Abstract
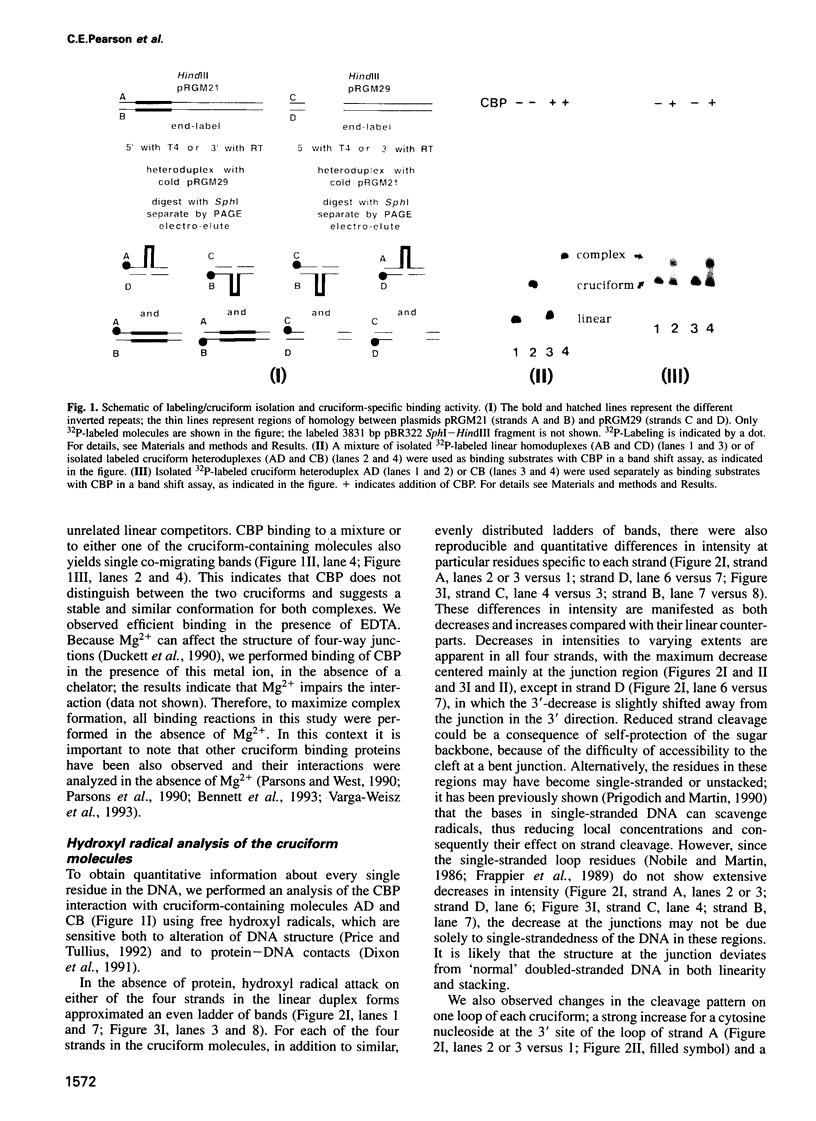
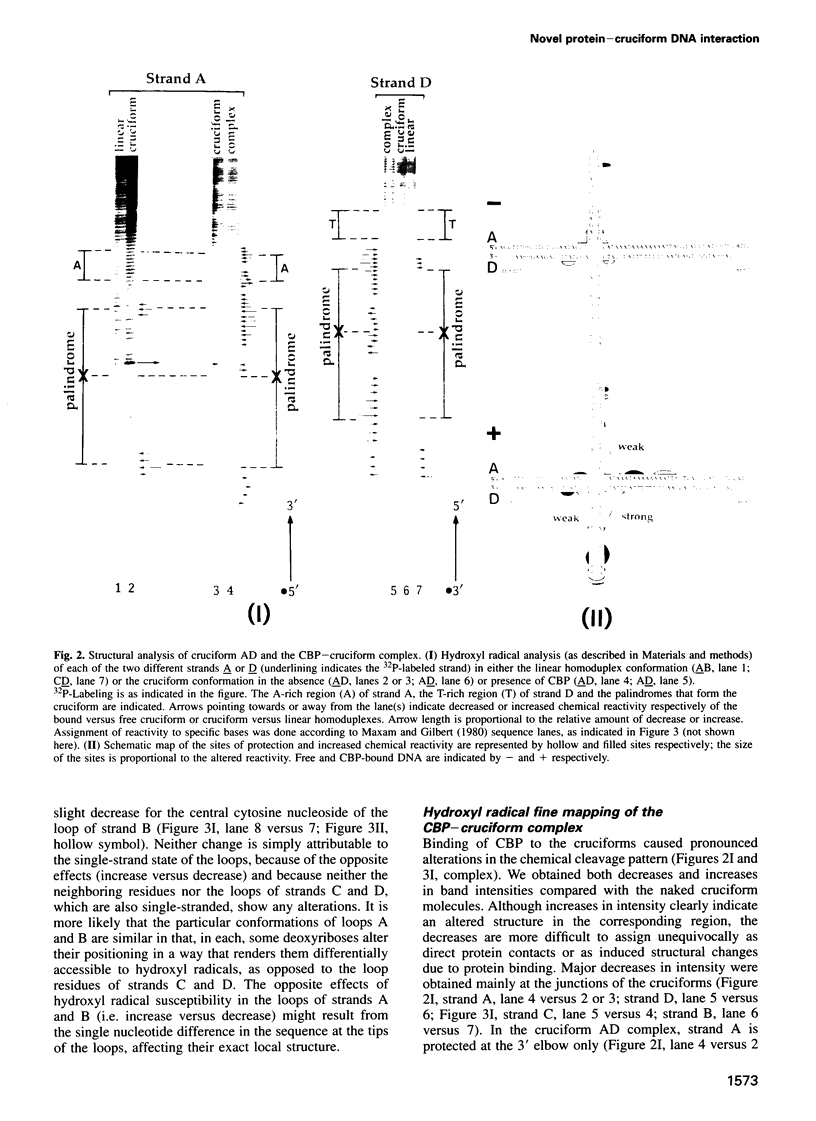
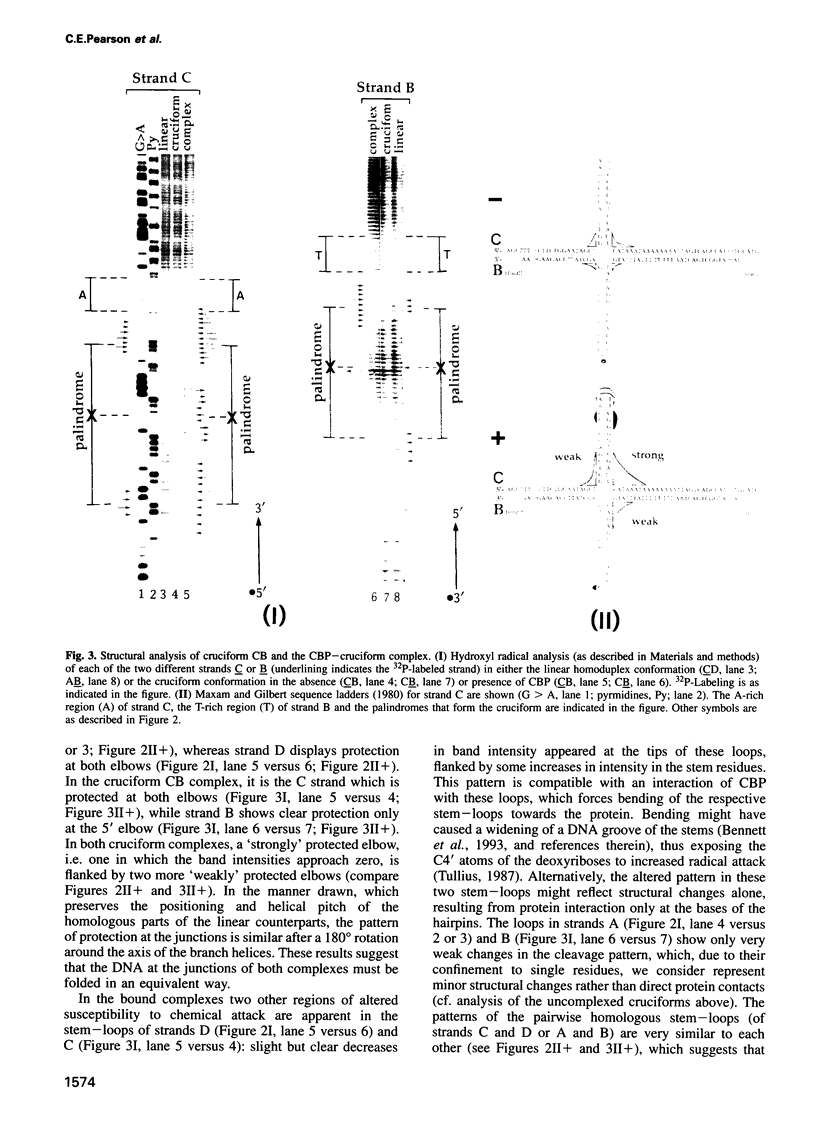
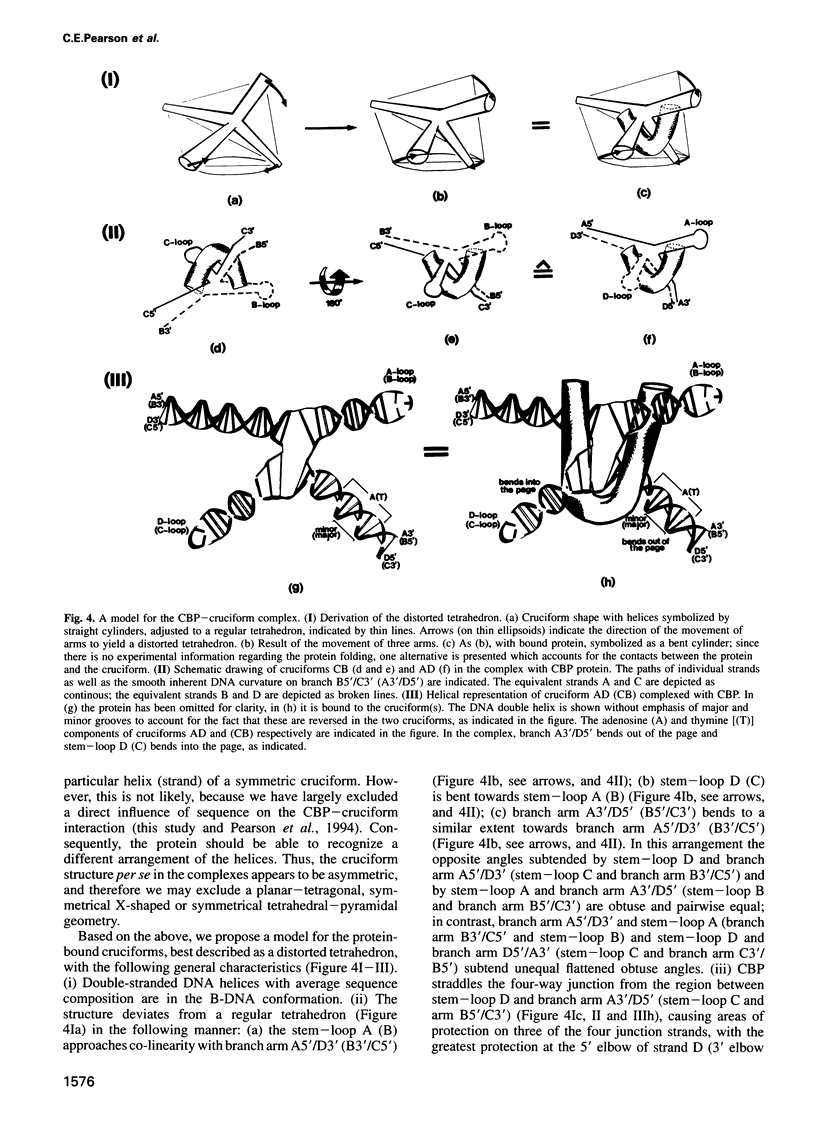
We recently identified and enriched a protein (CBP) from HeLa cells with binding specificity for cruciform-containing DNA. We have now studied the interaction of CBP with stable cruciform DNA molecules containing the 27 bp palindrome of SV40 on one strand and an unrelated 26 bp palindrome on the other strand by hydroxyl radical footprinting. The CBP-DNA interaction is localized to the four-way junction at the base of the cruciforms. CBP appears to interact with the elbows of the junctions in an asymmetric fashion. Upon CBP binding, structural distortions were observed in the cruciform stems and in a DNA region adjacent to the junction. These features distinguish CBP from other cruciform binding proteins, which bind symmetrically and display exclusively either contacts with the DNA backbone or structural alterations in the DNA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell D., Sabloff M., Zannis-Hadjopoulos M., Price G. Anti-cruciform DNA affinity purification of active mammalian origins of replication. Biochim Biophys Acta. 1991 Jul 23;1089(3):299–308. doi: 10.1016/0167-4781(91)90169-m. [DOI] [PubMed] [Google Scholar]
- Bennett R. J., Dunderdale H. J., West S. C. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993 Sep 24;74(6):1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A., Murchie A. I., von Kitzing E., Diekmann S., Kemper B., Lilley D. M. Model for the interaction of DNA junctions and resolving enzymes. J Mol Biol. 1991 Oct 20;221(4):1191–1207. doi: 10.1016/0022-2836(91)90928-y. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Beltrame M., Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- Blommers M. J., van de Ven F. J., van der Marel G. A., van Boom J. H., Hilbers C. W. The three-dimensional structure of a DNA hairpin in solution two-dimensional NMR studies and structural analysis of d(ATCCTATTTATAGGAT). Eur J Biochem. 1991 Oct 1;201(1):33–51. doi: 10.1111/j.1432-1033.1991.tb16253.x. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 1988 Oct;7(10):3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. Structural details of an adenine tract that does not cause DNA to bend. Nature. 1988 Feb 4;331(6155):455–457. doi: 10.1038/331455a0. [DOI] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Chen J. H., Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. Construction and analysis of monomobile DNA junctions. Biochemistry. 1988 Aug 9;27(16):6032–6038. doi: 10.1021/bi00416a031. [DOI] [PubMed] [Google Scholar]
- Clegg R. M., Murchie A. I., Zechel A., Carlberg C., Diekmann S., Lilley D. M. Fluorescence resonance energy transfer analysis of the structure of the four-way DNA junction. Biochemistry. 1992 May 26;31(20):4846–4856. doi: 10.1021/bi00135a016. [DOI] [PubMed] [Google Scholar]
- Deb S., DeLucia A. L., Koff A., Tsui S., Tegtmeyer P. The adenine-thymine domain of the simian virus 40 core origin directs DNA bending and coordinately regulates DNA replication. Mol Cell Biol. 1986 Dec;6(12):4578–4584. doi: 10.1128/mcb.6.12.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon W. J., Hayes J. J., Levin J. R., Weidner M. F., Dombroski B. A., Tullius T. D. Hydroxyl radical footprinting. Methods Enzymol. 1991;208:380–413. doi: 10.1016/0076-6879(91)08021-9. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Lilley D. M. Effects of base mismatches on the structure of the four-way DNA junction. J Mol Biol. 1991 Sep 5;221(1):147–161. doi: 10.1016/0022-2836(91)80211-c. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Lilley D. M. The role of metal ions in the conformation of the four-way DNA junction. EMBO J. 1990 Feb;9(2):583–590. doi: 10.1002/j.1460-2075.1990.tb08146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einck L., Bustin M. The intracellular distribution and function of the high mobility group chromosomal proteins. Exp Cell Res. 1985 Feb;156(2):295–310. doi: 10.1016/0014-4827(85)90539-7. [DOI] [PubMed] [Google Scholar]
- Frappier L., Price G. B., Martin R. G., Zannis-Hadjopoulos M. Characterization of the binding specificity of two anticruciform DNA monoclonal antibodies. J Biol Chem. 1989 Jan 5;264(1):334–341. [PubMed] [Google Scholar]
- Frappier L., Price G. B., Martin R. G., Zannis-Hadjopoulos M. Monoclonal antibodies to cruciform DNA structures. J Mol Biol. 1987 Feb 20;193(4):751–758. doi: 10.1016/0022-2836(87)90356-1. [DOI] [PubMed] [Google Scholar]
- Germann M. W., Kalisch B. W., Lundberg P., Vogel H. J., van de Sande J. H. Perturbation of DNA hairpins containing the EcoRI recognition site by hairpin loops of varying size and composition: physical (NMR and UV) and enzymatic (EcoRI) studies. Nucleic Acids Res. 1990 Mar 25;18(6):1489–1498. doi: 10.1093/nar/18.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Bustin M. Hierarchy of binding sites for chromosomal proteins HMG 1 and 2 in supercoiled deoxyribonucleic acid. Biochemistry. 1985 Mar 12;24(6):1428–1433. doi: 10.1021/bi00327a022. [DOI] [PubMed] [Google Scholar]
- Hand R. Eucaryotic DNA: organization of the genome for replication. Cell. 1978 Oct;15(2):317–325. doi: 10.1016/0092-8674(78)90001-6. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Tullius T. D. The missing nucleoside experiment: a new technique to study recognition of DNA by protein. Biochemistry. 1989 Nov 28;28(24):9521–9527. doi: 10.1021/bi00450a041. [DOI] [PubMed] [Google Scholar]
- Klein H. L., Welch S. K. Inverted repeated sequences in yeast nuclear DNA. Nucleic Acids Res. 1980 Oct 24;8(20):4651–4669. doi: 10.1093/nar/8.20.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of the four-way junction in DNA. Annu Rev Biophys Biomol Struct. 1993;22:299–328. doi: 10.1146/annurev.bb.22.060193.001503. [DOI] [PubMed] [Google Scholar]
- Lin L. S., Meyer R. J. DNA synthesis is initiated at two positions within the origin of replication of plasmid R1162. Nucleic Acids Res. 1987 Oct 26;15(20):8319–8331. doi: 10.1093/nar/15.20.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Clegg R. M., von Kitzing E., Duckett D. R., Diekmann S., Lilley D. M. Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature. 1989 Oct 26;341(6244):763–766. doi: 10.1038/341763a0. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Portugal J., Lilley D. M. Cleavage of a four-way DNA junction by a restriction enzyme spanning the point of strand exchange. EMBO J. 1991 Mar;10(3):713–718. doi: 10.1002/j.1460-2075.1991.tb08001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U. R., Fitch W. M. Evolutionary selection for perfect hairpin structures in viral DNAs. Nature. 1982 Aug 5;298(5874):582–585. doi: 10.1038/298582a0. [DOI] [PubMed] [Google Scholar]
- Nobile C., Martin R. G. Stable stem-loop and cruciform DNA structures: isolation of mutants with rearrangements of the palindromic sequence at the simian virus 40 replication origin. Intervirology. 1986;25(3):158–171. doi: 10.1159/000149671. [DOI] [PubMed] [Google Scholar]
- Noirot P., Bargonetti J., Novick R. P. Initiation of rolling-circle replication in pT181 plasmid: initiator protein enhances cruciform extrusion at the origin. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8560–8564. doi: 10.1073/pnas.87.21.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofverstedt L. G., Hammarström K., Balgobin N., Hjertén S., Pettersson U., Chattopadhyaya J. Rapid and quantitative recovery of DNA fragments from gels by displacement electrophoresis (isotachophoresis). Biochim Biophys Acta. 1984 Jun 16;782(2):120–126. doi: 10.1016/0167-4781(84)90014-9. [DOI] [PubMed] [Google Scholar]
- Parsons C. A., Kemper B., West S. C. Interaction of a four-way junction in DNA with T4 endonuclease VII. J Biol Chem. 1990 Jun 5;265(16):9285–9289. [PubMed] [Google Scholar]
- Parsons C. A., West S. C. Specificity of binding to four-way junctions in DNA by bacteriophage T7 endonuclease I. Nucleic Acids Res. 1990 Aug 11;18(15):4377–4384. doi: 10.1093/nar/18.15.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. E., Frappier L., Zannis-Hadjopoulos M. Plasmids bearing mammalian DNA-replication origin-enriched (ors) fragments initiate semiconservative replication in a cell-free system. Biochim Biophys Acta. 1991 Oct 8;1090(2):156–166. doi: 10.1016/0167-4781(91)90096-5. [DOI] [PubMed] [Google Scholar]
- Platt J. R. POSSIBLE SEPARATION OF INTERTWINED NUCLEIC ACID CHAINS BY TRANSFER-TWIST. Proc Natl Acad Sci U S A. 1955 Mar 15;41(3):181–183. doi: 10.1073/pnas.41.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue G. P., Hall T. C. The requirement for a 5' stem-loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J Virol. 1992 Feb;66(2):674–684. doi: 10.1128/jvi.66.2.674-684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. A., Tullius T. D. Using hydroxyl radical to probe DNA structure. Methods Enzymol. 1992;212:194–219. doi: 10.1016/0076-6879(92)12013-g. [DOI] [PubMed] [Google Scholar]
- Prigodich R. V., Martin C. T. Reaction of single-stranded DNA with hydroxyl radical generated by iron(II)-ethylenediaminetetraacetic acid. Biochemistry. 1990 Sep 4;29(35):8017–8019. doi: 10.1021/bi00487a003. [DOI] [PubMed] [Google Scholar]
- Schreck R., Zorbas H., Winnacker E. L., Baeuerle P. A. The NF-kappa B transcription factor induces DNA bending which is modulated by its 65-kD subunit. Nucleic Acids Res. 1990 Nov 25;18(22):6497–6502. doi: 10.1093/nar/18.22.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz P., van Holde K., Zlatanova J. Preferential binding of histone H1 to four-way helical junction DNA. J Biol Chem. 1993 Oct 5;268(28):20699–20700. [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Waga S., Mizuno S., Yoshida M. Chromosomal protein HMG1 removes the transcriptional block caused by the cruciform in supercoiled DNA. J Biol Chem. 1990 Nov 15;265(32):19424–19428. [PubMed] [Google Scholar]
- Ward G. K., McKenzie R., Zannis-Hadjopoulos M., Price G. B. The dynamic distribution and quantification of DNA cruciforms in eukaryotic nuclei. Exp Cell Res. 1990 Jun;188(2):235–246. doi: 10.1016/0014-4827(90)90165-7. [DOI] [PubMed] [Google Scholar]
- Ward G. K., Shihab-el-Deen A., Zannis-Hadjopoulos M., Price G. B. DNA cruciforms and the nuclear supporting structure. Exp Cell Res. 1991 Jul;195(1):92–98. doi: 10.1016/0014-4827(91)90503-m. [DOI] [PubMed] [Google Scholar]
- Zannis-Hadjopoulos M., Frappier L., Khoury M., Price G. B. Effect of anti-cruciform DNA monoclonal antibodies on DNA replication. EMBO J. 1988 Jun;7(6):1837–1844. doi: 10.1002/j.1460-2075.1988.tb03016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis-Hadjopoulos M., Kaufmann G., Martin R. G. Mammalian DNA enriched for replication origins is enriched for snap-back sequences. J Mol Biol. 1984 Nov 15;179(4):577–586. doi: 10.1016/0022-2836(84)90156-6. [DOI] [PubMed] [Google Scholar]
- Zheng G. X., Kochel T., Hoepfner R. W., Timmons S. E., Sinden R. R. Torsionally tuned cruciform and Z-DNA probes for measuring unrestrained supercoiling at specific sites in DNA of living cells. J Mol Biol. 1991 Sep 5;221(1):107–122. doi: 10.1016/0022-2836(91)80208-c. [DOI] [PubMed] [Google Scholar]
- Zhou N., Vogel H. J. Two-dimensional NMR and restrained molecular dynamics studies of the hairpin d(T8C4A8): detection of an extraloop cytosine. Biochemistry. 1993 Jan 19;32(2):637–645. doi: 10.1021/bi00053a032. [DOI] [PubMed] [Google Scholar]
- Zorbas H., Rein T., Winnacker E. L. Transfer-RNA interferes with the uniform cleavage pattern of DNA by hydroxyl radicals. Nucleic Acids Res. 1990 Oct 25;18(20):6160–6160. doi: 10.1093/nar/18.20.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorbas H., Rogge L., Meisterernst M., Winnacker E. L. Hydroxyl radical footprints reveal novel structural features around the NF I binding site in adenovirus DNA. Nucleic Acids Res. 1989 Oct 11;17(19):7735–7748. doi: 10.1093/nar/17.19.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]






