Abstract
Definitive correction of disease causing mutations in somatic cells by homologous recombination (HR) is an attractive therapeutic approach for the treatment of genetic diseases. However, HR-based somatic gene therapy is limited by the low efficiency of gene targeting in mammalian cells and replicative senescence of primary cells ex vivo, forcing investigators to explore alternative strategies such as retro- and lentiviral gene transfer, or genome editing in induced pluripotent stem cells. Here, we report correction of mutations at the LAMA3 locus in primary keratinocytes derived from a patient affected by recessive inherited Herlitz junctional epidermolysis bullosa (H-JEB) disorder using recombinant adenoassociated virus (rAAV)-mediated HR. We identified a highly recombinogenic AAV serotype, AAV-DJ, that mediates efficient gene targeting in keratinocytes at clinically relevant frequencies with a low rate of random integration. Targeted H-JEB patient cells were selected based on restoration of adhesion phenotype, which eliminated the need for foreign sequences in repaired cells, enhancing the clinical use and safety profile of our approach. Corrected pools of primary cells assembled functional laminin-332 heterotrimer and fully reversed the blistering phenotype both in vitro and in skin grafts. The efficient targeting of the LAMA3 locus by AAV-DJ using phenotypic selection, together with the observed low frequency of off-target events, makes AAV-DJ based somatic cell targeting a promising strategy for ex vivo therapy for this severe and often lethal epithelial disorder.
Introduction
Epidermolysis bullosa is a family of monogenic disorders characterized by severe blistering of the skin.1,2,3 The need for effective therapy is currently unmet and, despite attempts at protein,1,2,3,4 bone marrow transplant,5 cell6,7 and induced pluripotent stem cells–based treatments,8,9 palliative care remains the only option available. The most severe form of EB, often lethal by early childhood, is Herlitz-junctional EB (H-JEB), caused by mutations in the heterotrimer laminin-332, which plays a nonredundant role in epidermal–dermal adhesion by serving as a stable anchoring contact between the epidermal integrin receptors α3β1 and α6β6, and collagen VII, the main component of the dermal anchoring fibrils.2 Mutations in any of the three genes encoding the subunits of the heterotrimer, LAMA3, LAMB2, or LAMC3, give rise to H-JEB.
H-JEB is inherited in a recessive manner, thus gene therapy efforts have been focused on delivery of the therapeutic gene by vectors that integrate randomly or quasi-randomly.10,11,12 However, this therapy has several disadvantages, including variable site and frequency of integration, which can lead to insertional mutagenesis.13,14 An attractive alternative is precise genome editing at the endogenous site by homologous recombination (HR) between homologous sequences present in a donor vector and the genomic target, providing predictable integration site and endogenous regulation.15,16 Yet HR in mammalian cells is inefficient (one event per 105 to 107 treated cells), and random integration occurs orders of magnitude more frequently than HR (one cell per 102 to 104 treated cells),17 making random integration of transgenes preferred for somatic gene therapy, and limiting the use of HR to mammalian cells with unlimited expansion, such as transformed cell lines and induced pluripotent stem cells.18,19 Recombinant adenoassociated virus (rAAV) vectors containing single-stranded DNA efficiently transduce cells and can be used for targeted HR in vitro and in vivo at efficiencies several logs higher than classical HR, with low rates of random integration.20,21,22 Here, we identify a hybrid AAV serotype, AAV-DJ,23 that efficiently mediates HR in keratinocytes and use it to correct H-JEB primary keratinocytes derived from a patient with mutations in the LAMA3 gene. We employ restoration of adhesion in targeted cells as a selectable marker, eliminating the need of any transgene expression cassette and increasing the safety of our approach. Repaired cells, obtained in just 2 weeks, re-express laminin-332 from its endogenous promoter, reverse the blistering phenotype and regenerate the skin in vivo. Our data represent a feasible approach to somatic gene correction of H-JEB mutations and should allow rapid clinical implementation.
Results
The hybrid AAV serotype, AAV-DJ efficiently mediates HR at the LAMA3 locus inhuman keratinocytes
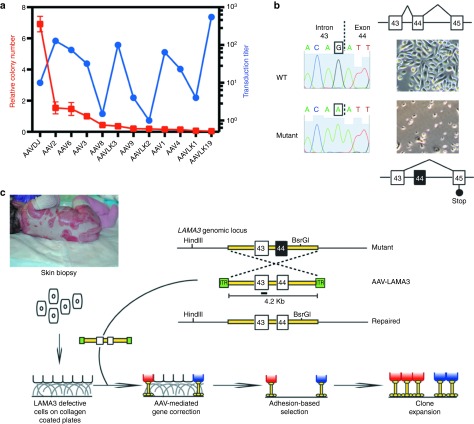
As part of our program for definitive genetic cell-based EB therapies, we performed a comprehensive screen of AAV serotypes for their potential to transduce and mediate HR at high efficiencies in primary keratinocytes. To determine the ability of the AAV serotypes to deliver DNA into the cells, we first transduced primary normal keratinocytes with self-complementary (sc)24 rAAV–enhanced green fluorescent protein (eGFP) viral preparations of seven naturally occurring and five hybrid AAV serotypes generated by DNA family shuffling technology,23 and measured fluorescence after 48 hours using fluorescence-activated cell sorting (Figure 1a in blue). As expected, transduction efficiencies varied significantly among the capsids tested, with AAV-LK1925 chimera outperforming the previously reported AAV2 and AAV6 serotypes26,27 by one log, and hybrid serotypes like AAV-DJ23 by two logs.
Figure 1.
AAV-DJ is a highly recombinogenic virus. (a) Relative colony number, a measure of homologous recombination (in red), and transduction titer as determined by green fluorescent protein (GFP)-based fluorescence-activated cell sorting (FACS) (in blue) of naturally occurring and hybrid adenoassociated virus (AAV) capsids in keratinocytes. Targeting was measured as the number of colonies stained with crystal violet after 10 days of adhesion selection on plastic and expressed relative to AAV3, which is the standard AAV with the highest homology to AAV-LK19,25 the most efficient capsid at functionally transducing keratinocytes. (b) Diagram depicting the mutation in the LAMA3 locus. The mutation causes incorrect splicing of exon 44, leading to a frame shift and an early termination codon. At right, light microscopy images of cells grown in culture. Mutant cells do not proliferate to plastic tissue culture dishes and attach normally. (c) Schematic overview of the targeting strategy based on adhesion selection for the LAMA3 locus. The targeting vector is a 4.2 Kb wild-type (WT) LAMA3 fragment encompassing exons 43 and 44. Southern probe is shown as a black line. ITR, inverse terminal repeats.
To determine recombination frequencies, we targeted keratinocytes derived from a 2-year-old H-JEB patient carrying a homozygous point mutation in the LAMA3 locus that produces a nonfunctional truncated protein (Figure 1b) and thus impairs normal assembly of the heterotrimer laminin-332.2 In vitro, the mutant cells exhibited an adhesion defect and failed to grow directly on plastic (Figure 1c). We hypothesized that reestablishment of adhesion after correction of the mutation could be used to enrich for HR events and dilute random integrants, as only cells with a targeted, and thus functional, LAMA3 allele would be able to attach to plastic, ameliorating the need for any additional selection transgene expression cassette. For this purpose, we constructed a single-stranded (ss)-rAAV-targeting construct consisting of a 4.2 Kb LAMA3 wild-type isogenic DNA fragment encompassing the mutation flanked by the inverse terminal repeats of AAV2 (Figure 1c). Because only cells with functional laminin-332 can adhere and proliferate on plastic, the number of colonies obtained after restoration of natural adhesion is a direct measure of the targeting efficiency at the LAMA3 locus in keratinocytes, allowing us to directly compare the ability of the AAV serotypes at mediating HR. To facilitate clonal analysis, we immortalized the LAMA3 deficient keratinocytes with HPV16 E6 and E7 proteins,28 and used the transduction titers obtained with sc-rAAV-eGFP vectors to normalize and deliver the same amount of ss-rAAV-LAMA3 DNA to these cells. This normalization was preferred over transducing the cells at the same multiplicity of infection (MOI) based on dot blot titer (Supplementary Figure S1a) given the different transduction efficiencies of each of the serotypes (Figure 1a in blue). In addition, using sc-rAAV-eGFP data for normalization removes the single to double strand DNA conversion variable. Interestingly, AAV-LK19 was the least competent at performing gene targeting as measured by the number of colonies obtained after 10 days of adhesion selection (Figure 1a in red). On the other hand, the AAV-DJ chimera, which did not perform as well as AAV-LK19 in the transduction studies, greatly outperformed all other serotypes (Figure 1a in red). We verified for the most recombinogenic capsids that transduction efficiencies in primary keratinocytes were similar upon delivery of scAAV-GFP or ssAAV-GFP viral preparations (Supplementary Figure S1b), indicating that cell entry29 is not affected by the single or double stranded nature of the AAV genome and pointing to intracellular trafficking and/or ssDNA to dsDNA29 conversion as the cause(s) of differential efficiency in transduction and targeting of chromosomal homologous sequences.
It has been proposed that the single-stranded nature of AAV and the presence of free DNA ends at the inverse terminal repeats contribute to efficient targeting by rAAV.30 Thus, we hypothesized that targeting efficiency was directly proportional to the time the rAAV genome remained as ssDNA. To test this possibility, we measured eGFP expression by fluorescence-activated cell sorting in primary keratinocytes transduced for 6 hours with ss-rAAV-GFP or sc-rAAV-GFP viral preparations. Since eGFP expression requires the transgene to be present as dsDNA, the level of GFP expression from ss-rAAV relative to sc-rAAV transduction can be used to evaluate the kinetics of ssDNA to dsDNA conversion. We compared several capsid variants that exhibit different targeting efficiencies, and interestingly, found no correlation between their ability to perform HR and ssDNA to dsDNA conversion, as measured as the ss/sc ratio of GFP positive cells (Supplementary Figure S1c). This suggests that other, currently unidentified, intracellular mechanisms/factors may influence the efficiency of gene targeting by rAAV. Based on our data showing high recombination frequencies and the ease in generating high titer vector preparations,23 our subsequent efforts focused on rAAV-DJ–mediated HR.
Efficient HR mediated by AAV-DJ is not limited to the LAMA3 locus
The targeting frequency for rAAV-DJ at the LAMA3 locus was calculated to be ~1% of the unselected population using a MOI of 2,000 in immortalized LAMA3 deficient keratinocytes. To investigate if this high targeting frequency at a relatively low MOI was unique for the targeted region in the LAMA3 gene, we generated an additional construct to target the COL7A1 locus in wild-type primary keratinocytes. We used puromycin resistance as a selectable marker (Supplementary Figure S2a) to compare with our LAMA3 results, and found the targeting frequency to be at 2.5%. Analysis of distinct clones using polymerase chain reaction (PCR) primers to detect both on and off-target events indicated that 92% of the clones were correctly targeted at the COL7A1 locus (Supplementary Figure S2b). Based on these data, the corrected targeting rate was recalculated to be 2.3%, slightly higher than the targeting frequency for the LAMA3 locus and showing that efficient AAV-DJ–mediated HR is not limited to a particular locus.
Restoration of laminin-332 expression rescues mutant cells
Laminin-332 is an extracellular protein complex that acts in a noncell autonomous manner to assemble into anchoring filaments2,6,7,31; thus, we expected rescue of some of the untargeted cells by deposited laminin-332 from the targeted cells. We determined the frequency of corrected and rescued cells in the adhesive cellular population using reconstitution of the wild-type SfcI restriction site upon HR within the LAMA3 locus (Figure 2a). Using this restriction fragment length polymorphism assay, we screened 70 AAV-DJ-LAMA3 clones grown on plastic derived from immortalized LAMA3 deficient keratinocytes. All of the clones contained a second smaller band (Figure 2b, for clarity only 13 clones are shown) indicating 100% gene targeting efficiency after selection, an anticipated finding given that the presence of wild-type laminin-α3 is required for laminin-332 assembly, adhesion and proliferation8,32 on regular cell culture dishes. However, most of the clones were a mosaic population of targeted and untargeted cells as quantified by the percentage of targeted allele by densitometry analysis on the SfcI generated bands (Figure 2c, all 70 clones are included). Assuming monoallelic gene targeting,33 the maximum percentage of corrected allele is 50%, only detected in clone 16, corresponding to 100% corrected cells. For the additional isolated clones, the repaired allele was as low as 2.5%, suggesting that at least in immortalized human keratinocytes, a corrected population of 5% produces enough laminin-α3 to assemble and deposit an appropriate amount of laminin-332 to rescue surrounding mutant cells. Our data suggest that local nonautonomous rescue of neighboring cells may greatly enhance the effectiveness of our rAAV-HR based strategy in H-JEB patients (Figure 2a).
Figure 2.
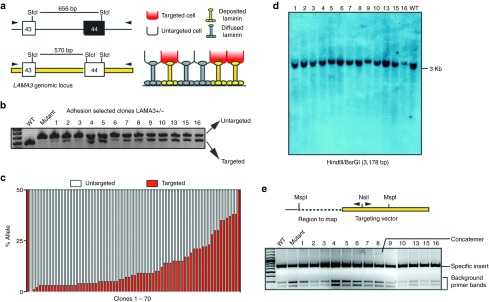
AAV-DJ-LAMA3 efficiently corrects somatic junctional EB cells with a low random integration frequency and leads to mosaicism. (a) Diagram depicting the restriction fragment length polymorphism (RFLP) assay based on the presence of SfcI restriction site. (b) RFLP assay on 13 clones selected based on restoration of adhesion. (c) Quantification of percentage of targeted and untargeted allele by densitometry on the SfcI produced bands on 70 clones. The presence of untargeted cells in the selected clones likely comes from rescue coming from deposited and diffusable laminin-332 from corrected cells (diagram on the right in a). (d,e) Detection of off-target events by (d) Southern blot and (e) inverse PCR. WT, wild-type.
AAV-DJ–mediated HR occurs with low rates of off-target events
Because the utility of HR techniques depends on both targeting efficiency and lack of random integration, we sought to determine the rate of off-target rAAV-DJ integration. Since our targeting vector does not confer function unless it integrates specifically at the LAMA3 locus, it is not possible to determine the absolute rate of random integration in the LAMA3-targeted population. Thus, to determine the absolute frequency of off-target events by rAAV-DJ–mediated targeting in a system that does not depend on restoration of a phenotype, we analyzed the primary keratinocyte clones targeted to the COL7A1 locus (Supplementary Figure S2a). In this targeting strategy, selection is based on resistance to puromycin driven by the phosphoglycerate kinase promoter, thus, both on- and off-target events will be detected. Out of 12 clones analyzed by Southern blot, only 1 had a random integration (8.3%) (Supplementary Figure S2c), the same clone that was previously determined as nonspecifically targeted by PCR (Supplementary Figure S2b). Importantly, none of the 11 targeted clones had random integrations, in agreement with previous reports indicating that the chance of having both specific and random integration in a same cell targeted by AAV is low.33
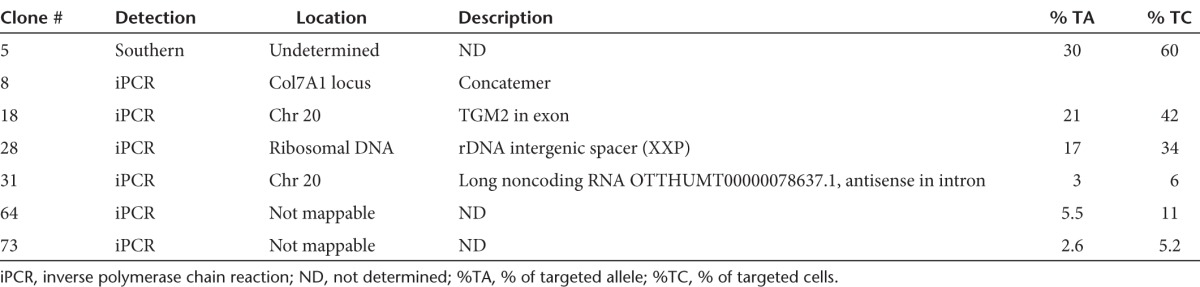
To estimate the rate of random integration of rAAV-DJ in the LAMA3-targeted cells, we carried out Southern blot and inverse PCR (iPCR) analysis34 to map the specific and random integration sites on DNA extracted from the rAAV-DJ–targeted isolated clones from HPV-immortalized LAMA3 cells. For Southern blot, we selected 27 of the clones with the highest percentage of corrected cells and observed the expected band at 3 Kb in all of the clones analyzed (Figure 2d, for clarity only 13 are shown). Only one additional faint band was detected in clone 5, indicating the presence of a substoichiometric population of off-targeted cells. On the other hand, iPCR analysis allowed us to detect more off-target events than Southern blot. Out of the 70 clones analyzed by iPCR, we detected one concatemerization event in clone 8 (Figure 2e) and five off-target integrations (Table 1), two of them mapped to intergenic regions, one to a ribosomal DNA sequence and two unmapped. Thus, 1.5% of the enriched targeted clones carried a population of cells with concatemerization events at the LAMA3 locus and 8.5% with off-target events. Since the clones were a mixed population and we could not detect the random integrations detected by iPCR using Southern blot in 5/6 clones, the events most likely occurred in small fraction of the rescued nontargeted cell within the clones (Table 1). Altogether, while 10% of the clones contained detectable concatemerization/random integration events, we interpret the iPCR and Southern data as consistent with random integration in subpopulations of cells being rescued by the targeted population.33
Table 1. rAAV-DJ nonhomologous and concatemer integration events in LAMA3 corrected clones.
Targeting laminin-α3 deficient primary cells corrects the blistering phenotype
A roadblock for targeting passage-limited somatic stem cells has been the need for serial clonal selection to identify low frequency events.35 To bypass the limited lifespan of somatic cells, we determined whether we could use pools of targeted somatic cells. LAMA3 deficient primary keratinocytes were transduced with AAV-DJ-LAMA3 and after 14 days of adhesion selection cells were pooled and used for DNA analysis, organotypic cultures and xenograft studies. Restriction fragment length polymorphism assay on this polyclonal population indicated targeting of up to 40% of the cells (Figure 3a). DNA sequencing confirmed correction of the mutation in the LAMA3 locus (Figure 3b), while native electrophoresis and western blot analysis on the secreted media showed restoration of laminin-332 assembly (Figure 3c). Interestingly, while we detected substoichiometric random integration events in immortalized LAMA3 clones, we could not detect such events either by iPCR (Figure 3d) or Southern blot (Figure 3e) analysis in the pools of corrected primary cells.
Figure 3.
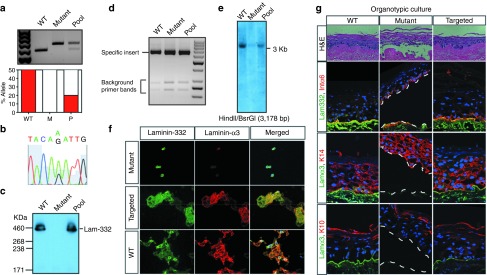
AAV-DJ-LAMA3 targeted primary keratinocytes correctly assemble laminin-332 and reverse the blistering phenotype. (a) Quantification of percentage of targeted allele by SfcI digest. Densitometry is shown below the gel. (b) Correction of the LAMA3 mutation in primary cell pools by DNA sequencing. (c) Restoration of laminin-332 assembly by native electrophoresis followed by western blot on the secreted media. (d,e) Detection of off-targeted events by (d) inverse PCR and (e) Southern blot. (f) Immunofluorescence staining on deposited laminin-α3 and laminin-332. (g) H&E and immunofluorescence staining of 2-week old organotypic cultures derived from wild-type (WT), mutant and targeted primary cells.
To determine whether our corrected cells made functional laminin-α3, we took advantage of the fact that cells deposit properly assembled and processed heterotrimeric laminin-332 onto plastic.31 We plated the targeted primary cells at limiting dilutions and performed immunohistochemical analysis of the deposited heterotrimer using antibodies specific for laminin-332 and the first globular domain of the laminin-α3 chain (Figure 3f).31 The extracellular matrix surrounding targeted cells stained brightly for laminin-α3 and for laminin-332, showing restoration of expression and correct heterotrimer assembly.
To determine if targeted primary cells could differentiate, stratify and form a normal epidermal basement membrane zone, we used organotypic cultures which provide an in vitro system to recapitulate normal epidermal tissue regeneration.36 Histological analysis of tissue regenerated in this setting for 2 weeks with the targeted pool of primary cells showed complete reversal of the blistering phenotype that was clearly visible in the mutant cells (Figure 3g, central panel). Immunofluorescence staining showed that corrected cells expressed laminin-α3 and laminin-332 at levels comparable to those in the wild-type control. Staining with the differentiation markers integrin α6, Keratin-14, and Keratin-10 was indistinguishable among wild-type, mutant and targeted cells, showing that gene targeting did not affect stratification.
Targeted primary cells have long-term graftability
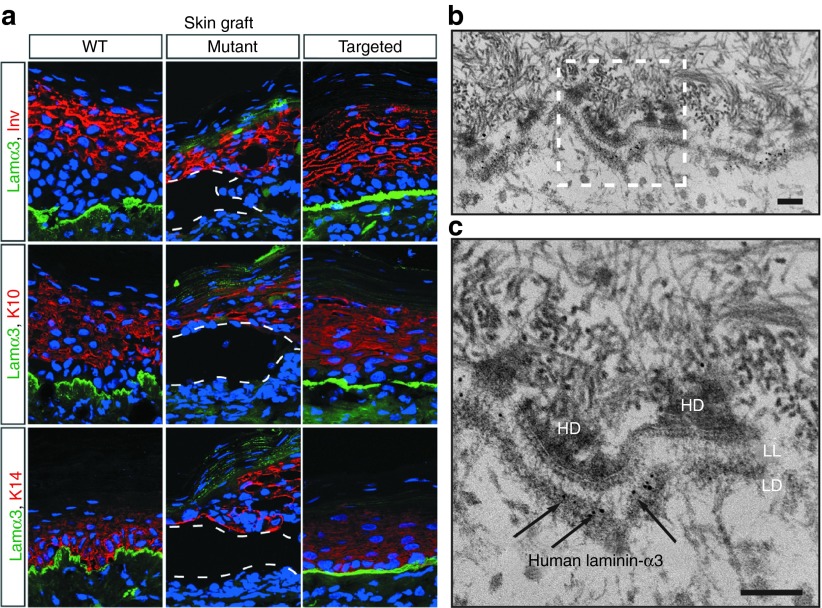
To test if the primary keratinocytes targeted with rAAV-DJ could regenerate and sustain the continuous renewal of the skin in vivo, we tested the corrected cells for graftability by transplanting them on the back of nude mice for several tissue turnover cycles.37 The histological and immunofluorescence analysis of five targeted and control grafts showed that the corrected cells were indistinguishable from normal control cells, with continuous linear staining at the dermal–epidermal junction, and normal stratification (Figure 4a). By contrast, mutant grafts lacked a basement membrane zone and tended to slough off after a few days. Immunoelectron micrographs of grafted skin using a human-specific laminin-α3 antibody showed a normal basement membrane zone, with localization of human laminin-α3 to anchoring filaments, below the basal-dense plate of hemidesmosomes, confirming functional gene correction (Figure 4b,c). Collectively, our data indicate that rAAV-DJ targets and effectively corrects the LAMA3 locus in the keratinocyte population that supports long-term renewal of the skin.38,39
Figure 4.
Targeted primary cells have long-term graftability in vivo. (a) Immunofluorescence staining of xenografts from wild-type (WT) (5 weeks), mutant (3 weeks) and targeted (5 weeks) cells. (b) Immunoelectron micrograph on ultrathin sections of a 5-week xenograft from targeted cells using an antibody against human laminin-α3. (c) A higher resolution micrograph. Immunostaining is pointed by the black arrows. Scale bar corresponds to 100 nm. HD, hemidemosome, LL, lamina lucida, LD, lamina densa.
Discussion
Here we demonstrate the use of the highly recombinogenic rAAV-DJ variant for correction of pools of somatic progenitors without the need for drug selection. AAV has been shown to mediate HR at high efficiencies.20 However, cell transduction by AAV is serotype dependent and human keratinocytes are not transduced efficiently by the most commonly used AAV serotypes.40 Previous studies demonstrated the feasibility of using the naturally occurring serotype AAV6 for disruption of a dominant negative disorder in primary human keratinocytes.23,27 The former approach required high MOIs to achieve clinically relevant numbers of disrupted keratinocytes and the concatemerization of the targeting vector under these conditions was frequently observed. While concatemerization represents an advantage for dominant negative disorders, recessive conditions require precise editing to restore expression of the missing gene. In this study, we have identified a hybrid AAV serotype that mediates HR at high efficiencies in human keratinocytes, and have used it to correct the LAMA3 locus in primary cells using a selection strategy based on restoration of natural adhesion.
Our data provide the first direct comparison between AAV transduction efficiency and HR frequency. We found a wide variation in both transduction and recombination frequencies among the capsid variants tested, suggesting that the processes are controlled by distinct genetic mechanisms. Interestingly, high functional transduction does not correlate directly with the HR efficiency, as rAAV-LK19 was very efficient at transducing keratinocytes but performed poorly at HR even when delivered at the same MOI (Supplementary Figure S1a). Since all of the AAV-viral preparations contain the same genetic information with inverse terminal repeats derived from AAV2, the observed discrepancy must arise from differences in cell entry, cell trafficking, nuclear entry, or persistence of the vector as a single strand DNA molecule as a function of the capsid proteins. Our results suggest that the persistence of the vector as ssDNA is not the rate-limiting factor. We observed that AAV6 has the fastest kinetics of eGFP expression from the sc-rAAV construct, yet is the slowest in ss to dsDNA genome conversion (Supplementary Figure S1c). Similar trends were observed for AAV2 and AAV3. On the other hand, highly recombinogenic rAAV-DJ was the fastest at the ssDNA to dsDNA genome conversion, while a slightly lower ss/sc ratio was observed for rAAV-LK3, which mediated low frequency of HR. While elucidating the mechanism of gene targeting by AAV will require further studies, it is an intriguing possibility that the capsid protein determines the protein-DNA interactions required to perform efficient HR. The discrepancy between the gene targeting and transduction shows that selection of AAV variants for HR applications requires functional tests for HR and not a reliance on transgene expression screening.
AAV-DJ, the AAV variant we found to be highly recombinogenic in human keratinocytes, mediates HR at lower MOIs than AAV6 and with lower rates of concatemerization events (1.5% of the analyzed clones). We found that between 1 and 2% of the transduced cells undergo HR. This high frequency was not linked to a particular locus as both the LAMA3 and COL7A1 loci were targeted efficiently. Epidermal grafts in our gene therapy protocol require about 10 million keratinocytes, which we calculate we could easily obtain using AAV-DJ–mediated HR from a starting population of about 1 million mutant keratinocytes obtained from 2 biopsies. These estimates demonstrate that AAV-DJ–mediated correction could provide a practical number of cells for clinical applications.
Although phenotypic correction of the LAMA3 locus does not allow us to quantify the absolute rate of random integration, we estimated for the COL7A1 locus that 1/12 clones (8.3% of the population) contained unanticipated integration events. Accordingly, we identified 6/70 (8.5%) clones carried a population of off-targeted cells. AAV-DJ vector integration sites included two open reading frames and one repetitive ribosomal DNA sequence, in agreement with the insertional mutagenesis profile of the AAV2 genome.2,41 The observation of random integrations was likely due to rescue by corrected cells, as laminin-332 is deposited extracellularly and thus restores adhesion noncell autonomously. In immortalized cells, we found that isolated clones had an average of 25% of the cells corrected (assuming monoallelic correction),28,33 with some clones exhibiting as low as 5%. On the other hand, pooled primary cells had 40% of the cells corrected. The lower percentage observed in immortalized keratinocytes is presumably associated with the immortal nature of the cells. It is possible that some of the observed random integrations are not from the rescued cells but instead present in the same targeted cell. However, if present, this population likely represents a small fraction of the corrected cells given that, first, we were unsuccessful at subcloning the off-targeted population to determine the fraction within a clone; second, we could not detect most of the off-target events by Southern blot, even though some of the clones with identified off-target events had high percentage of correction (Table 1); and third, none of the COL7A1-targeted clones had random integrations, in agreement with several reports that suggest that the likelihood of having both off- and on-target events in the same cell is low.29,33 Accordingly, we could not detect random integrations in the pool of corrected primary keratinocytes as most likely they are too diluted among the population.
Presence of uncorrected and off-targeted cells together with targeted cells does not represent a problem to translate this therapy into the clinic as probably the adhesive corrected population undergoes further selection in vivo given its capacity to adhere and maintain the graft. In addition, random integrations, which do not lead to the correction of LAMA3 gene, would be lost over time. On the other hand, having an apparent higher number of corrected cells represents an advantage for somatic gene therapy as cells could be transplanted sooner for further proliferation and selection in vivo.
While other methods for HR such as those based on site-specific endonucleases are reported to achieve higher HR frequencies,29,42,43 the use of AAV has several advantages. First, delivery of AAV-based vectors to primary cells is simple and has no associated toxicity in keratinocytes. On the other hand, in the alternative endonucleases-based approaches, nucleases, and donor vectors have to be delivered by nucleofection or lipid-based methods, which cause toxicity and cell death in primary keratinocytes affecting targeting efficiency. For instance, similar targeting efficiencies to AAV-DJ were achieved by two independent CRISPRs designed to promote double strand breaks around the mutation using the same plasmid as donor template (E. Bashkirova, S. Melo, and A. Oro; unpublished data). Second, characterization of off-target effects requires extensive cloning, as the likelihood of having random double strand breaks and targeted integrations in the same cell are higher,9,30 limiting its use in somatic cells. Finally, several human trials using AAV variants for gene delivery have already demonstrated efficacy, providing reassuring safety data from related variants.23,44
In summary, the use of an appropriately selected AAV variant, in the case of primary keratinocytes: AAV-DJ, in conjunction with a selectable phenotype, overcomes the two major roadblocks for therapy and allows pooled targeting at relatively low MOI prior to the loss of self-renewing capacity. Because induced pluripotent stem cell–mediated organ regeneration is a complex, expensive, and challenging process, our strategy is useful to generate fully repaired, and not just gene disrupted, cell populations whenever there is a selectable phenotypic advantage. While in our study we used LAMA3 for simplicity, in practice any visible (pigment) or cellular (metabolite or differentiation state) phenotype could be employed. Corrected cells under our approach would have no foreign or antibiotic resistance gene sequences left behind, raising the safety of the strategy to the level of the so called “natural gene therapy,” based on naturally occurring mutations that are able to revert the phenotype.45 In conjunction with previous studies demonstrating ex vivo gene therapy for the treatment of JEB,12 our data suggest a permanent, safe, and rapid stem cell correction for genodermatoses.
Materials and Methods
Cell culture. Keratinocytes were cultured in 50:50 keratinocyte serum free media, a 1:1 mixture of Keratinocyte-SFM supplemented with Bovine Pituitary Extract and EGF (Gibco/Invitrogen, Carlsbad, CA) and Media 154CF supplemented with HKGS and 0.05 mM calcium (Gibco/Invitrogen, Carlsbad, CA). 293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 4 g of glucose per liter (Gibco/Invitrogen), 10% heat-inactivated bovine serum, penicillin, and streptomycin.
Plasmid and DNA analysis. The LAMA3 targeting vector was constructed by PCR amplification of a 6.0 Kb portion of the gene (from position 202536 to 208553), followed by digestion with the enzymes EcoRI and AseI to produce a 4.1 Kb fragment. This piece was blunted and inserted between inverse terminal repeat sequences cloned from AAV2 isolate. The packaging plasmids used for viral preparation have been described before.23 Plasmid and genomic DNA were isolated using an Endofree maxiprep kit (Qiagen, Valencia, CA) and phenol-chloroform extraction, respectively. Southern blot analysis was performed using standard radioactive techniques. Inverse PCR34 was carried out using MspI and NsiI enzymes. Bands separated in a 3% agarose gel were gel-purified or reamplified using stab-PCR for DNA sequencing analysis. For restriction fragment length polymorphism analysis, primers were used to PCR amplify a 653 bp product followed by digestion with SfcI. Products were separated in a 2% agarose gel and bands were quantified by densitometry.
AAV-viral preparation. All AAV vectors used in the study (scAAV-RSV-GFP, ssAAV-RSV-GFP-Luc-pA and ssAAV-LAMA3) were produced by Ca3(PO4)2 triple transfection and purified by CsCl gradient centrifugation as previously reported.23 Viral DNA was extracted by the sodium iodide method, using a DNA Extractor kit (Wako Chemicals USA, Richmond, VA) and titrated by a quantitative dot blot assay.
AAV transduction of human keratinocytes. Foreskin keratinocytes were transduced by seeding 6 × 104 cells to multiple wells of a 24-well plate in 50:50 keratinocyte serum free media. The following day, scAAV-RSV-GFP or ssAAV-RSV-GFP-Luc-pA viral preparations were added to the media at MOIs of 200, 2,000, and 20,000. After 48 hours of transduction, cells were trypsinized and analyzed by fluorescence-activated cell sorting. For gene targeting, LAMA3 deficient keratinocytes cells were transduced by seeding 0.25 × 106 cells each to multiple type I collagen-coated wells (0.003% Type 1 rat tail collagen (BD, Franklin Lakes, NJ), 0.01% BSA, 0.02 mol/l HEPES pH6.5 in HBSS) of a six-well plate in 50:50 keratinocyte serum free media. After cell attachment, cells were transduced with ssAAV-LAMA3 viral preparations at several MOIs. After 2 days of transduction, cells were trypsinized and seeded onto regular 10-cm culture dishes. Following 10 days of expansion, colonies were ring-cloned, or trypsinized and combined, for DNA analysis, organotypic cultures and xenografts. One of the plates from each transduction was stained with crystal violet following standard protocols. Primary human keratinocytes were immortalized and analyzed for nonhomologous integrations through infection with a retrovirus expressing human papilloma virus E6-E7 protein.28
Organotypic cultures of human skin equivalents and xenografts. Between 0.5 and 1 × 106 cells were seeded and grown for 5 days onto a 1.5 cm2 piece of devitalized human dermis and then raised to air liquid interface for 2 weeks, for organotypic cultures, or grafted onto the back of nude mice for xenografts as previously described (ref. 27). The grafts were performed at least in triplicate and were collected at 3 weeks for LAMA3 deficient cells (grafts older than 3 weeks would slough the nonadherent cells) or 5 weeks for corrected LAMA3 and wild-type keratinocytes. All mouse studies were approved by and conformed to the policies and regulations of the Institutional Animal Care and Use Committees at Stanford University.
Histology and immunofluorescence. Immunofluorescence was carried out overnight on 7 µm frozen sections of skin tissue directly embedded in optimal cutting temperature media (OCT) and then postfixed in 4% paraformaldehyde for 10 minutes. Alternatively, cells grown in four-chamber slides were washed with phosphate-buffered saline and fixed with paraformaldehyde for 10 minutes. Hybridomas for mouse BM165 that specifically recognizes the human laminin-α3 chain (1:100) and rabbit pAb to laminin-331 (pKaL, 1:500) were generous gifts of M.P.M. (Stanford, CA). Additional primary antibodies used were as follows: CD49f (Millipore, Billerica, MA, MAB1378, 1:200), Keratin-14 (Covance, Princeton, NJ, PRB-115P, 1:2,000), Keratin-10 (Covance, PRB-159P-100, 1:500) and human involucrin (Abcam, ab27495, 1:200). Secondary antibodies used were goat anti-mouse AF488 (Invitrogen, A11029), goat anti-rabbit AF488 (Invitrogen, A11034), goat anti-rabbit AF546 (Invitrogen, A11035), and goat anti-rat AF546 (Invitrogen, A11081) all at 1:500. Hoechst dye was used at 1:10,000. ProLong Gold Anti- fade reagent (Invitrogen) was used for mounting.
Western blot analysis. Supernatants collected from cultured keratinocytes were concentrated using 50 K Centrifugal Filter Units (Millipore) and subjected to immunoprecipitation using mouse monoclonal BM165 antibody (7 µg). Immunoprecipitates were resolved by Native-PAGE followed by western using rabbit pAb pKaL (1:1,000) and anti-rabbit HRP (Cell Signaling Technology, Boston, MA; 7074S, 1:1,000).
Electron microscopy. Skin tissue was rinsed in DMEM and immersed in human-specific monoclonal antibody BM165 (1:5) overnight at 4 °C. The tissues were rinsed extensively in DMEM, then immersed in Goat-anti-mouse 0.8 nm gold (Aurion Ultrasmall, EMS, Hatfield, PA, 1:3) overnight at 4 °C. Following an extensive rinse in DMEM, the tissues were gold enhanced (Nanoprobes, EMS), rinsed again in DMEM, fixed in 1.5% glutaraldehyde/1.5% paraformaldehyde with 0.05% tannic acid then in 1% OsO4, and finally dehydrated and embedded in Spurr's epoxy. Ultrathin sections mounted on formvar coated single hole grids were stained in uranyl acetate and lead citrate, then examined using an FEI G2 transmission electron microscope operated at 120 KV.
SUPPLEMENTARY MATERIAL Figure S1. Transduction Efficiency of AAV Capsids. Figure S2. AAV-DJ targets the COL7A1 locus efficiently with a low rate of random integration.
Acknowledgments
The authors thank P Khavari, M Porteus, and Z Suprashvilli for their comments on the manuscript. This work was funded by grants to A.E.O. (California Institute for Regenerative Medicine, CHRI), M.A.K. (NIH HL064274/HL092096), and M.P.M. (Debra International, Epidermolysis Bullosa Medical Research Foundation, R01 AR055914, ORD, Palo Alto VA Medical Center). The authors declare no conflict of interest.
Supplementary Material
Transduction Efficiency of AAV Capsids.
AAV-DJ targets the COL7A1 locus efficiently with a low rate of random integration.
References
- Fine JD. Inherited epidermolysis bullosa: recent basic and clinical advances. Curr Opin Pediatr. 2010;22:453–458. doi: 10.1097/MOP.0b013e32833bb74f. [DOI] [PubMed] [Google Scholar]
- Hashmi S, Marinkovich MP. Molecular organization of the basement membrane zone. Clin Dermatol. 2011;29:398–411. doi: 10.1016/j.clindermatol.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Berk DR, Jazayeri L, Marinkovich MP, Sundram UN, Bruckner AL. Diagnosing epidermolysis bullosa type and subtype in infancy using immunofluorescence microscopy: the Stanford experience. Pediatr Dermatol. 2013;30:226–233. doi: 10.1111/j.1525-1470.2012.01880.x. [DOI] [PubMed] [Google Scholar]
- Woodley DT, Keene DR, Atha T, Huang Y, Lipman K, Li W, et al. Injection of recombinant human type VII collagen restores collagen function in dystrophic epidermolysis bullosa. Nat Med. 2004;10:693–695. doi: 10.1038/nm1063. [DOI] [PubMed] [Google Scholar]
- Wagner JE, Ishida-Yamamoto A, McGrath JA, Hordinsky M, Keene DR, Woodley DT, et al. Bone marrow transplantation for recessive dystrophic epidermolysis bullosa. N Engl J Med. 2010;363:629–639. doi: 10.1056/NEJMoa0910501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto J. Cell-based therapy for RDEB: how does it work. J Invest Dermatol. 2011;131:1597–1599. doi: 10.1038/jid.2011.125. [DOI] [PubMed] [Google Scholar]
- Wong T, Gammon L, Liu L, Mellerio JE, Dopping-Hepenstal PJ, Pacy J, et al. Potential of fibroblast cell therapy for recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2008;128:2179–2189. doi: 10.1038/jid.2008.78. [DOI] [PubMed] [Google Scholar]
- Petrova A, Ilic D, McGrath JA. Stem cell therapies for recessive dystrophic epidermolysis bullosa. Br J Dermatol. 2010;163:1149–1156. doi: 10.1111/j.1365-2133.2010.09981.x. [DOI] [PubMed] [Google Scholar]
- Osborn MJ, Starker CG, McElroy AN, Webber BR, Riddle MJ, Xia L, et al. TALEN-based gene correction for epidermolysis bullosa. Mol Ther. 2013;21:1151–1159. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins PB, Lin Q, Goodnough JB, Tian H, Chen X, Khavari PA. In vivo restoration of laminin 5 beta 3 expression and function in junctional epidermolysis bullosa. Proc Natl Acad Sci USA. 2001;98:5193–5198. doi: 10.1073/pnas.091484998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Urda S, Lin Q, Yant SR, Keene D, Kay MA, Khavari PA. Sustainable correction of junctional epidermolysis bullosa via transposon-mediated nonviral gene transfer. Gene Ther. 2003;10:1099–1104. doi: 10.1038/sj.gt.3301978. [DOI] [PubMed] [Google Scholar]
- Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, Recchia A, et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med. 2006;12:1397–1402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Mermel CH, Lu J, Lu CW, Golub TR, Daley GQ. microRNA expression during trophectoderm specification. PLoS ONE. 2009;4:e6143. doi: 10.1371/journal.pone.0006143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrouzi A, Glimm H, von Kalle C, Schmidt M. Retroviral vectors: post entry events and genomic alterations. Viruses. 2011;3:429–455. doi: 10.3390/v3050429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus M. Homologous recombination-based gene therapy for the primary immunodeficiencies. Ann N Y Acad Sci. 2011;1246:131–140. doi: 10.1111/j.1749-6632.2011.06314.x. [DOI] [PubMed] [Google Scholar]
- Kmiec EB. Targeted gene repair – in the arena. J Clin Invest. 2003;112:632–636. doi: 10.1172/JCI19777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc Natl Acad Sci USA. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simara P, Motl JA, Kaufman DS. Pluripotent stem cells and gene therapy. Transl Res. 2013;161:284–292. doi: 10.1016/j.trsl.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW, Hirata RK. Human gene targeting by viral vectors. Nat Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DG, Wang PR, Petek LM, Hirata RK, Sands MS, Russell DW. Gene targeting in vivo by adeno-associated virus vectors. Nat Biotechnol. 2006;24:1022–1026. doi: 10.1038/nbt1231. [DOI] [PubMed] [Google Scholar]
- Deyle DR, Russell DW. Adeno-associated virus vector integration. Curr Opin Mol Ther. 2009;11:442–447. [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Lee JS, Wang L, Desai T, Akache B, Storm TA, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM. Self-complementary AAV vectors; advances and applications. Mol Ther. 2008;16:1648–1656. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- Lisowski L, Dane AP, Chu K, Zhang Y, Cunningham SC, Wilson EM, et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature (in press) [DOI] [PMC free article] [PubMed]
- Braun-Falco M, Eisenried A, Büning H, Ring J. Recombinant adeno-associated virus type 2-mediated gene transfer into human keratinocytes is influenced by both the ubiquitin/proteasome pathway and epidermal growth factor receptor tyrosine kinase. Arch Dermatol Res. 2005;296:528–535. doi: 10.1007/s00403-005-0547-y. [DOI] [PubMed] [Google Scholar]
- Petek LM, Fleckman P, Miller DG. Efficient KRT14 targeting and functional characterization of transplanted human keratinocytes for the treatment of epidermolysis bullosa simplex. Mol Ther. 2010;18:1624–1632. doi: 10.1038/mt.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon TJ, Flotte TR. Recombinant adeno-associated virus vectors for gene therapy. Expert Opin Biol Ther. 2004;4:1093–1101. doi: 10.1517/14712598.4.7.1093. [DOI] [PubMed] [Google Scholar]
- Vasileva A, Jessberger R. Precise hit: adeno-associated virus in gene targeting. Nat Rev Microbiol. 2005;3:837–847. doi: 10.1038/nrmicro1266. [DOI] [PubMed] [Google Scholar]
- Rousselle P, Lunstrum GP, Keene DR, Burgeson RE. Kalinin: an epithelium-specific basement membrane adhesion molecule that is a component of anchoring filaments. J Cell Biol. 1991;114:567–576. doi: 10.1083/jcb.114.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill KJ, Paller AS, Jones JC. Adhesion and migration, the diverse functions of the laminin alpha3 subunit. Dermatol Clin. 2010;28:79–87. doi: 10.1016/j.det.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IF, Hirata RK, Russell DW. AAV-mediated gene targeting methods for human cells. Nat Protoc. 2011;6:482–501. doi: 10.1038/nprot.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou EP, Sadelain M. Derivation of genetically modified human pluripotent stem cells with integrated transgenes at unique mapped genomic sites. Nat Protoc. 2011;6:1274–1289. doi: 10.1038/nprot.2011.362. [DOI] [PubMed] [Google Scholar]
- Sedivy JM, Sharp PA. Positive genetic selection for gene disruption in mammalian cells by homologous recombination. Proc Natl Acad Sci USA. 1989;86:227–231. doi: 10.1073/pnas.86.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Oro AE, Scott MP, Khavari PA. Induction of basal cell carcinoma features in transgenic human skin expressing Sonic Hedgehog. Nat Med. 1997;3:788–792. doi: 10.1038/nm0797-788. [DOI] [PubMed] [Google Scholar]
- Alonso L, Fuchs E. Stem cells of the skin epithelium. Proc Natl Acad Sci USA. 2003;100 (suppl. 1):11830–11835. doi: 10.1073/pnas.1734203100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Jensen KB. Epidermal stem cell diversity and quiescence. EMBO Mol Med. 2009;1:260–267. doi: 10.1002/emmm.200900033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnoux-Palacios L, Hervouet C, Spirito F, Roques S, Mezzina M, Danos O, et al. Assessment of optimal transduction of primary human skin keratinocytes by viral vectors. J Gene Med. 2005;7:1178–1186. doi: 10.1002/jgm.768. [DOI] [PubMed] [Google Scholar]
- Miller DG, Trobridge GD, Petek LM, Jacobs MA, Kaul R, Russell DW. Large-scale analysis of adeno-associated virus vector integration sites in normal human cells. J Virol. 2005;79:11434–11442. doi: 10.1128/JVI.79.17.11434-11442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multiplex genome engineering using CRISPR/Cas systems—Supplementary materials . < http://www.sciencemag.org/content/suppl/2013/01/02/science.1231143.DC1 >.
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokan A, Schaffer DV, Samulski RJ. The AAV vector toolkit: poised at the clinical crossroads. Mol Ther. 2012;20:699–708. doi: 10.1038/mt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Akker PC, Nijenhuis M, Meijer G, Hofstra RM, Jonkman MF, Pasmooij AM. Natural gene therapy in dystrophic epidermolysis bullosa. Arch Dermatol. 2012;148:213–216. doi: 10.1001/archdermatol.2011.298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transduction Efficiency of AAV Capsids.
AAV-DJ targets the COL7A1 locus efficiently with a low rate of random integration.