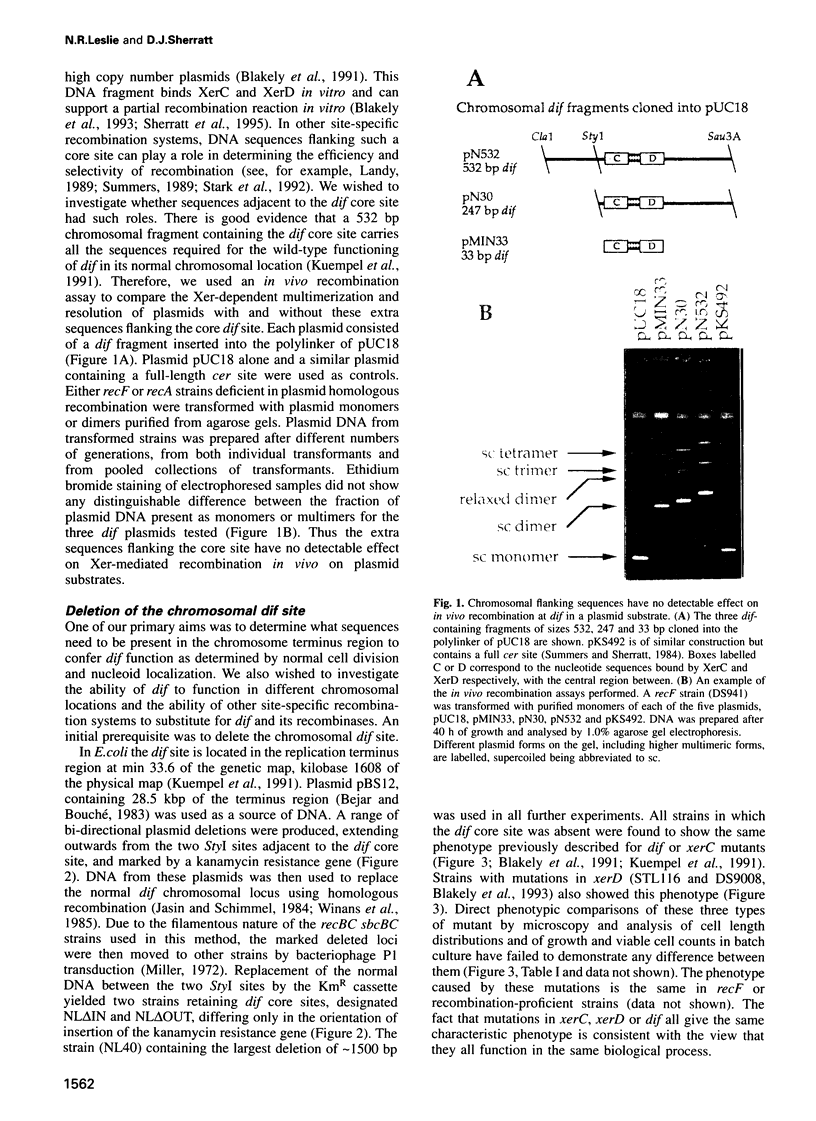
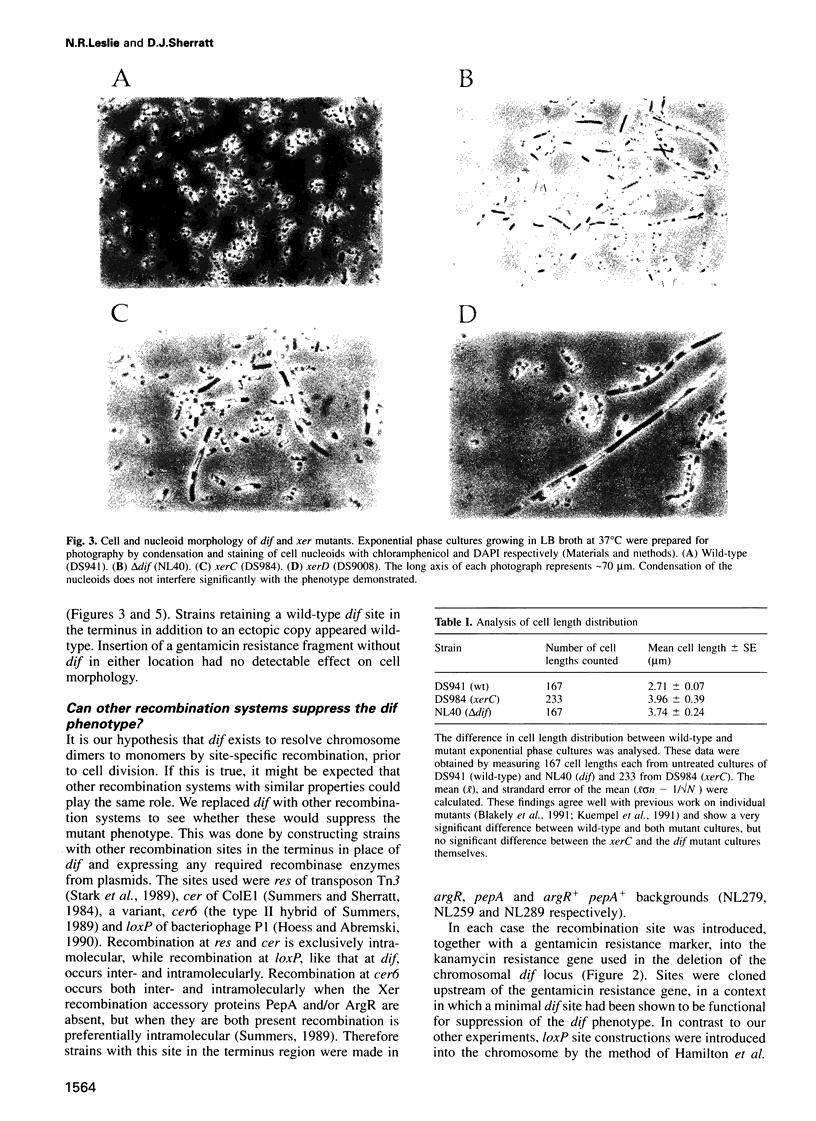
Abstract
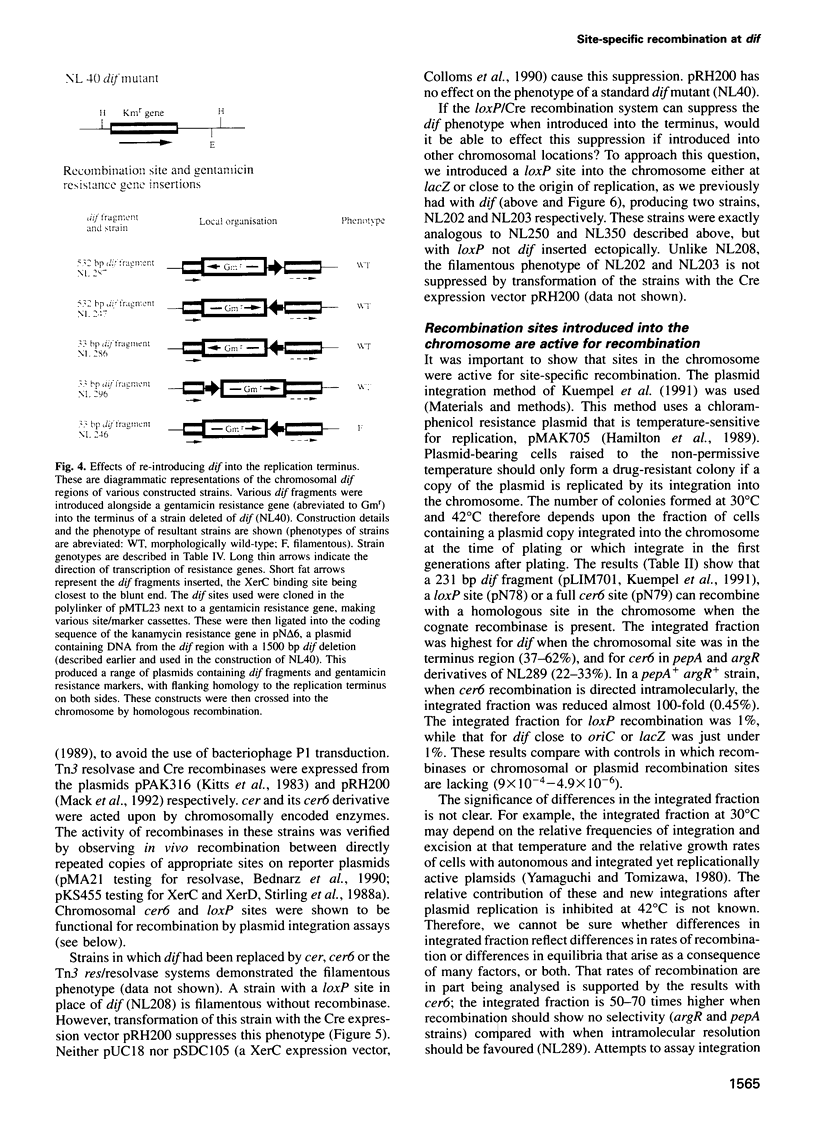
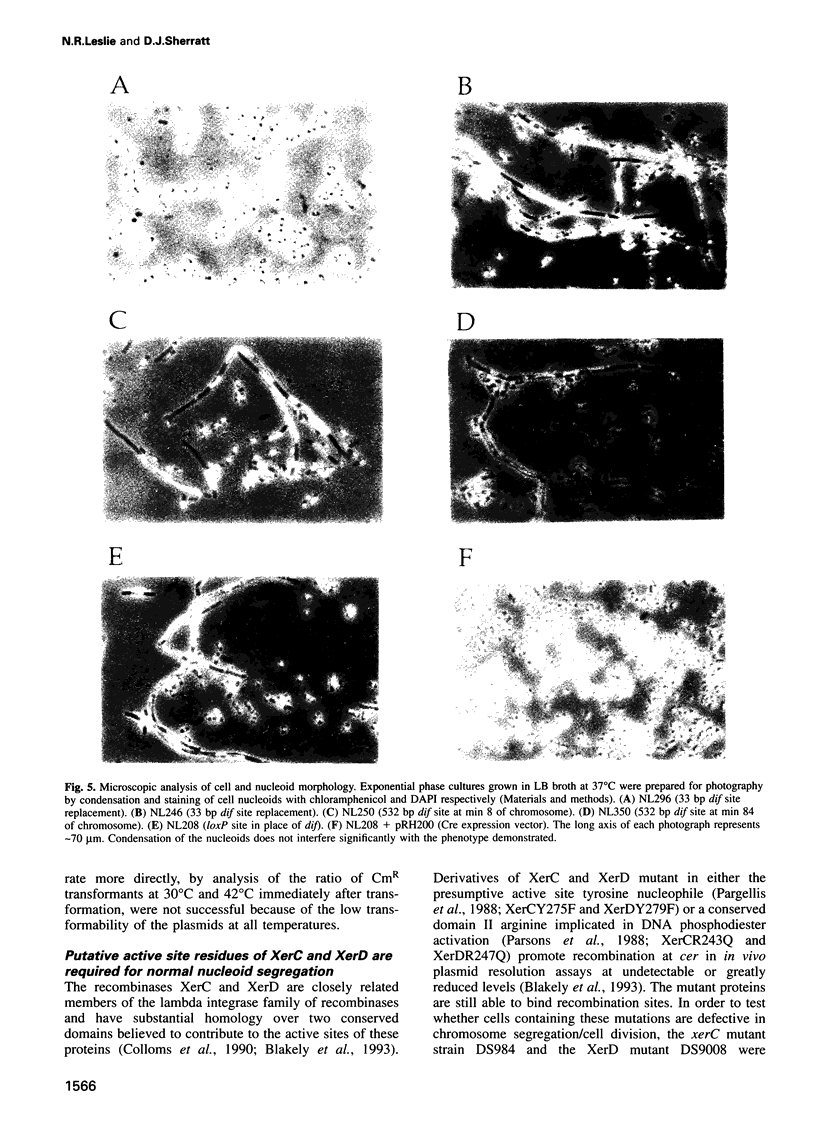
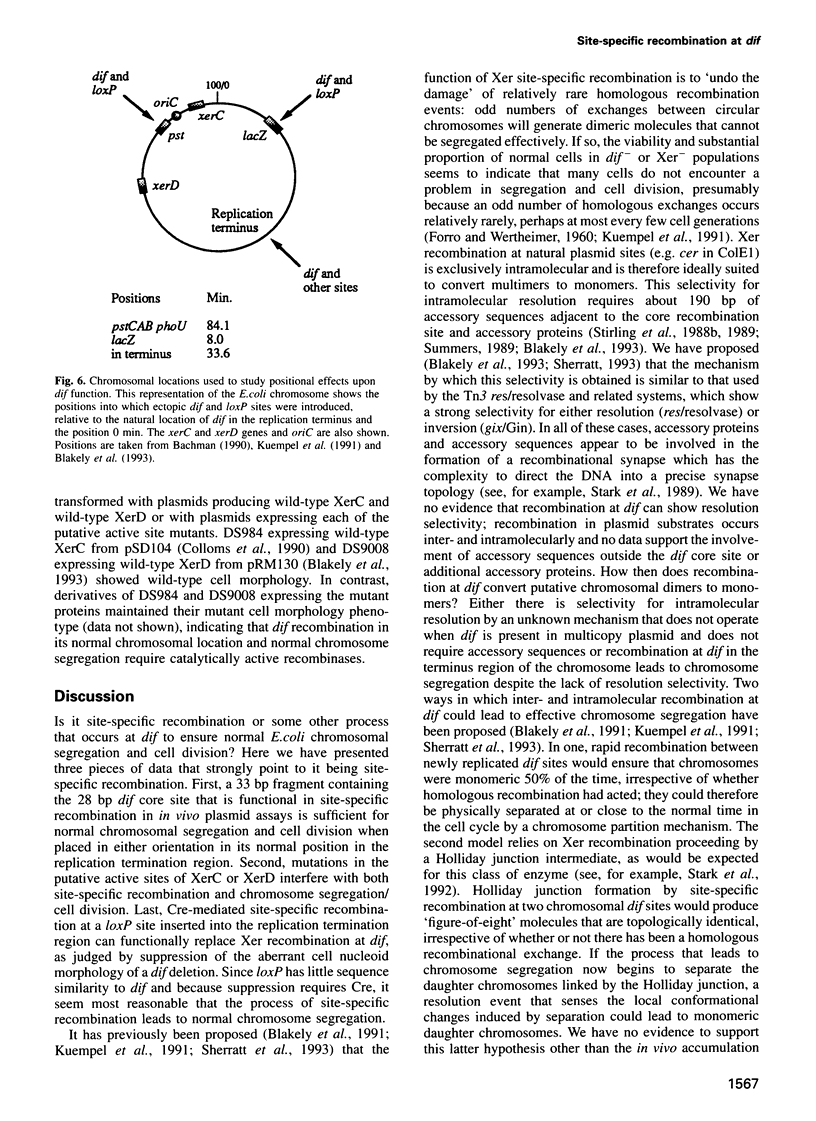
The replication terminus region of the Escherichia coli chromosome encodes a locus, dif, that is required for normal chromosome segregation at cell division. dif is a substrate for site-specific recombination catalysed by the related chromosomally encoded recombinases XerC and XerD. It has been proposed that this recombination converts chromosome multimers formed by homologous recombination back to monomers in order that they can be segregated prior to cell division. Strains mutant in dif, xerC or xerD share a characteristic phenotype, containing a variable fraction of filamentous cells with aberrantly positioned and sized nucleoids. We show that the only DNA sequences required for wild-type dif function in the terminus region of the chromosome are contained within 33 bp known to bind XerC and XerD and that putative active site residues of the Xer recombinases are required for normal chromosome segregation. We have also shown that recombination by the loxP/Cre system of bacteriophage P1 will suppress the phenotype of a dif deletion strain when loxP is inserted in the terminus region. Suppression of the dif deletion phenotype did not occur when either dif/Xer or loxP/Cre recombination acted at other positions in the chromosome close to oriC or within lacZ, indicating that site-specific recombination must occur within the replication terminus region in order to allow normal chromosome segregation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amemura M., Shinagawa H., Makino K., Otsuji N., Nakata A. Cloning of and complementation tests with alkaline phosphatase regulatory genes (phoS and phoT) of Escherichia coli. J Bacteriol. 1982 Nov;152(2):692–701. doi: 10.1128/jb.152.2.692-701.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S., Ziese M., Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981 Sep;25(3):729–736. doi: 10.1016/0092-8674(81)90180-x. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarz A. L., Boocock M. R., Sherratt D. J. Determinants of correct res site alignment in site-specific recombination by Tn3 resolvase. Genes Dev. 1990 Dec;4(12B):2366–2375. doi: 10.1101/gad.4.12b.2366. [DOI] [PubMed] [Google Scholar]
- Blakely G., Colloms S., May G., Burke M., Sherratt D. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991 Aug;3(8):789–798. [PubMed] [Google Scholar]
- Blakely G., May G., McCulloch R., Arciszewska L. K., Burke M., Lovett S. T., Sherratt D. J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993 Oct 22;75(2):351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- Béjar S., Bouché J. P. Molecular cloning of the region of the terminus of Escherichia coli K-12 DNA replication. J Bacteriol. 1983 Feb;153(2):604–609. doi: 10.1128/jb.153.2.604-609.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S. P., Prior S. E., Barstow D. A., Minton N. P. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988 Aug 15;68(1):139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- Colloms S. D., Sykora P., Szatmari G., Sherratt D. J. Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J Bacteriol. 1990 Dec;172(12):6973–6980. doi: 10.1128/jb.172.12.6973-6980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson A., Bernander R., Dasgupta S., Nordström K. Direct visualization of plasmid DNA in bacterial cells. Mol Microbiol. 1992 Jan;6(2):165–170. doi: 10.1111/j.1365-2958.1992.tb01997.x. [DOI] [PubMed] [Google Scholar]
- FORRO F., Jr, WERTHEIMER S. A. The organization and replication of deoxyribonucleic acid in thymine-deficient strains of Escherichia coli. Biochim Biophys Acta. 1960 May 6;40:9–21. doi: 10.1016/0006-3002(60)91310-x. [DOI] [PubMed] [Google Scholar]
- Hamilton C. M., Aldea M., Washburn B. K., Babitzke P., Kushner S. R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989 Sep;171(9):4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S., Niki H., Ogura T., Ichinose C., Mori H., Ezaki B., Jaffé A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989 Mar;171(3):1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M., Schimmel P. Deletion of an essential gene in Escherichia coli by site-specific recombination with linear DNA fragments. J Bacteriol. 1984 Aug;159(2):783–786. doi: 10.1128/jb.159.2.783-786.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitts P. A., Symington L. S., Dyson P., Sherratt D. J. Transposon-encoded site-specific recombination: nature of the Tn3 DNA sequences which constitute the recombination site res. EMBO J. 1983;2(7):1055–1060. doi: 10.1002/j.1460-2075.1983.tb01545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuempel P. L., Henson J. M., Dircks L., Tecklenburg M., Lim D. F. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 1991 Aug;3(8):799–811. [PubMed] [Google Scholar]
- Kuempel P. L., Pelletier A. J., Hill T. M. Tus and the terminators: the arrest of replication in prokaryotes. Cell. 1989 Nov 17;59(4):581–583. doi: 10.1016/0092-8674(89)90001-9. [DOI] [PubMed] [Google Scholar]
- Kushner S. R., Nagaishi H., Templin A., Clark A. J. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc Natl Acad Sci U S A. 1971 Apr;68(4):824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Buckman C. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J Bacteriol. 1985 Nov;164(2):836–844. doi: 10.1128/jb.164.2.836-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack A., Sauer B., Abremski K., Hoess R. Stoichiometry of the Cre recombinase bound to the lox recombining site. Nucleic Acids Res. 1992 Sep 11;20(17):4451–4455. doi: 10.1093/nar/20.17.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch R., Coggins L. W., Colloms S. D., Sherratt D. J. Xer-mediated site-specific recombination at cer generates Holliday junctions in vivo. EMBO J. 1994 Apr 15;13(8):1844–1855. doi: 10.1002/j.1460-2075.1994.tb06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B., Pospiech A., Schairer H. U. Rapid isolation of genomic DNA from gram-negative bacteria. Trends Genet. 1992 Oct;8(10):332–333. doi: 10.1016/0168-9525(92)90269-a. [DOI] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Parsons R. L., Prasad P. V., Harshey R. M., Jayaram M. Step-arrest mutants of FLP recombinase: implications for the catalytic mechanism of DNA recombination. Mol Cell Biol. 1988 Aug;8(8):3303–3310. doi: 10.1128/mcb.8.8.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt D. J., Arciszewska L. K., Blakely G., Colloms S., Grant K., Leslie N., McCulloch R. Site-specific recombination and circular chromosome segregation. Philos Trans R Soc Lond B Biol Sci. 1995 Jan 30;347(1319):37–42. doi: 10.1098/rstb.1995.0006. [DOI] [PubMed] [Google Scholar]
- Stark W. M., Boocock M. R., Sherratt D. J. Catalysis by site-specific recombinases. Trends Genet. 1992 Dec;8(12):432–439. [PubMed] [Google Scholar]
- Stark W. M., Boocock M. R., Sherratt D. J. Site-specific recombination by Tn3 resolvase. Trends Genet. 1989 Sep;5(9):304–309. doi: 10.1016/0168-9525(89)90113-3. [DOI] [PubMed] [Google Scholar]
- Stirling C. J., Colloms S. D., Collins J. F., Szatmari G., Sherratt D. J. xerB, an Escherichia coli gene required for plasmid ColE1 site-specific recombination, is identical to pepA, encoding aminopeptidase A, a protein with substantial similarity to bovine lens leucine aminopeptidase. EMBO J. 1989 May;8(5):1623–1627. doi: 10.1002/j.1460-2075.1989.tb03547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling C. J., Stewart G., Sherratt D. J. Multicopy plasmid stability in Escherichia coli requires host-encoded functions that lead to plasmid site-specific recombination. Mol Gen Genet. 1988 Sep;214(1):80–84. doi: 10.1007/BF00340183. [DOI] [PubMed] [Google Scholar]
- Summers D. K., Beton C. W., Withers H. L. Multicopy plasmid instability: the dimer catastrophe hypothesis. Mol Microbiol. 1993 Jun;8(6):1031–1038. doi: 10.1111/j.1365-2958.1993.tb01648.x. [DOI] [PubMed] [Google Scholar]
- Summers D. K. Derivatives of ColE1 cer show altered topological specificity in site-specific recombination. EMBO J. 1989 Jan;8(1):309–315. doi: 10.1002/j.1460-2075.1989.tb03378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. K., Sherratt D. J. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984 Apr;36(4):1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- Summers D. K., Sherratt D. J. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 1988 Mar;7(3):851–858. doi: 10.1002/j.1460-2075.1988.tb02884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wanner B. L., Latterell P. Mutants affected in alkaline phosphatase, expression: evidence for multiple positive regulators of the phosphate regulon in Escherichia coli. Genetics. 1980 Oct;96(2):353–366. doi: 10.1093/genetics/96.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C., Elledge S. J., Krueger J. H., Walker G. C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985 Mar;161(3):1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Tomizawa J. Establishment of Escherichia coli cells with an integrated high copy number plasmid. Mol Gen Genet. 1980;178(3):525–533. doi: 10.1007/BF00337857. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]