SUMMARY
Cnm, a collagen- and laminin-binding protein present in a subset of Streptococcus mutans strains, mediates binding to extracellular matrices (ECM), intracellular invasion and virulence in the Galleria mellonella model. Antibodies raised against Cnm were used to confirm expression and the cell surface localization of Cnm in the highly invasive OMZ175 strain. Sequence analysis identified two additional genes (cnaB and cbpA) encoding putative surface proteins immediately upstream of cnm. Inactivation of cnaB and cbpA in OMZ175, individually or in combination, did not decrease the ability of this highly invasive and virulent strain to bind to different ECM proteins, invade human coronary artery endothelial cells (HCAEC), or kill G. mellonella. Similarly, expression of cnaB and cbpA in the cnm− strain UA159 revealed that these genes did not enhance Cnmrelated phenotypes. However, integration of cnm in the chromosome of UA159 significantly increased its ability to bind to collagen and laminin, invade HCAEC, and kill G. mellonella. Moreover, the presence of antibodies against Cnm nearly abolished the ability of OMZ175 to bind to collagen and laminin and invade HCAEC, and significantly protected G. mellonella against OMZ175 infection. We concluded that neither CnaB nor CbpA is necessary for the expression of Cnm-related traits. We also provided definitive evidence that Cnm is an important virulence factor and a suitable target for the development of novel preventive and therapeutic strategies to combat invasive S. mutans strains.
Keywords: Cnm, collagen-binding protein, intracellular invasion, Streptococcus mutans
INTRODUCTION
A major contributor in the development of dental caries, Streptococcus mutans has been the subject of extensive research and the mechanisms associated with its ability to colonize and thrive in the oral environment have been well documented (Loesche, 1986; Bowen & Koo, 2011). In addition, S. mutans can cause extra-oral infections such as infective endocarditis (Mylonakis & Calderwood, 2001; Nagata et al., 2006; Nakano et al., 2008) and has been proposed to participate in the development of atherosclerosis (Nakano et al., 2006; Kesavalu et al., 2012). Strains of S. mutans are classified in four serotypes (c, e, f and k) based on the chemical composition of the cell surface rhamnose–glucose polysaccharide (Nakano et al., 2007a, 2008). In clinical samples, ~ 70% of S. mutans isolates from dental plaque belong to serotype c and nearly 20% to serotype e, while serotypes f and k comprise less than 5% each (Nakano et al., 2007a,b, 2008). Despite its well-established association in blood-borne infections, the mechanisms associated with S. mutans infection and persistence in extra-oral sites are still poorly understood.
The ability of oral streptococci to colonize extra-oral tissues, such as heart valves, depends on the expression of surface-associated adhesins that mediate bacterial binding to the extracellular matrix (ECM) or other host components (Burnette-Curley et al., 1995; Lee et al., 2001; Xiong et al., 2008; Seo et al., 2010). A small number of adhesins involved in ECMbinding have been identified in the S. mutans core genome (Nobbs et al., 2009). The wall associated protein A (WapA) was shown to bind to collagen to modulate sucrose-independent biofilm formation, and to participate in cell–cell aggregation (Han et al., 2006; Zhu et al., 2006). SpaP (also known as Antigen I/II or PA) was shown to mediate cell binding to saliva- coated tooth surfaces, and to specifically interact with collagen, fibronectin and fibrinogen in vitro (Beg et al., 2002; Brady et al., 2010). Binding to fibronectin is also mediated by FBP-130 and FnB, illustrating the redundant ability of multiple adhesins in interactions with ECM components (Chia et al., 2000). A number of studies have revealed that 10–20% of S. mutans clinical isolates express a collagen (and laminin) binding protein named Cnm (Sato et al., 2004; Nomura et al., 2009; Nakano et al., 2010). The Cnm protein is composed of a conserved collagen-binding domain (A domain), a threonine-rich repeat region (B domain) that is followed by a C-terminal LPXTG motif for anchoring to the cell wall (Sato et al., 2004). Interestingly, Cnm is found at higher frequency among strains belonging to the rarer serotypes e, f and k (Nakano et al., 2010), and a correlation between these less frequent serotypes and systemic infections has been reported (Nakano et al., 2007b, 2010, 2011). In addition to Cnm, a second surface protein with strong collagen-binding properties, named Cbm, which is absent in OMZ175 and UA159, is frequently detected in strains belonging to the rare serotype k (Nomura et al., 2012).
Previously, we demonstrated that the ability of S. mutans strains to invade human coronary artery endothelial cells (HCAEC) was dependent on the expression of Cnm (Abranches et al., 2011). Among the 34 strains tested so far, only cnm+ strains were able to invade HCAEC and showed increased virulence in Galleria mellonella (Abranches et al., 2009, 2011). Inactivation of cnm abolished the ability of S. mutans strains to attach to and invade HCAEC, and significantly attenuated virulence in G. mellonella (Abranches et al., 2011). Recently, high coverage, whole genome shotgun sequencing of 57 genetically diverse S. mutans isolates, which included the highly invasive Cnm+ serotype f OMZ175 strain, became available (Cornejo et al., 2013). In addition to OMZ175, cnm was found in three other strains, V1996 and SF14 both serotype c and the serotype e U2A (Palmer et al., 2013). In a separate study, the complete sequence of the cnm+ serotype k strain LJ23 was also obtained (Aikawa et al., 2012). By analyzing the cnm region of the sequenced S. mutans strains, we noted that, in all cases, two additional genes, named cnaB and cbpA (Palmer et al., 2013), encoding putative surface proteins with conserved collagen-binding-like domains were located immediately upstream of the cnm gene. Hence, it is possible that, in addition to Cnm, CnaB and CbpA might also play a role in ECM binding and invasion of host cells thereby contributing to the virulence of cnm+ strains.
In the present study, we assessed the contributions of cnaB and cbpA to several phenotypes previously associated with Cnm. Deletion of cnaB, cbpA or both in OMZ175 and expression of these two genes in a non-invasive cnm− strain indicated that CnaB and CbpA are not required for the manifestation of Cnm-dependent traits. On the other hand, expression of Cnm in a hitherto cnm− strain was sufficient to recapitulate every major phenotype (e.g. HCAEC invasion) previously described in OMZ175. Finally, we provided direct evidence that Cnm is a suitable target for the development of novel preventive and therapeutic strategies to combat infections caused by invasive strains of S. mutans.
METHODS
Bacterial strains and culture conditions
All S. mutans strains used in this study are listed in Table 1. Escherichia coli strains were routinely grown in Luria–Bertani medium at 37°C. When required, 100 µg ml−1 ampicillin or 100 µg ml−1 kanamycin was added to Luria–Bertani broth or agar plates. Strains of S. mutans were routinely cultured in brain–heart infusion (BHI) medium at 37°C in a humidified 5% CO2 atmosphere. When required, 1 mg ml−1 kanamycin or 10 µg ml−1 erythromycin was added to BHI broth or plates.
Table 1.
Streptococcus mutans strains used in this study
| Strain | Serotype | Source |
|---|---|---|
| OMZ175 | F | Abranches et al. (2011) |
| ΔcnaB | f | This study |
| ΔcbpA | f | This study |
| ΔcnaB/cbpA | f | This study |
| Δcnm | f | Abranches et al. (2011) |
| UA159 | c | Abranches et al. (2011) |
| UA159-pBGE | c | This study |
| UA159-cnaB | c | This study |
| UA159-cbpA | c | This study |
| UA159-cnm | c | This study |
| B14 | e | Abranches et al. (2011) |
| 11060 | e | Abranches et al. (2011) |
| LM7 | e | Abranches et al. (2011) |
| OM50E | f | Abranches et al. (2011) |
Genetic manipulation of S. mutans strains
Isogenic strains were generated in S. mutans by insertion of a non-polar kanamycin marker (Kremer et al., 2001) using standard techniques and the primers listed in Table 2. For cloning purposes, E. coli DH10B cells were used throughout this study. Briefly, for cnaB and cnaB/cbpA inactivation, two polymerase chain reaction (PCR) fragments were obtained containing the 5′ and the 3′ regions of each gene to introduce artificial restriction sites. After amplification, the 5′ DNA fragments were digested and ligated to pGEM-z5F(−) (Promega, Madison, WI) and the resulting plasmid was propagated in DH10B cells. Then, the 3′ DNA fragments were introduced into pGEM-z5F(−) already harboring the 5′ fragment. After a PstI digestion, the coding region of each target gene cloned onto pGEM-z5F(−) was interrupted by introducing a PstI-digested non-polar kanamycin-resistance cassette into the middle of the gene. For cbpA inactivation, a single PCR product containing a natural HindIII site was obtained and cloned into pGEM-z5F(−). After HindIII digestion, the coding region of cbpA cloned into pGEM-z5F(−) was disrupted by introducing a HindIII-digested non-polar kanamycin-resistance cassette into the middle of the gene. The resulting plasmids were transformed into S. mutans OMZ175 and positive transformants were selected on BHI plates containing kanamycin. The desired mutations were confirmed by PCR sequencing of the insertion site and flanking regions. To express CnaB, CbpA and Cnm in UA159, the cnaB, cbpA and cnm genes containing their respective non-coding upstream regions were amplified using the primers listed in Table 2. The amplified products were digested with XbaI and BsrGI and cloned onto the integration vector pBGE (Zeng & Burne, 2009) that had been previously digested with the same enzymes. The resulting plasmids were used to transform S. mutans UA159 and transformants were selected on BHI plates containing erythromycin. Genomic integration of cnaB, cbpA and cnm at the gtfA locus was confirmed by PCR and sequence analysis.
Table 2.
Primers used in this study
| Primer | Sequence | Analysis |
|---|---|---|
| cnaB F1 | Forward 5′-TGTCATCAGCCATGGACGCATTAG-3′ | Inactivation of cnaB |
| cnaB R1 | Reverse 5′-TCCAGTTTCTGCAGCTATCCAGCC-3′ | |
| cnaB F2 | Forward 5′-GGCTGGATAGCTGCAGAAACTGGA-3′ | |
| cnaB R2 | Reverse 5′-GCCTCTATACATATGTTCGGTCGT-3′ | |
| cbpA F1 | Forward 5′-CAGATGCTTACCATGGCACCTATGA-3′ | Inactivation of cbpA |
| cbpA R1 | Reverse 5′-CTTCAGTTGGCGGCCGCCTTTCAGTC-3′ | |
| cnaBcbpA F1 | Forward 5′-CATAGACAACCATGGTCATGACAG-3′ | Inactivation of cnaB and cbpA |
| cnaB R1 | Reverse 5′-TCCAGTTTCTGCAGCTATCCAGCC-3′ | |
| cnaBcbpA F2 | Forward 5′-GATCAGTCTCTGCAGCGAACTAGC-3′ | |
| cnaBcbpA R2 | Reverse 5′-GATTGCATCTCATATGGTCGTTGTA-3′ | |
| UAcnaB F | Forward 5′-GACATAAGTCTAGAATTCCTC-3′ | Expression of cnaB in UA159 |
| UAcnaB R | Reverse 5′-GTCAGTTCTGTACATAAGACTTAAC-3′ | |
| UAcbpA F | Forward 5′-GAAAGCATCTCTAGAAAGTCTTAG-3′ | Expression of cbpA in UA159 |
| UAcbpA R | Reverse 5′-TAGCTTAGTGTACATTAACGCTG-3′ | |
| UAcnm F | Forward 5′-AGCGTTAATCTAGACTAACGTAAATC-3′ | Expression of cnm in UA159 |
| UAcnm R | Reverse 5′-CCTATTTTTAATGTACATCAGTTATG-3′ | |
| rCnmA F | Forward 5′-ACTAAGGCTCATATGAGTGATGTC-3′ | Recombinant expression of CBD in E. coli |
| rCnmA R | Reverse 5′-TCCACACCGGATCCGGCATTAAC-3′ | |
| cnaB F3 | Forward 5′-CTGGAATCATTCATTATCAA-3′ | RT-PCR |
| cnaB R3 | Reverse 5′-TCGGCATAACATTTC-3′ | |
| cnaBcbpA F3 | Forward 5′-CGACAAGCGGAAAGCTG-3′ | RT-PCR |
| cnaBcbpA R3 | Reverse 5′-CTCAAACAGTGTCATTATAGA-3′ | |
| cbpA F3 | Forward 5′-ACTGCTTTTCTGGGT-3′ | RT-PCR |
| cbpA R3 | Reverse 5′-CAGTACCAGTTCTAAAACC-3′ | |
| cbpAcnm F1 | Forward 5′-CTTCAAGCCAGTCATCTGCT-3′ | RT-PCR |
| cbpAcnm R1 | Reverse 5′-GTTTTACTACCGTTGCCAAGG-3′ | |
| Cnm F1 | Forward 5′-TGGAACCTTGCCATCA-3′ | RT-PCR |
| Cnm R1 | Reverse 5′-CGACCATTGAACCTTCGAC-3′ |
CBD, collagen-binding domain; E. coli, Escherichia coli; RT-PCR, reverse transcription–polymerase chain reaction. Restriction sites that were introduced for cloning purposes are underlined.
Purification of the Cnm collagen-binding domain and antibody generation
To express and purify a portion of Cnm, the fragment encoding amino acids 32–319 comprising the collagen- binding A domain of Cnm was amplified from OMZ175 using the primers listed in Table 2. To avoid toxic effects to E. coli, the N-terminal secretion signal (amino acids 1–31) and the threonine-rich B domain (amino acids 320–465) were not included. The amplified rCnmA fragment and the expression vector pET-16b (Clontech, Mountain View, CA) were digested with NdeI and BamHI, ligated and transformed into E. coli BL21 DE3 cells. The E. coli strain harboring the pET16b-rCnmA plasmid was grown in Luria–Bertani broth containing ampicillin to an optical density at 600 nm ~ 0.5 and the expression of the His-tagged fusion protein was induced by the addition of 0.5 mm isopropyl-β-d-thiogalactopyranoside for 4 h. The recombinant protein was purified under native conditions using the Ni-NTA Protein Purification Kit (Qiagen, Valencia, CA) according to the supplier’s instructions. Identity and purity of rCnmA were confirmed by liquid chromatography-mass spectrometry analysis (data not shown). For generation of anti-rCnmA polyclonal antibodies, adult rabbits were immunized with 1 mg ml−1 of rCnmA with Freund’s adjuvant intravenously at days 1, 21 and 42 (Lampire Biological Laboratories, Ottsville, PA). After 50 days, the animals were bled, the titer of polyclonal antibodies in the serum against Cnm was determined by enzymelinked immunosorbent assay (data not shown), and the antibody specificity was confirmed by Western blotting.
RNA isolation and analysis by reverse transcription-PCR
RNA was extracted from S. mutans cultures grown in BHI to mid-exponential phase (optical density at 600 nm ~ 0.5) using a previously described protocol (Kajfasz et al., 2010). Complementary DNA was synthesized using a high-capacity cDNA reverse transcriptase kit containing random primers (Applied Biosystems, Foster, CA). Specific primers for cnaB, cbpA and cnm coding regions were used to determine the expression and transcriptional organization of these genes (Table 2). Transcriptional organization was determined using a forward primer with homology to the 3′ end of the upstream gene, and a reverse primer with homology to the 5′ start of the downstream gene.
Western blot analysis
Whole cell protein lysates were obtained by homogenization in the presence of glass beads using a bead-beater (Biospec, Bartlesville, OK). Protein lysates were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA). Cnm detection was performed using rabbit anti-rCnmA polyclonal antibody diluted 1 : 2000 in 1× phosphate-buffered saline (PBS) + 0.1% Tween 20 and anti-rabbit horseradish peroxidase-coupled antibody (Sigma, St Louis, MO) using standard techniques.
Immunoelectron microscopy
Cultures of S. mutans were grown overnight in BHI, washed once in 1× PBS pH 7.2, and incubated with anti-rCnmA antibodies diluted 1 : 250 in PBS at room temperature for 1 h. After incubation with primary antibody, cells were washed in PBS and incubated with anti-rabbit immunoglobulin G coupled with 12-nm colloidal gold particles diluted 1 : 25 in PBS (Jackson ImmunoResearch Laboratories, West Grove, PA) at room temperature for 1 h. Next, cells were washed with PBS, fixed with 2% glutaraldehyde for 30 min followed by a rinse with 0.5× PBS pH 7.2. Upon fixation, samples were embedded for sectioning before loading them onto grids. The grids were examined using a Hitachi 7650 transmission electron microscope and the images were captured using the attached Gatan Erlangshen 11 megapixel digital camera at the University of Rochester Medical Center Electron Microscopy core facility.
Binding of S. mutans strains to ECM proteins
Collagen, laminin, fibronectin and fibrinogen binding assays were performed as previously described, with some modifications (Sato et al., 2004). Briefly, 40 µg ml−1 type I collagen from rat tail (Sigma-Aldrich), 50 µg ml−1 mouse laminin (Becton-Dickinson, Franklin Lakes, NJ), 50 µg ml−1 human fibronectin (Becton-Dickinson) or 50 µg ml−1 bovine fibrinogen (MP Biomedicals, Solon, OH) were immobilized in 96-well plates for 18 h at 4°C. Wells were washed with PBS before and after being blocked with 5% bovine serum albumin for 1 h at 37°C. Overnight bacterial cultures were washed twice in PBS (pH 7.2), and resuspended in PBS to obtain bacterial suspensions containing approximately 1 × 109 colony-forming units (CFU) ml−1. One hundred microliters of bacterial suspensions was added to each well containing immobilized matrices and the plates were incubated for 3 h at 37°C. Unbound cells were removed by a series of washes using PBS; cells that remained adhered to the matrices were stained with 0.05% crystal violet solution. The crystal violet was extracted with 7% acetic acid and optical density was measured at 575 nm. To determine the effect of anti-rCnmA in ECM-binding, experiments were performed as described above but bacteria were pre-incubated with increasing concentrations of antirCnmA (1 : 200, 1 : 100, 1 : 50 or 1 : 20 dilutions in PBS) for 30 min before exposure to ECM proteins.
HCAEC invasion assay
Antibiotic protection assays were performed to assess the capacity of the mutant strains to invade HCAEC (Abranches et al., 2011). Briefly, primary HCAEC (Lonza, Alledale, NJ) suspensions containing 0.5 × 105 endothelial cells were seeded into the wells of 24-well flat-bottomed tissue culture plates and incubated in the presence of gentamicin and endothelial growth factor supplements (Lonza SingleQuots) at 37°C in a 5% CO2 atmosphere until they reached 80–90% confluence. Overnight bacterial cultures were washed twice in PBS (pH 7.2) and resuspended in endothelial cell basal medium (EBM-2) (Lonza) containing 2% fetal bovine serum (FBS) without antibiotics. One milliliter of 2% FBS-EBM-2 medium containing 1 × 107 CFU ml−1 of S. mutans was used to infect HCAEC-containing wells, for 2 h in the absence of antibiotics followed by 3 h of incubation in 1 ml of 2% FBS-EBM-2 medium containing 300 µg ml−1 gentamicin and 50 µg ml−1 penicillin G to kill extracellular bacteria. After incubation with antibiotics, HCAEC were lysed with 1 ml of sterile water and the mixture of lysed HCAEC and S. mutans was plated onto tryptic soy agar (TSA) to determine the number of intracellular bacteria. The percentage of invasion for each strain was calculated based on the initial inoculum. To determine the effect of anti-rCnmA in S. mutans invasion experiments were performed as described above, but bacteria were pre-incubated for 30 min with a 1 : 10 dilution of anti-rCnmA before coincubation with HCAEC.
Galleria mellonella infection
The G. mellonella infection model with S. mutans has been previously described (Abranches et al., 2011). Briefly, 5-µl aliquots containing 1 × 108 CFU ml−1 of overnight-grown cultures washed and resuspended in sterile saline were injected into the hemocoel of each larva via the last left proleg. Groups injected with heatinactivated S. mutans strains (30 min at 80°C) were used as controls in each experiment. After injection, larvae were kept in the dark at 37°C, and larval survival was recorded at selected intervals. To determine the effect of anti-rCnmA in S. mutans virulence experiments were performed as described above, but bacteria were (i) pre-incubated for 30 min with a 1 : 10 dilution of anti-rCnmA and injected into the larvae or, (ii) injected alone followed by a second injection 4 h post-infection with a 1 : 10 dilution of anti-rCnmA. Controls included injection of anti-rCnmA alone and OMZ175 plus pre-immune serum. Experiments were performed in triplicate.
Statistical analysis
A one-way analysis of variance was performed to verify the significance of the growth curves, ECM binding and HCAEC invasion. For the G. mellonella studies, Kaplan–Meier killing curves were plotted, and estimations of differences in survival were compared using the log rank test. In all cases, P-values ≤ 0.05 were considered significant.
RESULTS
Detection and localization of Cnm in OMZ175
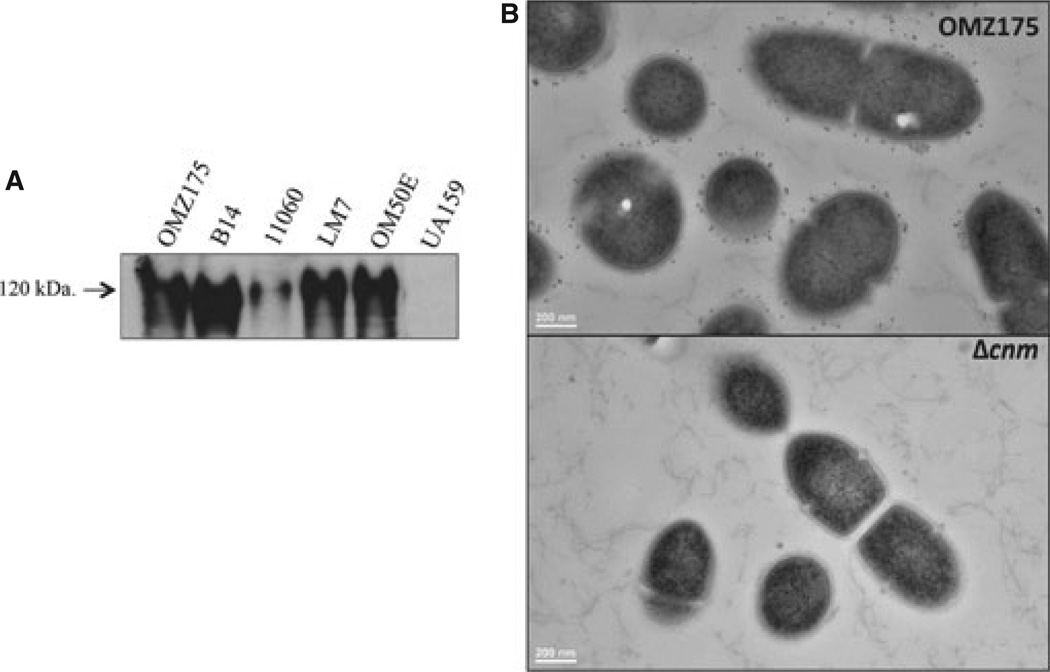
Western blot analysis with anti-rCnmA antibodies (see Methods) was used to confirm the expression of Cnm in our previously characterized cnm+ strains (Abranches et al., 2011). As shown in Fig. 1A, the expression of Cnm was confirmed in all cnm+ strains but not in the serotype c type strain UA159. The low level of Cnm observed in strain NCTC 11060 is in agreement with the low mRNA level previously described in this strain (Abranches et al., 2011). Localization of Cnm at the cell surface of the highly invasive strain OMZ175 (Abranches et al., 2009) was carried out by immunoelectron microscopy using anti-rCnmA. The immunoelectron microscopy images clearly show that, as predicted based on the presence of the LPXTG cell wall-anchoring motif, Cnm is localized at the cell surface (Fig. 1B).
Figure 1.
Cnm is produced and localized at the cell surface in Streptococcus mutans OMZ175. (A) Cnm production by invasive (OMZ175, B14, 11060, LM7, OM50) and non-invasive (UA159) strains of S. mutans was evaluated by Western blot analysis using 10 µg of whole cell protein lysate and detected with anti-rCnmA antiserum (1 : 2000). (B) Immunoelectron microscopy analysis of S. mutans cells using anti-rCnmA antiserum (1 : 250) showing the presence of Cnm on the surface of OMZ175 but not in the Δcnm strain.
Transcriptional organization of cnaB, cbpA and cnm
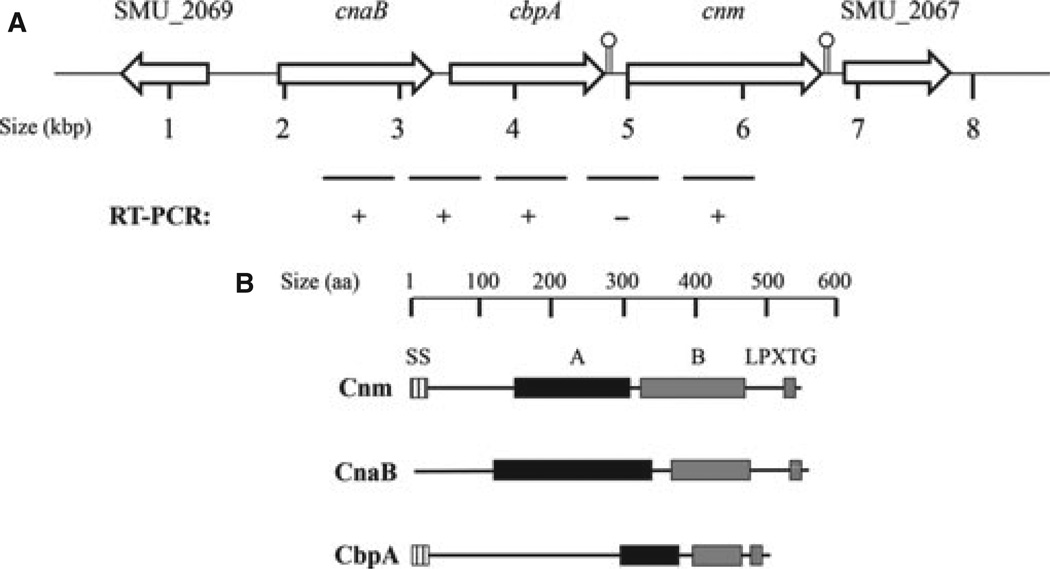
In all cnm+ strains sequenced to date (Aikawa et al., 2012; Song et al., 2012; Palmer et al., 2013), in silico analyses revealed that cnm is part of a unique 5-kilobase region located in the intergenic region between the core genes SMU_2067 and SMU_2069 (Fig. 2A). Further analysis of this region revealed the presence of two previously uncharacterized open reading frames upstream of cnm that were named cnaB and cbpA (Palmer et al., 2013). Notably, the region containing these three unique genes has a higher GC content (41.9%) than the whole genome of the type strain UA159 (36.8%) (Ajdic et al., 2002), suggesting that this region may have been acquired by horizontal gene transfer. We used PCR sequencing analysis to determine the localization and genetic arrangement of our collection of cnm+ strains and found that, in all cases, cnm was downstream to cnaB and cbpA and that the cnaB-cbpA-cnm cluster was located between SMU_2067 and SMU_2069 (Fig. 2A). Interestingly, the putative proteins encoded by cnaB and cbpA were structurally similar to Cnm. Like Cnm, CnaB and CbpA possess putative collagen-binding domains (A domain) and a unique threonine-rich repeat region (B domain) followed by a cell-wall anchoring LPXTG motif (Fig. 2B). Both Cnm and CbpA contain a conserved signal secretion signal whereas CnaB does not. We used reverse transcription (RT) -PCR to confirm the transcription of cnaB and cbpA and to show that both genes are co-transcribed. Conversely, a transcript encompassing cbpA and cnm was not detected (Fig. 2A). To support the RT-PCR analysis, in silico analysis using the ARNOLD software (http://rna.igmors.u-psud.fr/toolbox/arnold/) revealed the presence of a strong Rho-independent transcriptional terminator after cbpA but not downstream of cnaB (Fig. 2A).
Figure 2.
The cnm genomic locus in Streptococcus mutans contains two additional putative surface proteins. (A) In all cnm+ strains of S. mutans, cnm is located within a unique 5-kbp region between the core genes SMU_2067 and SMU_2069. SMU_2067 encodes a putative glycosyltransferase whereas SMU_2069 encodes the integral membrane zinc transporter ZupT. Two additional open reading frames (cnaB and cbpA) were identified upstream of cnm. Semi-quantitative RT-PCR analysis showed the co-transcription of cnaB with cbpA, whereas cnm was not part of this transcript. Presence of putative Rho-independent transcriptional terminator is indicated with a lollypop symbol. (B) Predicted domains of CnaB, CbpA and Cnm: SS, secretion signal; A, collagen-binding domain; B, threonine-rich repeats; LPXTG, cell-wall anchor motif.
Characterization of ΔcnaB, ΔcbpA and ΔcnaB/ΔcbpA strains
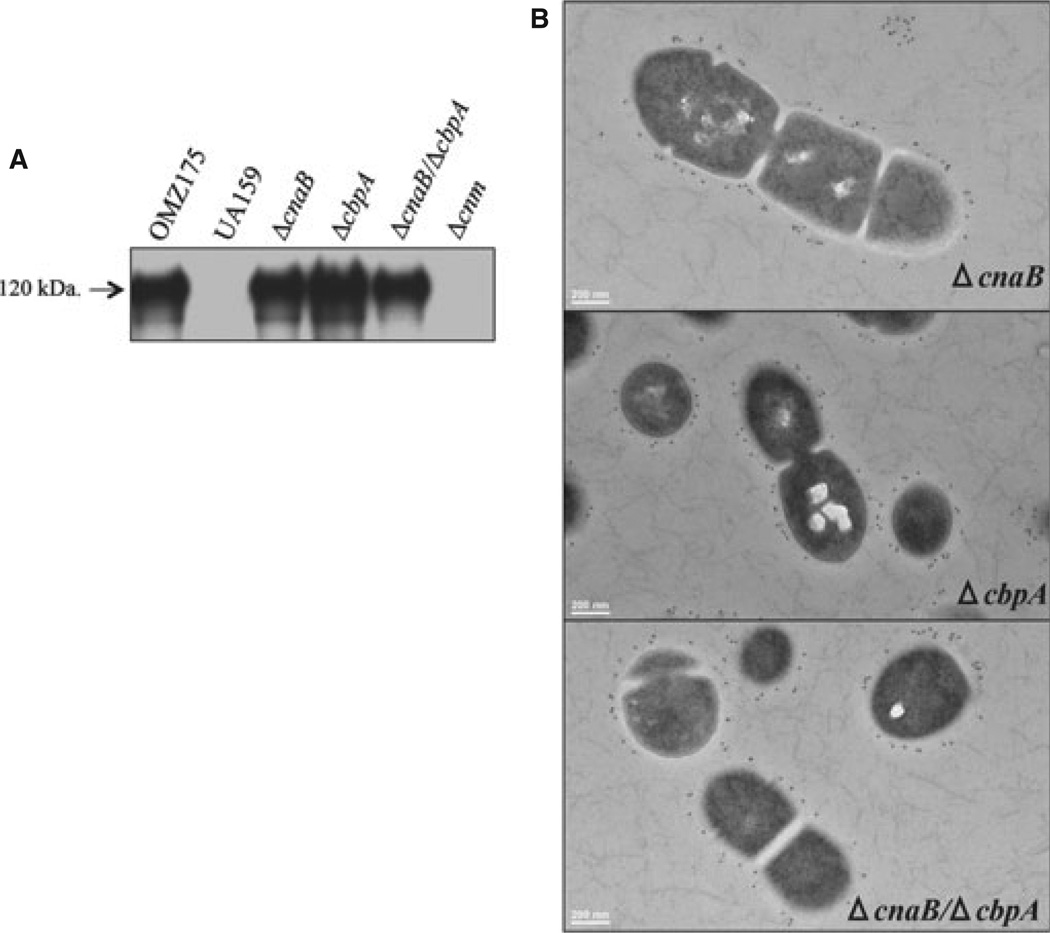
To explore the role of CnaB and CbpA in S. mutans, the cnaB, cbpA or both genes were replaced by a non-polar kanamycin-resistance cassette in OMZ175 as described in the Methods section. Western blot and immunoelectron microscopy analyses indicated that expression and localization of Cnm were not affected in the ΔcnaB, ΔcbpA and ΔcnaB/ΔcbpA strains (Fig. 3). However, cells of ΔcnaB were larger and elongated indicating a defect in cell division, whereas no obvious morphological differences were observed in the ΔcbpA strain when compared with the parent strain (Fig. 3B). Under standard growth conditions (e.g. BHI at 37°C in a 5% CO2 atmosphere), ΔcnaB grew as well as the parent strain (92 ± 4.2 and 88 ± 6 min, respectively), whereas the ΔcbpA strain grew significantly more slowly (109 ± 1.9 min) than OMZ175 (P < 0.05). Interestingly, the double mutant ΔcnaB/ΔcbpA strain had no apparent growth defects (91 ± 2.2 min), or differences in cell morphology when compared with OMZ175 (Fig. 3B).
Figure 3.
Cnm production and localization is not affected in ΔcnaB, ΔcbpA and ΔcnaB/cbpA mutant strains. (A) Cnm production by ΔcnaB, ΔcbpA and ΔcnaB/cbpA mutant strains was evaluated by Western blot analysis using 10 µg of whole cell protein lysate and detected with anti-rCnmA antiserum (1 : 2000). (B) Immunoelectron microscopy analysis and surface localization of Cnm in the ΔcnaB, ΔcbpA and ΔcnaBΔcbpA strains using anti-rCnmA antiserum (1 : 250).
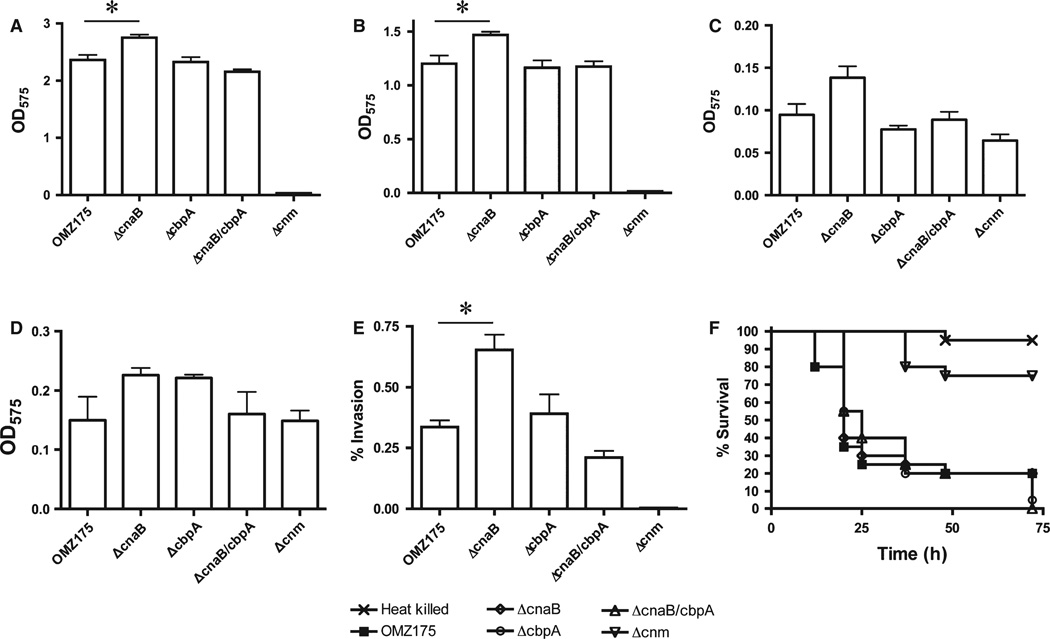
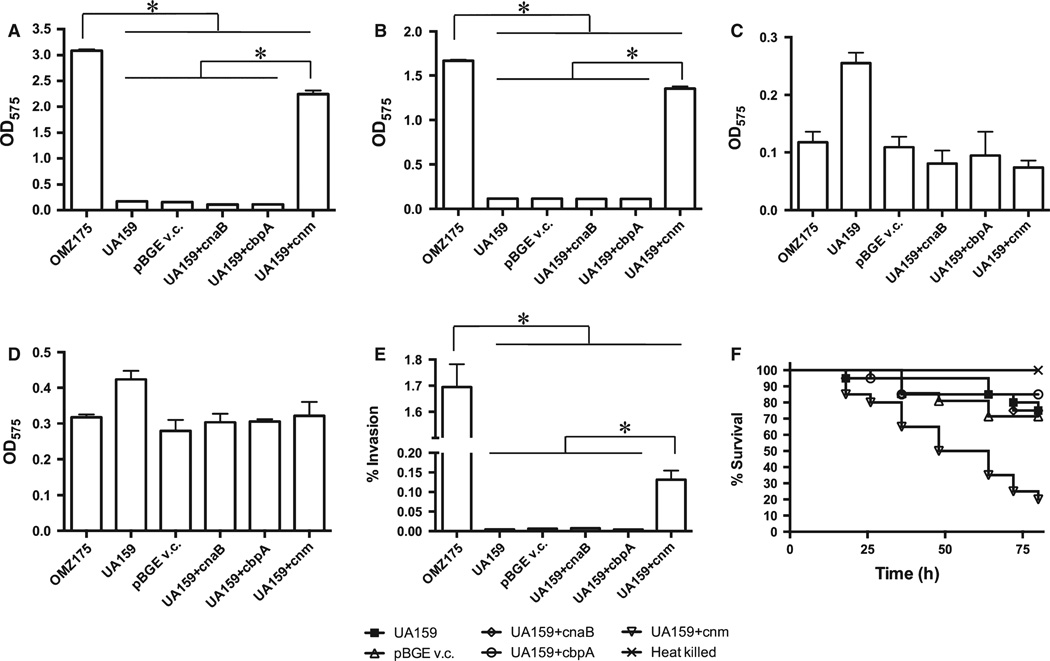
To evaluate whether CnaB and CbpA play a role in Cnm-dependent phenotypes, we tested the ability of ΔcnaB, ΔcbpA and ΔcnaB/ΔcbpA to: (i) bind different types of ECM proteins (collagen, laminin, fibrinogen or fibronectin), (ii) invade HCAEC, and (iii) kill G. mellonella. Unexpectedly, a significant enhanced binding to both collagen and laminin was observed in ΔcnaB, whereas no significant differences were observed in ΔcbpA or ΔcnaB/cbpA strains (Fig. 4A,B). No significant difference in fibrinogen or fibronectin binding was detected for any of the mutant strains (Fig. 4C,D). In agreement with the increased collagen- and laminin-binding phenotypes, ΔcnaB displayed increased rates of HCAEC invasion whereas ΔcbpA and ΔcnaB/ΔcbpA showed no significant differences when compared with the parent strain (Fig. 4E). Finally, the ΔcnaB, ΔcbpA or ΔcnaB/cbpA strains were able to kill G. mellonella as efficiently as the parent strain (Fig. 4F).
Figure 4.
Phenotypic characterization of ΔcnaB, ΔcbpA and ΔcnaB/cbpA strains of Streptococcus mutans OMZ175. (A–D) Binding of S. mutans strains to different types of extracellular matrix proteins: (A) collagen type I, (B) laminin, (C) fibrinogen, (D) fibronectin. (E) Antibiotic protection assay showing the percent of human coronary artery endothelial cell (HCAEC) invasion for each strain after 5 h of infection. (F) Killing of Galleria mellonella larvae infected with S. mutans strains *P < 0.05.
Expression of cnaB, cbpA and cnm genes in the non-invasive S. mutans strain UA159
In an attempt to assign a functional role for CnaB and CbpA and to evaluate whether the expression of Cnm alone was sufficient to enhance ECM-binding, HCAEC invasion and virulence in G. mellonella, each gene was individually amplified from OMZ175 and integrated at the gtfA locus of the non-invasive cnm− strain UA159 using the pBGE integration vector (Zeng & Burne, 2009). Transcription of cnaB, cbpA or cnm in UA159 was confirmed by RT-PCR and, in the case of cnm, by Western blotting as well (data not shown). The resulting recombinant UA159 strains were tested for their ability to bind to collagen, laminin, fibrinogen or fibronectin, invade HCAEC and kill G. mellonella. As shown in Fig. 5, expression of CnaB or CbpA in UA159 did not enhance ECM binding, invasion of HCAEC or virulence in G. mellonella. On the other hand, expression of cnm in UA159 (UA159-cnm+) significantly enhanced collagen and laminin binding properties to levels that were almost identical to those attained in OMZ175 (Fig. 5A,B). A similar trend was observed in the ability of UA159- cnm+ to invade HCAEC albeit the invasion rates were not as high as those observed in OMZ175 (Fig. 5E). Finally, UA159-cnm+ was significantly more virulent in G. mellonella when compared with UA159 transformed with the empty pBGE vector (Fig. 5F).
Figure 5.
Expression of cnm, but not cnaB or cbpA, by Streptococcus mutans UA159 increases its collagen- and laminin-binding ability, invasion to human coronary artery endothelial cells and virulence in Galleria mellonella. Extracellular matrix-binding assays were performed in the presence of (A) collagen type I, (B) laminin, (C) fibrinogen or (D) fibronectin. (E) Antibiotic protection assay showing the percentage of human coronary artery endothelial cell invasion (HCAEC) for each strain after 5 h of infection. (F) Killing of G. mellonella larvae infected with each S. mutans strain. *P < 0.05.
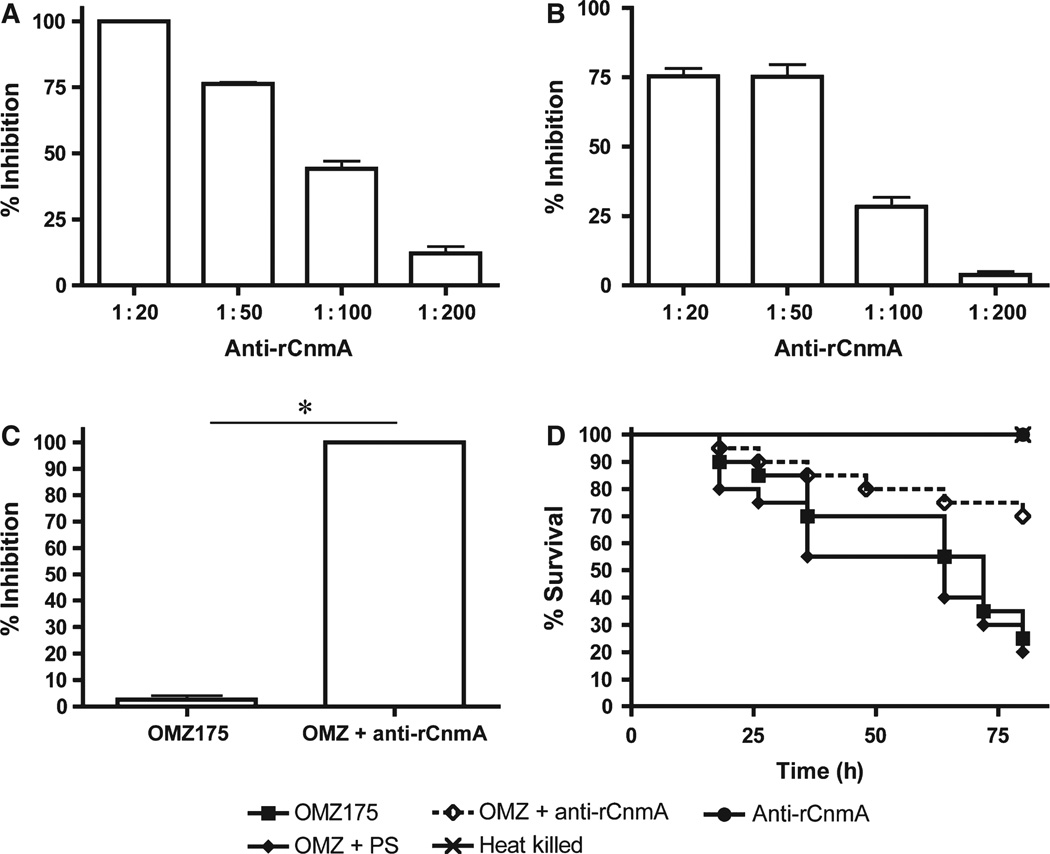
Anti-rCnmA antibodies inhibited ECM-binding, HCAEC invasion and G. mellonella killing by OMZ175
The results obtained with the cnaB and cbpA mutants strongly suggest that Cnm is the only product in the cnaB-cbpA-cnm gene cluster involved in the binding to the tested ECMs, intracellular invasion and virulence phenotypes. To confirm that these functional activities were directly attributable to Cnm, blocking assays in the presence of anti-rCnmA polyclonal antiserum were performed. Binding of OMZ175 to laminin was inhibited by approximately 75% in the presence of the anti-rCnmA antiserum whereas collagen binding was nearly abolished (Fig. 6A,B). Invasion of HCAEC by OMZ175 was also significantly inhibited by the anti-rCnmA antibody (Fig. 6C). The addition of pre-immune serum did not affect ECM-binding and invasion properties of OMZ175. Finally, injection of a 1 : 10 mix containing 1 × 107 CFU OMZ175 and anti-rCnmA antibody or pre-immune serum resulted in the increased survival(~ 50%) of animals infected with the mix containing the anti-rCnmA antibody but not the pre-immune serum (Fig. 6D). Next, to further confirm the protective effects of the anti-rCnmA antibody, we infected G. mellonella with OMZ175 followed by injection of the anti-rCnmA antibody 4 h post-infection. Despite the obvious signs of active infection, such as increased melanization, which appeared during the initial 4 h, antibody administration significantly (P < 0.05) increased the survival rates of G. mellonella at 72 h post-infection by approximately 25% when compared with larvae that received pre-immune serum (data not shown).
Figure 6.
Inhibition of Cnm using anti-rCnmA antibody. Inhibition of OMZ175 binding to (A) collagen type I or (B) laminin by anti-rCnmA antiserum. (C) The effect of the anti-rCnmA antiserum (1 : 10 dilution) in human coronary artery endothelial cell invasion by Streptococcus mutans OMZ175 was evaluated using the antibiotic protection assay. (D) Killing of Galleria mellonella larvae infected with OMZ175 with or without anti-rCnmA (1 : 10 dilution). Refer to Methods section for additional details. *P < 0.05.
DISCUSSION
Bacterial adhesion to components of host ECM such as collagen, fibronectin and laminin are known to contribute to the pathogenesis of a variety of diseases (Hienz et al., 1996; Lamont & Jenkinson, 2000; Chhatwal, 2002; Mintz, 2004; Nobbs et al., 2009; Konkel et al., 2010; Edwards & Butler, 2011). In this study, we investigated the role of an infrequent gene cluster (cnaB-cbpA-cnm) found in a subset of S. mutans strains encoding three putative surface-associated proteins with conserved collagen binding domains in each of them. Sequencing analysis revealed that the genetic arrangement and genomic location of cnaB-cbpA-cnm was conserved among all invasive strains in our collection as well as in strains with full genome sequences available (Aikawa et al., 2012; Song et al., 2012; Palmer et al., 2013). Previous studies revealed that expression of cnm is directly linked to the ability of S. mutans to avidly bind to collagen and laminin and to adhere to and invade endothelial cells (Sato et al., 2004; Abranches et al., 2011; Nomura et al., 2012; Lapirattanakul et al., 2013).
Given the genetic proximity of cnaB and cbpA with cnm and the fact that all three genes are predicted to encode proteins with conserved collagen-binding-like domains, we hypothesized that CnaB and CbpA were functionally linked to Cnm. However, phenotypic characterization of strains lacking cnaB, cbpA or both suggests that CnaB or CbpA, if they are produced, do not contribute to ECM binding, HCAEC invasion or virulence in G. mellonella. Likewise, expression of cnaB and cbpA in the non-invasive cnm− UA159 strain did not enhance ECM-binding, HCAEC invasion or virulence in G. mellonella. Unexpectedly, the ΔcnaB strain showed enhanced collagen- and laminin- binding properties as well as increased ability to invade HCAEC. At this time, the reason for the ΔcnaB phenotypes and their amelioration in the ΔcnaBΔcbpA double mutant raises the possibility of balancing regulatory functions between these two genes. In contrast, expression of cnm in UA159 resulted in enhanced collagen-binding and invasive phenotypes as well as increased virulence in G. mellonella.
Despite the absence of Cnm-related phenotypes, the ΔcbpA mutant grew significantly more slowly than the parent strain whereas the ΔcnaB strain formed larger and elongated cells. Interestingly, the growth defect and cell morphology phenotypes were no longer observed in a ΔcnaBΔcbpA double mutant strain. Although the specific roles of CnaB and CbpA remain unclear, our results strongly suggest that these two putative proteins are not functionally associated with Cnm. Nevertheless, the invariable presence of the cnaB-cbpA-cnm cluster at a conserved site in the genome of invasive S. mutans strains raises an important evolutionary question that merits further investigation. Hence, it is possible that CnaB and CbpA are associated with functions that were not analysed in our study.
Collectively, our results indicate that Cnm is the major mediator of ECM binding and intracellular invasion of HCAEC, and that strains expressing Cnm are potentially better armed to infect non-oral sites of the human host. Immunoelectron microscopy analysis confirmed that Cnm is localized at the cell surface and, therefore, behaves as a typical cell wall-associated adhesin. By expressing cnm in the non-invasive UA159 strain and by using anti-rCnmA antibodies to block the Cnm-related phenotypes, we were able to provide further evidence that Cnm is an important virulence factor. Our results show that intracellular invasion of HCAEC and virulence of Cnm+ strains in G. mellonella can be significantly attenuated by co-inoculation with the anti-rCnmA antibody, or when the antibody was supplied 4 h post-infection. Along these lines, immunizations with closely related collagen-binding proteins from Enterococcus faecalis and Staphylococcus aureus, Ace and Cna, respectively, have been successfully used to protect rodents against subsequent infection with each corresponding organism (Nilsson et al., 1998; Singh et al., 2010). Therefore, Cnm may also be a suitable target for the development of preventive strategies against invasive strains of S. mutans.
ACKNOWLEDGEMENTS
This study was supported by the American Heart Association grant 10GRNT4210049 and by NIH-NIDCR R01 DE022559. AAR was supported by the NIDCR training grant in oral sciences T90 DE021985.
REFERENCES
- Abranches J, Zeng L, Belanger M, et al. Invasion of human coronary artery endothelial cells by Streptococcus mutans OMZ175. Oral Microbiol Immunol. 2009;24:141–145. doi: 10.1111/j.1399-302X.2008.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches J, Miller JH, Martinez AR, Simpson-Haidaris PJ, Burne RA, Lemos JA. The collagen-binding protein Cnm is required for Streptococcus mutans adherence to and intracellular invasion of human coronary artery endothelial cells. Infect Immun. 2011;79:2277–2284. doi: 10.1128/IAI.00767-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa C, Furukawa N, Watanabe T, et al. Complete genome sequence of the serotype k Streptococcus mutans strain LJ23. J Bacteriol. 2012;194:2754–2755. doi: 10.1128/JB.00350-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, McShan WM, McLaughlin RE, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg AM, Jones MN, Miller-Torbert T, Holt RG. Binding of Streptococcus mutans to extracellular matrix molecules and fibrinogen. Biochem Biophys Res Commun. 2002;298:75–79. doi: 10.1016/s0006-291x(02)02390-2. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Maddocks SE, Larson MR, et al. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol Microbiol. 2010;77:276–286. doi: 10.1111/j.1365-2958.2010.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette-Curley D, Wells V, Viscount H, et al. FimA, a major virulence factor associated with Streptococcus parasanguis endocarditis. Infect Immun. 1995;63:4669–4674. doi: 10.1128/iai.63.12.4669-4674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhatwal GS. Anchorless adhesins and invasions of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol. 2002;10:205–208. doi: 10.1016/s0966-842x(02)02351-x. [DOI] [PubMed] [Google Scholar]
- Chia JS, Yeh CY, Chen JY. Identification of a fibronectin binding protein from Streptococcus mutans. Infect Immun. 2000;68:1864–1870. doi: 10.1128/iai.68.4.1864-1870.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo OE, Lefébure T, Pavinski-Bitar PD, et al. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol Biol Evol. 2013;30:881–893. doi: 10.1093/molbev/mss278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JL, Butler EK. The Pathobiology of Neisseria gonorrhoeae Lower Female Genital Tract Infection. Front Microbiol. 2011;2:102. doi: 10.3389/fmicb.2011.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TK, Zhang C, Dao ML. Identification and characterization of collagen-binding activity in Streptococcus mutans wall-associated protein: a possible implication in dental root caries and endocarditis. Biochem Biophys Res Commun. 2006;343:787–792. doi: 10.1016/j.bbrc.2006.03.025. [DOI] [PubMed] [Google Scholar]
- Hienz SA, Schennings T, Heimdahl A, Flock JI. Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. J Infect Dis. 1996;174:83–88. doi: 10.1093/infdis/174.1.83. [DOI] [PubMed] [Google Scholar]
- Kajfasz JK, Rivera-Ramos I, Abranches J, et al. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol. 2010;192:2546–2556. doi: 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavalu L, Lucas AR, Verma RK, et al. Increased atherogenesis during Streptococcus mutans infection in ApoE-null mice. J Dent Res. 2012;91:255–260. doi: 10.1177/0022034511435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel ME, Larson CL, Flanagan RC. Campylobacter jejuni FlpA binds fibronectin and is required for maximal host cell adherence. J Bacteriol. 2010;192:68–76. doi: 10.1128/JB.00969-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer BH, van der Kraan M, Crowley PJ, Hamilton IR, Brady LJ, Bleiweis AS. Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J Bacteriol. 2001;18:2543–2552. doi: 10.1128/JB.183.8.2543-2552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol Immunol. 2000;15:341–349. doi: 10.1034/j.1399-302x.2000.150601.x. [DOI] [PubMed] [Google Scholar]
- Lapirattanakul J, Nomura R, Nemoto H, Naka S, Ooshima T, Nakano K. Multilocus sequence typing of Streptococcus mutans strains with the cbm gene encoding a novel collagen-binding protein. Arch Oral Biol. 2013;58:989–996. doi: 10.1016/j.archoralbio.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Lee SY, Kim KK, Choe SJ. Binding of oral streptococci to human fibrinogen. Oral Microbiol Immunol. 2001;16:88–93. doi: 10.1034/j.1399-302x.2001.016002088.x. [DOI] [PubMed] [Google Scholar]
- Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz KP. Identification of an extracellular matrix protein adhesin, EmaA, which mediates the adhesion of Actinobacillus actinomycetemcomitans to collagen. Microbiology. 2004;150:2677–2688. doi: 10.1099/mic.0.27110-0. [DOI] [PubMed] [Google Scholar]
- Mylonakis E, Calderwood SB. Infective endocarditis in adults. N Engl J Med. 2001;345:1318–1330. doi: 10.1056/NEJMra010082. [DOI] [PubMed] [Google Scholar]
- Nagata E, Okayama H, Ito HO, Yamashita Y, Inouw M, Oho T. Serotype-specific polysaccharide of Streptococcus mutans contributes to infectivity in endocarditis. Oral Microbiol Immunol. 2006;21:420–423. doi: 10.1111/j.1399-302X.2006.00317.x. [DOI] [PubMed] [Google Scholar]
- Nakano K, Inaba H, Nomura R, et al. Detection of cariogenic Streptococcus mutans in extirpated heart valve and atheromatous plaque specimens. J Clin Microbiol. 2006;44:3313–3317. doi: 10.1128/JCM.00377-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Lapirattanakul J, Nomura R, et al. Streptococcus mutans clonal variation revealed by multilocus sequence typing. J Clin Microbiol. 2007a;45:2616–2625. doi: 10.1128/JCM.02343-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Nemoto H, Nomura R, et al. Serotype distribution of Streptococcus mutans a pathogen of dental caries in cardiovascular specimens from Japanese patients. J Med Microbiol. 2007b;56:551–556. doi: 10.1099/jmm.0.47051-0. [DOI] [PubMed] [Google Scholar]
- Nakano K, Nomura R, Ooshiao T. Streptococcus mutans and cardiovascular diseases. Jpn Dent Sci Rev. 2008;44:29–37. [Google Scholar]
- Nakano K, Nomura R, Taniguchi N, et al. Molecular characterization of Streptococcus mutans strains containing the cnm gene encoding a collagen-binding adhesin. Arch Oral Biol. 2010;55:34–39. doi: 10.1016/j.archoralbio.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Nakano K, Hokamura K, Taniguchi N, et al. The collagen-binding protein of Streptococcus mutans is involved in haemorrhagic stroke. Nat Commun. 2011;2:485–494. doi: 10.1038/ncomms1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson IM, Patti JM, Bremell T, Hook M, Tarkowski A. Vaccination with a recombinant fragment of collagen adhesin provides protection against Staphylococcus aureus-mediated septic death. J Clin Invest. 1998;101:2640–2649. doi: 10.1172/JCI1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura R, Nakano K, Taniguchi N, et al. Molecular and clinical analyses of the gene encoding the collagen-binding adhesin of Streptococcus mutans. J Med Microbiol. 2009;58:469–475. doi: 10.1099/jmm.0.007559-0. [DOI] [PubMed] [Google Scholar]
- Nomura R, Nakano K, Naka S, et al. Identification and characterization of a collagen-binding protein, Cbm, in Streptococcus mutans. Mol Oral Microbiol. 2012;27:308–323. doi: 10.1111/j.2041-1014.2012.00649.x. [DOI] [PubMed] [Google Scholar]
- Palmer SR, Miller JH, Abranches J, et al. Phenotypic heterogeneity of genomically-diverse isolates of Streptococcus mutans. PLoS ONE. 2013;8:e61358. doi: 10.1371/journal.pone.0061358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Okamoto K, Kagami A, Yamamoto Y, Igarashi T, Kizaki H. Streptococcus mutans strains harboring collagen-binding adhesin. J Dent Res. 2004;83:534–539. doi: 10.1177/154405910408300705. [DOI] [PubMed] [Google Scholar]
- Seo HS, Xiong YQ, Mitchell J, Seepersaud R, Bayer AS, Sullam PM. Bacteriophage lysin mediates the binding of Streptococcus mitis to human platelets through interaction with fibrinogen. PLoS Pathog. 2010;6:e1001047. doi: 10.1371/journal.ppat.1001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KV, Nallapareddy SR, Sillanpaa J, Murray BE. Importance of the collagen adhesin ace in pathogenesis and protection against Enterococcus faecalis experimental endocarditis. PLoS Pathog. 2010;6:e1000716. doi: 10.1371/journal.ppat.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Sudhakar P, Wang W, et al. A genome-wide study of two-component signal transduction systems in eight newly sequenced mutans streptococci strains. BMC Genomics. 2012;13:128–147. doi: 10.1186/1471-2164-13-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong YQ, Bensing BA, Bayer AS, Chambers HF, Sullam PM. Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis. Microb Pathog. 2008;45:297–301. doi: 10.1016/j.micpath.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Burne RA. Transcriptional regulation of the cellobiose operon of Streptococcus mutans. J Bacteriol. 2009;191:2153–2162. doi: 10.1128/JB.01641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Kreth J, Cross SE, Gimzewski JK, Shi W, Qi F. Functional characterization of cell-wall-associated protein WapA in Streptococcus mutans. Microbiology. 2006;152:2395–2404. doi: 10.1099/mic.0.28883-0. [DOI] [PubMed] [Google Scholar]