocampal mRNA expression and DNA methylation after contextual fear conditioning. The gray boxes indicate exposure to training context, the arrows indicate the foot shocks, and the white boxes show the time until the mice were killed. Groups were studied at: C0.5, conditioned and killed 0.5 h after conditioning; C24.5, conditioned and killed 24.5 h after conditioning; R0.5, conditioning, reexposed 24 h after conditioning and killed 0.5 h after reexposure. (b) The freezing score during reexposure, 24 h after the first exposure is shown. Contextual fear conditioning induced freezing to context 24 h after conditioning (R0.5)(males; n=5, females; n=5). Data are means ± SEM; **p<0.01.
Nr4a1 mRNA expression in the hippocampus is regulated after contextual fear conditioning and re-exposure to the context
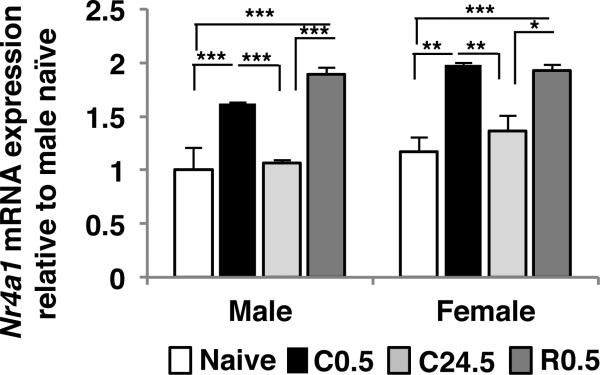
Nr4a1 (also known as nur77 or NGFI-B) was previously identified as context-shock-specific immediate early gene (von Hertzen & Giese 2005). We tested Nr4a1 mRNA expression using qPCR in four groups of mice (naïve, C0.5, C24.5, R0.5) (Fig. 2). Because two-way ANOVA showed that data were not normally distributed, pairwise t-tests or Mann-Whitney Rank Sum test were performed between sexes and showed a significant sex difference in Nr4a1 mRNA expression. Nr4a1 mRNA expression was higher in females than in males 0.5 h after contextual fear conditioning (t8=15.04, p<0.001), but there were no significant sex differences for the other groups (naïve; t8=0.98, p=0.36; C24.5; T8=36 p=0.10; R0.5; t8=0.35 p=0.74). Therefore, the data were analyzed separately for each sex. Female data were analyzed by one-way ANOVA which showed significant differences after contextual fear conditioning (F3,16=14.14, p<0.001). Post hoc analysis showed that Nr4a1 mRNA was significantly up-regulated 0.5 h after contextual fear conditioning and 0.5 h after memory reactivation compared to naïve and 24.5 h after contextual fear conditioning (naïve vs C0.5: p<0.001, naïve vs R0.5: p<0.001, naïve vs C24.5: p=0.15, C24.5 vs C0.5: p=0.004, C24.5 vs R0.5: p=0.003, R0.5 vs C0.5=0.77). Male data were analyzed by Kruskal-Wallis one-way ANOVA on ranks. Analysis showed that Nr4a1 mRNA was significant differences after contextual fear conditioning (H3,16=16.16 p<0.05). Post hoc analysis showed that Nr4al mRNA was significantly up-regulated 0.5 h after contextual fear conditioning and 0.5 h after memory reactivation compared to naïve (p<0.05). Furthermore, 24.5 h after contextual fear conditioning, Nr4a1 mRNA expression had returned to the baseline level. Nr4a1 mRNA expression was up-regulated when the memory had been reactivated 24 h after contextual fear conditioning (p<0.05) same pattern as females. Previously an up-regulation of Nr4a1 mRNA expression after reactivation of memory was not observed (von Hertzen & Giese 2005): this difference could be due to duration of re-exposure, because we re-exposured for 10 min rather than 5 min. Nr4a1 is one of three members of the NR4A family of transcription factors and is an immediate early gene induced by a variety of stimuli, and especially by the activation of transcription factor cAMP-response element binding protein (Hawk & Abel 2011) and after contextual fear conditioning (von Hertzen & Giese 2005). Therefore, our Nr4a1 mRNA expression analyses showed that our experimental conditions were suitable for the analysis of gene expression after contextual fear conditioning.
Figure 2.
Nr4a1 mRNA expression in hippocampus is up-regulated after contextual fear conditioning and reconsolidation. qPCR showed that Nr4a1 mRNA expression was up-regulated in the hippocampus 0.5 h after contextual fear conditioning and 0.5 h after context-shock memory reactivation compared with the naïve and 24.5 h after contextual fear conditioning groups in males (a) and in females (b). Data are means ± SEM; *p<0.05; **p<0.01; ***p<0.001.
Nr4a1 DNA methylation in hippocampus after contextual fear conditioning
We quantified DNA methylation across two Nr4a1 CpG islands located up-stream and relative to the transcription start site of exon I as shown in Fig. 3a in the hippocampus after contextual fear conditioning. None of the observed showed a significant difference in DNA methylation between groups, although we observed a trend for decreased DNA methylation 0.5 h and 24.5 h after contextual fear conditioning at a CpG site (CpG 7) in the promoter CpG island assay (naïve vs C0.5; t8=2.07, p=0.073, naïve vs C24.5; t7=2.23, p=0.061) (Fig. 3b). Differences in DNA methylation at both time points do not mirror the mRNA expression changes, which occur only at 0.5 h after contextual fear conditioning.
Figure 3.
Nr4a1 DNA methylation in hippocampus is altered after contextual fear conditioning. (a) Nr4a1B exons are indicated by boxes: the black boxes indicate the coding region of the gene. The position of two CpG islands (CpG55 and CpG65) are shown by gray boxes. Individual bisulfite-PCR amplicons are labeled NR4A1 A and NR4A1 B and shown under each CpG island. (b) Assay NR4AI77A and (c) Assay NR4AI77B : Sequenom EpiTYPER analysis showed that CpG7 in CpG island 55 was relatively hypomethylated in the hippocampus 0.5 h and 24.5 h after contextual fear conditioning compared with the naïve group shown in (b).
Regulation of Bdnf gene expression in hippocampus during contextual fear memory conditioning
We studied whether contextual fear memory conditioning associated alterations in Bdnf gene expression might involve isoform-specific transcription. We quantified exon-specific Bdnf mRNA using assays specific to 6 exons (exon I, III, IV, VI, VII, and IX) in hippocampus using qPCR across four groups of mice (naïve, C0.5, C24.5, R0.5). Bdnf gene transcripts and the location of the qPCR primers used in this study are illustrated in Fig. 4a.
Figure 4.
Bdnf exon I and exon VI mRNA expression in hippocampus are up-regulated after contextual fear conditioning. (a) Bdnf xons (I to IX with older nomenclature displayed above in bracket) indicated by boxes. The gray box indicates the coding region of Bdnf gene. qRCR analysis was performed by using primer pairs specific for exon I, III, IV, VI, VII and IX shown by the black arrow. qPCR showed that bdnf exon I (b) and exon VI (c) mRNA expression were up-regulated in the hippocampus 0.5 h and 24.5 h after contextual fear conditioning compared the naïve group and not additionally up-regulated 0.5 h after context-shock memory reactivation. Data are means ± SEM; *p<0.05; **p<0.01.
Bdnf exon I and exon VI mRNA expression in the hippocampus is up-regulated for at least 24 h after contextual fear conditioning
Two-way ANOVA showed that there was no significant sex difference in Bdnf exon I and VI mRNA expression (exon I; F1,32=0.781 and p=0.384, exon VI; F1,32=1.11 and p=0.30), so subsequent analyses were performed on data pooled across both sexes. One-way ANOVA showed significant differences after contextual fear conditioning (exon I; F3,36=4.42 and p=0.01, exon VI; F3,36=7.19 and p<0.001), as shown in Fig. 4b and c. Post hoc analysis showed that Bdnf exon I and VI mRNA were significantly up-regulated 0.5 h and 24.5 h after contextual fear conditioning and 0.5 h after memory reactivation compared to naïve animals (exon I; C0.5: p=0.038, C24.5: p=0.013, R0.5: p=0.011, exon VI; C0.5: p=0.049, C24.5: p=0.002, R0.5: p=0.001). However, there was no significant difference in expression between 0.5 h and 24.5 h after contextual fear conditioning (exon I; p=0.54, exon VI; p=0.14) or between 0.5 h after contextual fear conditioning and 0.5 h after memory reactivation (exon I; p=0.37, exon VI; p=0.07) or between 24.5 h after contextual fear conditioning and 0.5 h after memory reactivation (exon I; p=0.87. exon VI; p=0.94). Therefore, Bdnf exon I and exon VI mRNA appears to be up-regulated at 0.5 h and maintained at this level for at least 24 h after contextual fear conditioning, and memory reactivation does not further up-regulated the expression.
Bdnf exon IV, exon VII and exon IX mRNA expression in the hippocampus is up-regulated for at least 24 h after contextual fear conditioning in females but not males
Two-way ANOVA revealed significant differences in training and sex for Bdnf exon IV, VII and IX mRNA expression (exon IV: training F3,32=7.36 and p<0.001, sex F1,32=10.25 and p=0.003, training and sex interaction F3,32=0.715 and p=0.55, exon VII: training F3,32=8.89 and p<0.001, sex F1,32=4.99 and p=0.033, training and sex interaction F3,32=0.94 and p=0.43, exon IX: training F3,32=9.90 and p<0.001, sex F1,32=6.41 and p=0.016, training and sex interaction F3,32=0.97 and p=0.42), as shown in Fig. 5a-c. Post hoc analysis showed that Bdnf exon IV, VII and IX mRNA were significantly up-regulated 0.5 h and 24.5 h after contextual fear conditioning and 0.5 h after memory reactivation compared to naïve (C0.5: exon IV; p=0.003, exon VII; p=0.031, exon IX; p=0.008, C24.5: exon IV; p=0.039, exon VII; p=0.005, exon IX; p<0.001, R0.5: exon IV; p=0.003, exon VII; p<0.001, exon IX; p<0.001). There was a significant higher expression of Bdnf exon IV, VII and IX mRNA at 24.5 h after contextual fear conditioning in females than in males (exon IV; p=0.01, exon VII; p=0.034, exon IX; p=0.011). These results suggest that Bdnf exon IV, VII and IX mRNA is up-regulated and maintained for at least 24 h after contextual fear conditioning in females, but not in males.
Figure 5.
Bdnf exon IV, VII and IX mRNA expression is up-regulated after contextual fear conditioning and reconsolidation, but not exon III mRNA. qPCR showed that Bdnf exon IV (a), VII (b) and IX (c) mRNA expression was up-regulated in the hippocampus 0.5 h and 24.5 h after contextual fear conditioning and 0.5 h after context-shock memory reactivation compared to the naïve group. There is a sex difference in the up-regulation 24.5 h after contextual fear conditioning. Bdnf exon VII (b) mRNA expression was up-regulated 0.5 h after context-shock memory reactivation compared with 0.5 h after contextual fear conditioning. (d) qPCR showed that Bdnf exon III mRNA expression was not up-regulated in the hippocampus after contextual fear conditioning and context-shock memory reactivation compared with the naïve group. Bdnf exon III mRNA was higher in females compared to males 0.5 h and 24.5 h after contextual fear conditioning. Data are means ± SEM; *p<0.05; **p<0.01; ***p<0.001.
Bdnf exon III mRNA expression in the hippocampus is not altered after contextual fear conditioning
Two-way ANOVA identified significant differences in sex and sex/training interaction, but not in training, in Bdnf exon III mRNA expression (sex: F1,32=7.96 and p=0.008, training: F3,32=1.59 and p=0.21, training and sex interaction: F3,32=3.06 and p=0.042), as shown in Fig. 5d. Post hoc analysis demonstrates that Bdnf exon III mRNA is higher in females than in males 0.5 h and 24.5 h after contextual fear conditioning (C0.5: p=0.022, C24.5: p=0.002). There was a significantly higher expression of Bdnf exon III mRNA at 24.5 h after contextual fear conditioning in females compared with the female naïve (p=0.024).
Bdnf DNA methylation in the hippocampus is altered after contextual fear conditioning
We studied DNA methylation in the vicinity of four CpG islands associated with the Bdnf gene in the hippocampus after contextual fear conditioning. The location of these four Bdnf CpG islands located relative to the transcription start site of exon I, III, VI and IX are shown in Fig. 6a. Among the amplicons tested (BDNFA-BDNFI) we found significant differences at several CpG sites particularly in the vicinity of CpG island 2. In particular, CpG6 in the BDNFB amplicon (CpG island 2) was significantly hypomethylated 0.5 h and 24.5 h after contextual fear conditioning compared to the naïve (Fig. 6b) (naïve vs C0.5; t17=2.31 p=0.034, naïve vs C24.5; t16=2.23 p=0.04, naïve vs R0.5; t16=1.30 p=0.21). Furthermore, there was a significant difference in average hypomethylation across this amplicon (t17=2.30 p=0.035) at C0.5 compared to naive. These data suggest that specific CpG sites in Bdnf CpG island 2 are hypomethylated 0.5 h after contextual fear conditioning with levels maintained up to 24 h although memory reactivation seems to return DNA methylation back to the baseline levels.
Figure 6.
Bdnf DNA methylation in hippocampus is altered after contextual fear conditioning. (a) Bdnfexons (I to IX with older nomenclature displayed above in bracket) are indicated by boxes; the gray box indicates the coding region of the gene. The position of four Bdnf CpG islands are shown by solid lines and indicated relative to transcription site of exon I, III, IV and IX. Individual bisulfite-PCR amplicons are labeled BDNFA to BDNFI and shown under each CpG islands. (b) Assay BDNFB: Sequenom EpiTYPER analysis showed that CpG6 in CpG island 2 was relatively hypomethylated in the hippocampus 0.5 h and 24.5 h after contextual fear conditioning compared with the naïve group. *p<0.05.
Discussion
We studied whether contextual fear memory formation is associated with alterations in Bdnf gene expression involving isoform-specific transcription and changes in DNA methylation at relevant CpG islands. We chose contextual fear conditioning for the experiments, because the task is hippocampus dependent and long-term memory can form by a single training session. Furthermore, Bdnf mRNA expression is known to be altered after contextual fear conditioning (Hall et al. 2000; Mizuno et al. 2006). We found differential transcription of Bdnf exons and altered DNA methylation at specific CpG sites in hippocampus after contextual fear conditioning.
We measured exon-specific Bdnf mRNA (exon I, III, IV, VI, VII and IX) levels in the hippocampi of four group of mice, naïve, 0.5 h and 24.5 h after contextual fear conditioning and 0.5 h after memory had been reactivated 24 h after contextual fear conditioning. We observed three types of expression patterns in exon-specific Bdnf mRNA (Table 1): 1) where mRNA expression was up-regulated at 0.5 h and was maintained at least 24 h after contextual fear conditioning with no further up-regulation following memory reactivation. (Bdnf exon I and VI) 2) where mRNA expression was up-regulated at 0.5 h in both sexes, and maintained at least 24 h only in females after contextual fear conditioning. (Bdnf exon IV, VII and IX). 3) where there was no differential regulation of Bdnf by conditioning in males. (Bdnf exon III) To our knowledge we describe for the first time transcriptional changes in the hippocampus 24 h after training. Generally, it is believed that memory consolidation is completed within 24 h (Dudai 2004). Therefore, transcriptional changes at 24 h of contextual fear conditioning might contribute to ongoing storage mechanisms of long-term memory.
Our findings are in agreement with recent studies examining the differential usage of Bdnf exons in hippocampus and amygdala during consolidation of fear learning in male rats (Lubin et al. 2008; Rattiner et al. 2004; Ou & Gean 2007). We show that Bdnf exon I and VI are up-regulated by contextual fear conditioning. Our experimental design could not determine whether this regulation is specific for the context-shock association or whether it is due to context exposure per se. Consistent with our findings, Lubin et al. found that Bdnf exon I and VI are up-regulated in the contextual fear conditioning task (Lubin et al. 2008). Their control experiment determined that this alteration is due to learning a novel environment rather than associative learning between context and shock, as these exons are up-regulated after context exposure alone and not context-shock association. Additionally, Lubin et al. found that Bdnf exon IV mRNA is up-regulated in hippocampus 2 h after training in male rats. Consistently, our data show that Bdnf exon IV mRNA is up-regulated at 30 min after training in male mice. Lubin et al. did not find a regulation at this time point for total Bdnf exon IX transcripts in male rats, which we also did not find in male mice at this time point. However, we investigated Bdnf regulation 24 h after training in more detail than previous studies. We found that in male mice Bdnf exon 1 and VI expression is still up-regulated 24 h after conditioning. We also demonstrate a long-lasting regulation of these two Bdnf transcripts in female mice. Additionally, in female mice Bdnf exon IV and IX mRNA expression is still up-regulated 24 h after conditioning. In the current experiments we could not specify what causes this long-lasting up-regulation in females, although we hypothesize it could be either due to novelty or the learned association between context and shock, or other factors such as fear (Dalla & Shors 2009). Another limitation of this study is that we were only able to specifically assess the transcription of six of the nine Bdnf exons. However, the data presented here highlight that some, but not all, Bdnf exons are regulated in a long lasting manner. Future analyses will build upon these data, and include longer time points and the analysis of additional exons.
Previously, we found sex differences in Bdnf exon IX expression after contextual fear conditioning, demonstrating that Bdnf mRNA was up-regulated 0.5 h after training in males, but not females (Mizuno et al., 2006). This is in contrast with the results for the females presented here. Various parameters differed between these two experiments, including the strain of mice (hybrid vs C57BL/6J), training paradigm (1 shock vs 3 shocks), and handling (without handling vs with handling). These differences can potentially account for different results, although further investigations are warranted.
A previous study suggested that BDNF mRNA expression is up-regulated during reconsolidation (Kirtley and Thomas, 2010), although such transcription may not be essential for maintaining the long-term memory (Lee et al., 2004). Therefore, we also investigated BDNF mRNA expression after reactivation of long-term memory. We found that BDNF mRNA expression is not regulated by memory reactivation in either males and females. Our finding concurs with the notion that BDNF has a role in consolidation but not reconsolidation, as proposed by Lee et al., 2004.
We found that DNA methylation is altered at specific CpG sites in the Bdnf gene after contextual fear conditioning. In particular, we identified a significant and persistent DNA hypomethylation at a CpG site in the vicinity of CpG island 2, which is located at the end of exon III. This hypomethylation was detected 0.5 h and maintained up to 24 h after contextual fear conditioning. It is notable that the sites of differential DNA methylation appear to be quite restricted in this area, since we did not detect significant change of DNA methylation in the other nearby CpGs. This hypomethylation up to 24 h after contextual fear conditioning corresponds with the increase in exon I and VI mRNA expression in both sexes and IV, VII and IX mRNA expression in females. Although we have no mechanistic insight about how reduced DNA methylation upstream of exon IV influences the expression of these exons, the observation suggests that demethylation of CpG sites within CpG islands might promote transcription of exon-specific Bdnf gene after contextual fear conditioning. Previously, it was shown that contextual fear conditioning decreases DNA methylation within the Bdnf exon IV promoter in rat hippocampus CA1, and that this is correlated with the up-regulation of contextual learning specific Bdnf exon IV mRNA expression (Lubin et al. 2008). Our findings, although preliminary, represent the first demonstration of training-induced hypomethylation related to altered transcription in the mouse Bdnf gene.
We found that several Bdnf transcripts are up-regulated after contextual fear conditioning, although all encode the same protein. Recently, the functional consequence of multiple transcripts of BDNF is suggested in the “spatial code hypothesis of Bdnf transcripts” (Tongiorgi 2008). In this model, different transcripts represent a spatial code used by neurons to selectively target the effects of BDNF to distinct dendritic compartments (Baj et al. 2011). Thus, BDNF might have spatially restricted effects upon dendritic complexity after contextual fear conditioning.
Previously, it was reported that hippocampal DNA methylation is associated with memory formation, however these hippocampal changes are transient, returning to basal levels within 24 h after conditioning (Miller & Sweatt 2007). Instead persistent changes in cortical DNA methylation were proposed to underlie long-lasting memory formation (Miller et al. 2010). However, our study suggests that there may be longer lasting alterations to DNA methylation in the hippocampus. As contextual fear memory has been suggested to become hippocampus-independent within 4 weeks after training (Frankland & Bontempi 2005), future studies will need to establish how long any dynamic changes to DNA methylation in the Bdnf gene lasts.
The hippocampus plays an important role in acquisition and protein synthesis-dependent consolidation of new memories into long-term memory. For the longer-term storage of memory, so called system consolidation, the memory trace is believed to be transiently stored in the hippocampus and transferred to other brain areas such as cortical structures (Frankland & Bontempi 2005). However, it is not clear whether the hippocampus has a temporary or a permanent role in the storage and retrieval of memory (Sutherland & Lehmann 2011). It was shown that 12 h after contextual fear conditioning a novel protein synthesis-dependent phase and BDNF activity in the hippocampus is necessary for memory persistence but not for memory formation (Bekinschtein et al. 2007; 2008a; 2010). Here we show that transcriptional changes in the Bdnf gene can persist at least 24 h after conditioning. This suggests that BDNF is required for the establishment of long-lasting memory storage.
In sum, our findings support the recent idea that long-term memories are established and maintained in the hippocampus, in parallel with multiple extra hippocampus networks (Sutherland & Lehmann 2011).
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the National Institute of Health (RO1 MH-097463-01A1) for KPG and JM.
Footnotes
The authors declare no conflicts of interest.
References
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baj G, Leone E, Chao MV, Tongiorgi E. Spatial segregation of BDNF transcripts enables BDNF to differentially shape distinct dendritic compartments. Proc Natl Acad Sci U S A. 2011;108:16813–16818. doi: 10.1073/pnas.1014168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LR, Izquierdo I, Medina JH. Persistence of long-term memory storage requires a late protein synthesis- and BDNF- dependent phase in the hippocampus. Neuron. 2007;53:261–277. doi: 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Izquierdo I, Medina JH. BDNF and memory formation and storage. Neuroscientist. 2008a;14:147–156. doi: 10.1177/1073858407305850. [DOI] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Katche C, Slipczuk L, Rossato JI, Goldin A, Izquierdo I, Medina JH. BDNF is essential to promote persistence of long-term memory storage. Proc Natl Acad Sci U S A. 2008b;105:2711–2716. doi: 10.1073/pnas.0711863105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekinschtein P, Katche C, Slipczuk L, Gonzalez C, Dorman G, Cammarota M, Izquierdo I, Medina JH. Persistence of long-term memory storage: new insights into its molecular signatures in the hippocampus and related structures. Neurotox Res. 2010;18:377–385. doi: 10.1007/s12640-010-9155-5. [DOI] [PubMed] [Google Scholar]
- Cunha C, Brambilla R, Thomas KL. A simple role for BDNF in learning and memory? Front Mol Neurosci. 2010;3:1. doi: 10.3389/neuro.02.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla C, Shors TJ. Sex differences in learning processes of classical and operant conditioning. Physiol Behav. 2009;97:229–238. doi: 10.1016/j.physbeh.2009.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudai Y. The neurobiology of consolidations, or, how stable is the engram? Annu Rev Psychol. 2004;55:51–86. doi: 10.1146/annurev.psych.55.090902.142050. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland PW, Bontempi B. The organization of recent and remote memories. Nat Rev Neurosci. 2005;6:119–130. doi: 10.1038/nrn1607. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Balogh SA, Wehner JM, Jones KR. Learning deficits in forebrain-restricted brain-derived neurotrophic factor mutant mice. Neuroscience. 2003;121:341–354. doi: 10.1016/s0306-4522(03)00426-3. [DOI] [PubMed] [Google Scholar]
- Hall J, Thomas KL, Everitt BJ. Rapid and selective induction of BDNF expression in the hippocampus during contextual learning. Nat Neurosci. 2000;3:533–535. doi: 10.1038/75698. [DOI] [PubMed] [Google Scholar]
- Hawk JD, Abel T. The role of NR4A transcription factors in memory formation. Brain Res Bull. 2011;85:21–29. doi: 10.1016/j.brainresbull.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirtley A, Thomas KL. The exclusive induction of extinction is gated by BDNF. Learn. Mem. 2010;17:612–619. doi: 10.1101/lm.1877010. [DOI] [PubMed] [Google Scholar]
- Lee JL, Everitt BJ, Thomas KL. Independent cellular processes for hippocampal memory consolidation and reconsolidation. Science. 2004;304:839–843. doi: 10.1126/science.1095760. [DOI] [PubMed] [Google Scholar]
- Liu IY, Lyons WE, Mamounas LA, Thompson RF. Brain-derived neurotrophic factor plays a critical role in contextual fear conditioning. J Neurosci. 2004;24:7958–7963. doi: 10.1523/JNEUROSCI.1948-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Giese KP. Towards a molecular understanding of sex differences in memory formation. Trends Neurosci. 2010;33:285–291. doi: 10.1016/j.tins.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Ris L, Sanchez-Capelo A, Godaux E, Giese KP. Ca2+/calmodulin kinase kinase alpha is dispensable for brain development but is required for distinct memories in male, though not in female, mice. Mol Cell Biol. 2006;26:9094–9104. doi: 10.1128/MCB.01221-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsey MS, Ota KT, Akingbade IF, Hong ES, Schafe GE. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLoS One. 2011;6:e19958. doi: 10.1371/journal.pone.0019958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou LC, Gean PW. Transcriptional regulation of brain-derived neurotrophic factor in the amygdala during consolidation of fear memory. Mol Pharmacol. 2007;72:350–358. doi: 10.1124/mol.107.034934. [DOI] [PubMed] [Google Scholar]
- Rattiner LM, Davis M, Ressler KJ. Differential regulation of brain-derived neurotrophic factor transcripts during the consolidation of fear learning. Learn Mem. 2004;11:727–731. doi: 10.1101/lm.83304. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Zoladz PR, Sweatt JD, Diamond DM. Epigenetic modification of hippocampal Bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. J Psychiatr Res. 2011;45:919–926. doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Giese KP. Plastic genes are in. Curr Opin Neurobiol. 1994;4:413–420. doi: 10.1016/0959-4388(94)90104-x. [DOI] [PubMed] [Google Scholar]
- Sutherland RJ, Lehmann H. Alternative conceptions of memory consolidation and the role of the hippocampus at the systems level in rodents. Curr Opin Neurobiol. 2011;21:446–451. doi: 10.1016/j.conb.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Josselyn SA, Frankland PW, Masushige S, Silva AJ, Kida S. Memory reconsolidation and extinction have distinct temporal and biochemical signatures. J Neurosci. 2004;24:4787–4795. doi: 10.1523/JNEUROSCI.5491-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. Experience-dependent epigenetic modifications in the central nervous system. Biol Psychiatry. 2009;65:191–197. doi: 10.1016/j.biopsych.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E. Activity-dependent expression of brain-derived neurotrophic factor in dendrites: facts and open questions. Neurosci Res. 2008;61:335–346. doi: 10.1016/j.neures.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Tyler WJ, Alonso M, Bramham CR, Pozzo-Miller LD. From acquisition to consolidation: on the role of brain-derived neurotrophic factor signaling in hippocampal-dependent learning. Learn Mem. 2002;9:224–237. doi: 10.1101/lm.51202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hertzen LS, Giese KP. Memory reconsolidation engages only a subset of immediate-early genes induced during consolidation. J Neurosci. 2005;25:1935–1942. doi: 10.1523/JNEUROSCI.4707-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Mizuno M, Nabeshima T. Role for brain-derived neurotrophic factor in learning and memory. Life Sci. 2002;70:735–744. doi: 10.1016/s0024-3205(01)01461-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.