Abstract
An investigation into the kinetics and regulatory properties of the type-1 phosphofructokinase (PFK) from the extreme thermophile Thermus thermophilus (TtPFK) reveals an enzyme that is inhibited by PEP and activated by ADP by modifying the affinity exhibited for the substrate fructose 6-phosphate (Fru-6-P) in a manner analogous to other prokaryotic PFKs. However, TtPFK binds both of these allosteric ligands significantly more tightly than do other bacterial PFKs while effecting a substantially more modest extent of inhibition or activation at 25°C, reinforcing the principle that binding affinity and effectiveness can be both independent and uncorrelated to one another. These properties have allowed us to rigorously establish that PEP only inhibits by antagonizing the binding of Fru-6-P, and not by influencing turnover – a conclusion that requires kcat be determined under conditions in which both inhibitor and substrate are saturating simultaneously. In addition, the temperature dependence of the allosteric effects on Fru-6-P binding indicate that the coupling free energies are entropy-dominated, as observed previously for PFK from B. stearothermophilus but not for PFK from E. coli, supporting the hypothesis that entropy-dominated allosteric effects may be a characteristic of enzymes derived from thermostable organisms. For such enzymes, the root cause of the allosteric effect may not be easily discerned from static structural information such as might be obtained from X-ray crystallography.
Phosphofructokinase-1 (PFK)1 catalyzes the phosphoryl transfer from MgATP to fructose-6-phosphate (Fru-6-P) forming fructose-1,6-bisphosphate and MgADP. This reaction is the first committed step of glycolysis and a critical control point of the glycolytic pathway, making PFK usually subject to rigorous allosteric regulation. The regulation of the PFKs from eukaryotic organisms is very complex since they have distinct activator and inhibitor binding sites and are regulated by a variety of allosteric effectors including fructose 1,6-bisphosphate, fructose 2,6-bisphosphate, citrate, AMP, and ATP (1, 2). Eukaryotic PFKs can also exist in multiple active oligomeric states (3, 4). In contrast, bacterial PFKs provide a relatively simple paradigm for allosteric regulation (5). The catalytically active form of bacterial PFK is a homotetramer that contains 4 identical active sites and 4 identical allosteric sites and is regulated by phosphoenolpyruvate (PEP) and MgADP, which compete for binding at the same effector sites. Both of these ligands are K-type effectors as they impact the binding affinity of the enzyme for the substrate Fru-6-P, while leaving the Vmax unchanged, although this conclusion has rarely been established in a rigorous manner as discussed below.
The PFKs from E. coli (EcPFK) and Bacillus stearothermophilus (BsPFK) have been extensively studied resulting in a wealth of kinetic, structural, and thermodynamic information (6–19). The crystal structures of these two enzymes with various ligand combinations bound show a high degree of similarity. However, substantial differences in the binding affinities for the substrate and the allosteric ligands, as well as in the magnitude of the allosteric responses, are evident. Another difference is that both the inhibition by PEP and activation by MgADP of EcPFK are enthalpically-driven (14), while the effects in BsPFK are entropically-driven (20). This observation raises the question of whether the entropy-dominated regulation might be in some way related to the thermostability of BsPFK, suggesting that it might also be observed in PFKs from other thermophiles. To further evaluate the potential relationship between thermal stability and the thermodynamic basis of allosteric regulation we have examined the allosteric properties of PFK from the extreme thermophile, Thermus thermophilus (TtPFK).
TtPFK was partially purified (to a specific activity of 0.49 U/mg) by Yoshida (21, 22) and characterized as extremely thermostable with minimal loss of activity after incubation at 70°C for more than 30 hours. TtPFK was reported to exhibit simple Michaelis-Menten kinetics when either Fru-6-P or MgATP were the variable substrate. Michaelis constants of 15 μM and 60 μM, respectively, were determined at 30°C and pH 8.4. The addition of 0.1 mM PEP resulted in a roughly 10-fold increase in the Km for Fru-6-P and a change in the Fru-6-P binding curve from hyperbolic to sigmoidal yielding a Hill number of 2. ADP was able to partially relieve the inhibition by PEP, and the effects of PEP and ADP were more pronounced at 75°C compared to 30°C.
In 1990 TtPFK was purified to homogeneity by Xu et al., and the specific activity was determined to be 57 U/mg at 25°C and pH 8.4 (23). TtPFK dissociation into dimers was observed when the enzyme was applied onto a gel filtration column that was equilibrated and eluted with buffer containing 0.1 mM PEP. This process could be reversed by adding Fru-6-P or by removing PEP from the buffer. The authors suggested that this association-dissociation plays a role in the regulation of the activity of the enzyme.
Although these studies confirmed the allosteric regulation of TtPFK by PEP and ADP, the extent of PEP inhibition and ADP activation was not quantified. The present study offers a more comprehensive analysis of the allosteric effects of PEP and ADP on TtPFK using thermodynamic linkage analysis, a model-free approach that quantifies the magnitude of the allosteric response by comparing the difference in the substrate binding affinity for the enzyme in effector-free and effector-saturated forms.
Materials and Methods
Materials
All chemical reagents used in buffers, protein purifications, and enzymatic assays were of analytical grade, and were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Fair Lawn, NJ). The EPPS buffer used for fluorescence experiments was purchased from Acros Organics (Geel, Belgium). The sodium salt of Fru-6-P was purchased from Sigma-Aldrich or USB Corporation (Cleveland, OH). NADH and dithiothreitol were purchased from Research Products International (Mt. Prospect, IL). Creatine kinase and the ammonium sulfate suspension of glycerol-3-phosphate dehydrogenase were purchased from Roche Applied Sciences (Indianapolis, IN). The ammonium sulfate suspensions of aldolase and triosephosphate isomerase, as wells as, the sodium salts of phosphocreatine and PEP were purchased from Sigma-Aldrich. The sodium salt of ATP was purchased from Sigma-Aldrich and Roche Applied Sciences. The experiments involving quantifying the allosteric response of TtPFK to MgADP were conducted using the sodium salt of ATP purchased from Roche Applied Sciences. The coupling enzymes were dialyzed extensively against 50 mM MOPS-KOH, pH 7.0, 100 mM KCl, 5 mM MgCl2, and 0.1 mM EDTA before use.
Mutagenesis
The pALTER plasmid containing the wild type TtPFK gene was used as the starting template for mutagenesis (13). The L313W mutation was introduced using QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using the following primers:
GGACATCAACCGGGCCTGGTTGCGCCTATCGC (forward)
GCGATAGGCGCAACCAGGCCCGGTTGATGTCC (reverse)
The C111F/A273P mutations were introduced using the Altered Sites in vitro Mutagenesis System protocol (Promega, Madison, Wisconsin) using the following primers (complementary to the coding strand):
GTGCTCCTCCACGAGAAAAAGCGCCCCGC (C111F)
GGCCTCCACCGCGGGCGCCCCCAGGCG (A273P)
The resulting sequences were verified via DNA sequencing at the Gene Technology Laboratory at Texas A&M University.
Protein Expression and Purification
Thermus thermophilus HB8 cells were purchased from ATCC (Manassas, VA). Cells were propagated in ATCC medium 697 (0.4% yeast extract, 0.8% polypepetone, and 2% NaCl; pH 7.5). Genomic T. thermophilus DNA was purified using the Wizard Genomic DNA Purification Kit (Promega; Madison, WI). The isolated genomic DNA was digested with HindIII before subcloning. We first attempted to subclone the TtPFK gene into pLEAD4 (Ishida and Oshima (2002)). This vector is specially designed to express thermophilic bacterial genes containing high GC-content. While we were able to successfully subclone into pLEAD4 and see expression of TtPFK when using JM109 as the host strain, the plasmid was not compatible with our expression strain, RL257, which has the E. coli genes pfkA and pfkB deleted (24). Using PCR primers with the restriction enzymes appropriate for cloning into pALTER-1 (Promega), we amplified the TtPFK gene using the pLEAD4 construct as the template. The ligation products were screened via restriction enzyme digests and constructs containing the correct banding pattern were sequence verified.
In the process of cloning TtPFK from the genome, we found three single base differences in the sequence of the gene, relative to the published sequence of T. thermophilus HB8 (Accession number M71213.1 (25)). One of the differences is inconsequential to the protein product. However, the other two result in differences in the predicted amino acid sequence. Position 111 had been reported to be a phenylalanine while our results predict a cysteine, and position 273 was reported to be proline while we anticipate an alanine at that position. It should be noted that the sequence we determined is consistent with the more recently released, as yet unpublished, submission of the complete genome of T. thermophilus HB8 (Accession number YP_145228).
The RL257 cells containing the plasmid with the TtPFK gene were induced with IPTG from the start and grown at 30°C for 18 hours in Luria-Bertani media (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L sodium chloride) containing 15 μg/mL tetracycline. The cells were harvested by centrifugation in a Beckman J6 at 3600xg and frozen at −80°C for at least 2 hours before lysis. The cells were re-suspended in 20 mM Tris-HCl, 1 mM EDTA; pH 8.0 and sonicated with a Fisher 550 Sonic Dismembrator at 0°C for 8–10 min using a 15 second pulse/45 second rest sequence. The crude lysate was centrifuged using a Beckman J2-21 centrifuge at 22,500xg for 30 min at 4°C. The supernatant was heated in a 70°C water bath for 20 minutes, cooled on ice, and centrifuged at 22,500xg for 30 min at 4°C. The protein was then precipitated using 35% ammonium sulfate at 0°C and the suspension was again centrifuged at 22,500xg for 30 min at 4°C. The pellet was dissolved in a minimal volume of 20 mM Tris-HCl, pH8 and dialyzed several times against the same buffer. The protein solution was then applied to a MonoQ column (GE Life Sciences), which was equilibrated with 20 mM Tris-HCl, pH8 and eluted with a 0 to 1M NaCl gradient. Fractions containing PFK activity were analyzed for purity using SDS-PAGE, pooled and dialyzed against the 20 mM Tris-HCl, pH8 and stored at 4°C. The protein concentration was determined using the BCA assay (Pierce) using BSA as a standard.
Kinetic assays
Initial velocity measurements were carried out in 600 μL of buffer containing 50 mM EPPS-KOH, pH 8, 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 2 mM dithiothreitol, 0.2 mM NADH, 250 μg of aldolase, 50 μg of glycerol-3-phosphate dehydrogenase, 5 μg of triosephosphate isomerase, 0.5 mM ATP, and varying amount of Fru-6-P. PEP or MgADP were also included as indicated. To determine the Ka for MgATP initial velocity measurements were carried out in 600 μL of buffer containing 50 mM EPPS-KOH, pH 8, 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 2 mM dithiothreitol, 0.2 mM NADH, 250 μg of aldolase, 50 μg of glycerol-3-phosphate dehydrogenase, 5 μg of triosephosphate isomerase, 5 mM Fru-6-P, and varied concentrations of MgATP. 40 μg/mL of creatine kinase and 4 mM phosphocreatine were present in all assays performed in the absence of MgADP. When measuring the activation by MgADP, equimolar concentrations of MgATP and MgADP were added to overcome competitive product inhibition by MgADP expected at the active site. The reaction was initiated by adding 10 μL of TtPFK appropriately diluted into 50 mM EPPS (KOH) pH 8, 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA. The conversion of Fru-6-P to Fru-1,6-BP was coupled to the oxidation of NADH, which resulted in a decrease in absorbance at 340nm. The rate of the decrease in A340 was monitored using a Beckman Series 600 spectrophotometer.
Steady-state fluorescence assays
The fluorescence intensity measurements were performed using an ISS KOALA photon counting fluorometer (ISS, Inc. Champaign, IL). The titrations were performed in buffer containing 50 mM EPPS (KOH) pH 8, 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA. The enzyme concentration in the sample was 0.5 μM. The sample was excited at 295 nm, and the fluorescence intensity was detected using a 2-mm thick 335 nm Schott cut-on filter. Change in the intrinsic tryptophan fluorescence at 25°C was measured as a function of PEP concentration at varying concentrations of Fru-6-P.
Data analysis
Data were fit using the non-linear least-squares fitting analysis of Kaleidagraph software (Synergy). Initial velocity data were plotted against the concentration of Fru-6-P and fit to the following equation:
| (1) |
where v∘ is the initial velocity, [A] is the concentration of the substrate Fru-6-P, V is the maximal velocity, nH is the Hill coefficient, and Ka is the Michaelis constant defined as the concentration of substrate that gives one-half the maximal velocity. For a reaction in rapid equilibrium, Ka is equivalent to the geometric mean dissociation constant for the substrate from the enzyme-substrate complex. The rapid equilibrium assumption for the Fru-6-P interaction in TtPFK was verified by comparing the value of Qay obtained by the method described below to that obtained from PEP titrations as described in (26) (data not shown).
Data collected using steady-state fluorescence were plotted as a function of the PEP concentration. The data were fit using the equation:
| (2) |
where F is the relative intensity, F∘ is the relative intensity in the absence of PEP, [Y] is the concentration of PEP, Ky is the apparent dissociation constant for PEP, and nH is Hill number. The resulting values for the apparent dissociation constants for PEP were then plotted as a function of the Fru-6-P concentration and fit to equation 3b.
The Ka and Ky values obtained from the initial velocity and fluorescence experiments were plotted against effector or substrate concentrations and fit to equation 3a or 3b:
| (3a) |
| (3b) |
where is the dissociation constant for Fru-6-P in the absence of allosteric effector, Y is PEP, is the dissociation constant for PEP in the absence of Fru-6-P, and Qay is the coupling coefficient (27–29). When equation 3a is applied to the allosteric action of MgADP, the subscripts are changed from “y” to “x”, and MgADP is designated as “X”, to be consistent with the notation we have used previously (14).
Qay is defined as the coupling constant, which describes the effect of allosteric effector on the binding of the substrate (and vice versa) and is defined by equation 4:
| (4) |
where represents the dissociation constant for the substrate in the saturating presence of the allosteric effector, and represents the dissociation constant for the allosteric effector in the saturating presence of the substrate.
Based on its definition, Qay represents the equilibrium constant for the following disproportionation equilibrium (Scheme I):
Scheme I.
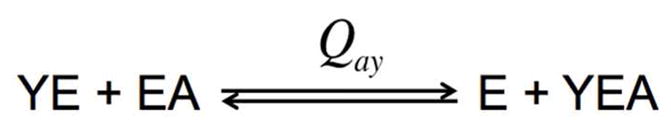
The coupling constant Qay is related to the coupling free energy (ΔGay) and its enthalpy (ΔHay) and entropy (ΔSay) components through the following relationship (30):
| (5) |
The coupling entropy and enthalpy components were determined by measuring the coupling constant as a function of temperature and the data were fit to equation 6a:
| (6a) |
where ΔGay is the coupling coefficient, ΔSay is the coupling enthalpy, ΔHay is coupling entropy, T is absolute temperature in K, and R is gas constant (R=1.99 cal·K−1·mol−1). The data that display non-linearity were fit to the modified van’t Hoff equation:
| (6b) |
where T° is the reference temperature, in this case 298K, and ΔCp is the change in the heat capacity (31).
Results
The TtPFK Ka for MgATP was determined to ensure that all subsequent measurements were performed at saturating concentrations of MgATP, and the data were fit well by equation 1. At saturating Fru-6-P (5 mM), the Hill number for MgATP binding is 0.8 ± 0.07 and the Ka for MgATP is 6.0 ± 0.5 μM, with the specific activity equal to 41 units/mg at pH 8 (a unit of activity is defined as the amount of enzyme required to produce 1 μmol of fructose 1,6-bisphosphate/min) and a kcat of 25 s−1 at 25°C (Table 1). Consequently subsequent assays were performed with 0.5 mM MgATP. The Ka for Fru-6-P under these conditions is 27.0 ± 0.6 μM. The Fru-6-P titration exhibited a slight positive homotropic cooperativity resulting in a Hill number of 1.58 ± 0.06. To address the possible consequences of the discrepancy between the sequence of TtPFK gene reported by Xu et al. (25) and the sequence we determined, we constructed the C111F/A273P variant and verified that the presence of these substitutions does not cause any dramatic changes in the properties of the enzyme (Table 2)
Table 1.
Summary of kinetic and thermodynamic parameters for TtPFK, BsPFK and EcPFK, at pH 8 and 25°Ca.
| TtPFK | BsPFKb | EcPFKc | ||
|---|---|---|---|---|
|
|
27.0 ± 0.6 | 31 ± 2 | 300 ± 10 | |
|
|
0.4 ± 1 | 19 ± 2 | 48 ± 2 | |
| Qax | 1.6 ± 0.1 | 1.70 ± 0.01 | 11.1 ± 0.2 | |
| ΔGax (kcal/mol) | −0.28 ± 0.04 | −0.314±0.003 | −1.42±0.01 | |
|
|
1.58 ± 0.07 | 93 ± 6 | 300 ± 10 | |
| Qay | 0.067 ± 0.002 | 0.0021 ± 0.0003 | 0.0080 ± 0.0003 | |
| ΔGay (kcal/mol) | 1.60 ± 0.02 | 3.67 ± 0.1 | 2.7 ± 0.1 | |
| SA (U/mg) | 41 | 163 | 240 | |
| kcat (s−1) | 25 | 91 | 142 | |
| kcat/Km (M−1 s−1) | 9.1×105 | 2.9×106 | 4.7×105 |
Table 2.
Summary of the kinetic and thermodynamic properties of the wild type and C11F/A273P and L313W variants of TtPFK at pH 8 and 25°C
| WT | C111F/A273P | L313W | L313W (fluor) | ||
|---|---|---|---|---|---|
|
|
27.0 ± 0.6 | 36.5± 0.7 | 14.4±0.03 | 0.063±0.009 | |
|
|
1.58 ± 0.07 | 4.5±0.2 | 1.15±0.07 | 0.66±0.06 | |
| Qay | 0.067 ± 0.002 | 0.063± 0.001 | 0.072± 0.002 | 0.068±0.006 | |
| SA (U/mg) | 41 | 34 | 54 | n/a |
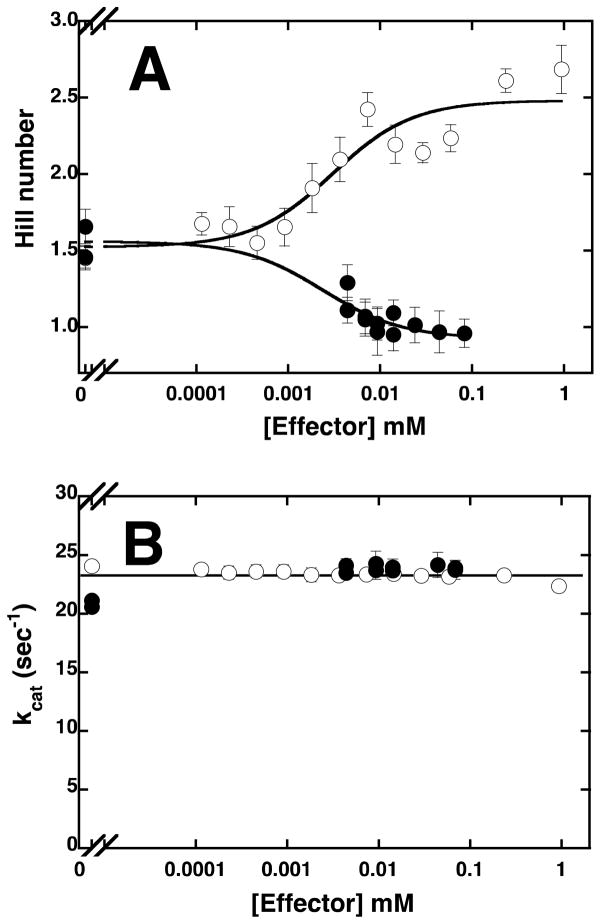
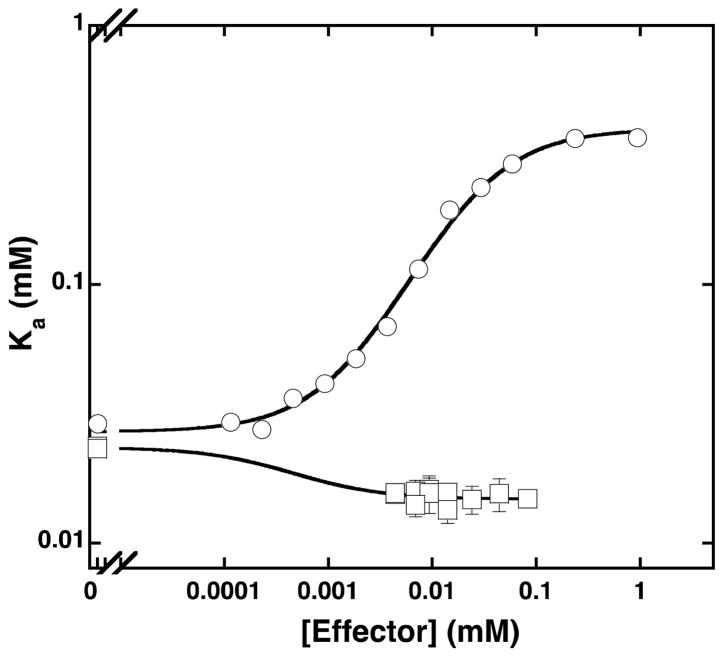
To measure the allosteric effects of PEP and MgADP, the Fru-6-P titrations were performed at increasing concentrations of either effector and the individual titration curves were fit to equation 1. Hill numbers and specific activity determined from these titrations are presented as a function of effector concentration in Figures 1A and 1B, respectively. The fits of the apparent dissociation constant for Fru-6-P as a function of PEP or MgADP concentration to equation 2 are shown in Figure 2. The resulting values of , Qay, and Qax are presented in Table 1 along with the values for these parameters pertaining to PFK from other sources for comparison. Increasing concentrations of PEP, as expected, lead to a decrease in the Fru-6-P binding affinity. This effect continues until PEP saturation is reached, after which no additional inhibition of Fru-6-P binding can be achieved by further increasing the concentration of PEP. The addition of PEP also resulted in heterotropically induced homotropic cooperativity (32) in Fru-6-P binding as evidenced by the increase of the Hill numbers from 1.6 to above 2.5 as [PEP] approaches saturation (Figure 1A). However, no effect on the apparent specific activity of the enzyme as a function of PEP concentration is evident (Figure 1B). Similarly, the addition of MgADP had little effect the specific activity of the enzyme, but did diminish the homotropic cooperativity of Fru-6-P binding, reducing the Hill numbers from 1.6 to 1 (Figures 1A and 1B). The magnitude of the activation by MgADP is much smaller, compared to the magnitude of the inhibition by PEP (Figure 2). The large error in the value of is a result of an experimental limitation, because the lowest attainable concentration of MgADP used in the assay (that of the MgADP contamination in the MgATP) is well above the estimated dissociation constant of MgADP.
Figure 1.
Figure 1A. The plot of the Hill number as a function of PEP (open circles) or MgADP (closed circles) concentration at pH 8 and 25°C.
Figure 1B. The plot of the relative maximal activity as a function of PEP (open circles) or MgADP (closed circles) concentration at pH 8 and 25°C
Figure 2.
The plot of apparent dissociation constants (Ka) for Fru-6-P as a function of PEP (circles) or MgADP (squares) concentration at pH 8 and 25°C. The data were fit to equation 3 to obtain the dissociation constants for PEP ( ) and MgADP ( ) and the coupling constants (Qay and Qax) reported in Tables 1 and 2.
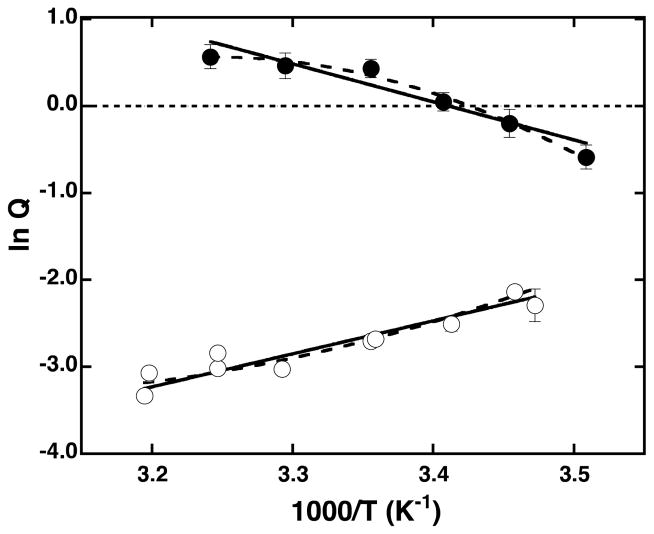
To determine the entropic and enthalpic components of inhibition by PEP and activation by MgADP in TtPFK at room temperature, the coupling coefficients (Qax and Qay) were measured at temperatures ranging between 10 to 35°C (Figure 3). The plots of lnQax or lnQay as a function of inverse temperature were fit to equations 6a and 6b. The values obtained from both the linear and non-linear fits are presented in Table 3. The data for the temperature dependence of lnQay are well described by a straight line with a positive slope. In contrast, the data for the temperature dependence of lnQax are fit better by the non-linear equation 6b, suggesting that the entropy and enthalpy components vary as a function of temperature. The overall trend implies greater inhibition and activation at higher temperature consistent with the data of Yoshida (22). It is interesting to note, that the values of ΔHay and TΔSay at 25°C obtained by the linear versus the non-linear fits are comparable (Table 3).
Figure 3.
Van’t Hoff plots of lnQay (open circle) and lnQax (closed circles) as a function of temperature. The data were fit to equation 6 (solid line) to obtain the enthalpy component of coupling free energy or to equation 6a (dashed line) to obtain the enthalpy and change in the heat capacity at 25°C.
Table 3.
Thermodynamic parameters for inhibition and activation of TtPFK at pH 8 and 25°Ca
| Linear Fit | Non-linear Fit | |
|---|---|---|
| Activation | ||
|
| ||
| ΔHax (kcal/mol) | +9.0 ± 1.0 | +8.0 ± 1.0 |
| -TΔSax (kcal/mol) | −9.3 ± 1.0 | −8.3 ± 1.0 |
| ΔCp (kcal/K mol) | n/a | −0.7 ± 0.3 |
| R | 0.939 | 0.990 |
|
| ||
| Inhibition | ||
|
| ||
| ΔHay (kcal/mol) | −7.5 ± 0.3 | −8.4 ± 0.4 |
| -TΔSay (kcal/mol) | +9.1 ± 0.3 | +10.0 ± 0.4 |
| ΔCp (kcal/K mol) | n/a | +0.3± 0.1 |
| R | 0.926 | 0.938 |
The values for the enthalpy and the change in heat capacity are obtained from the linear or non-linear fits of the temperature dependency of lnQay and lnQax to Equations 7 and 8. The values for the entropy and change in heat capacity were determined directly from the fit to the data. The values for TΔSay and TΔSax were calculated using the values for the coupling free energy obtained from the initial velocity experiments at 25° and the values for the enthalpy calculated from the slope on the van’t Hoff plots. R represents the correlation coefficient for the respective fits.
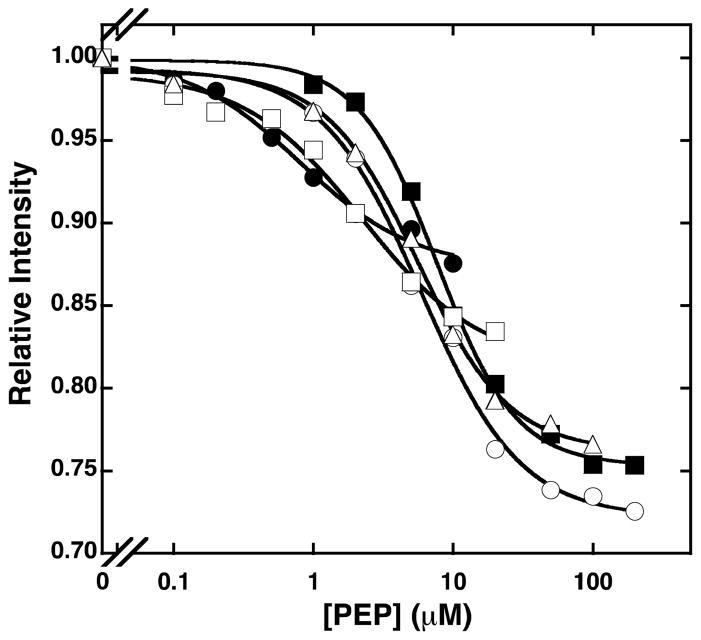
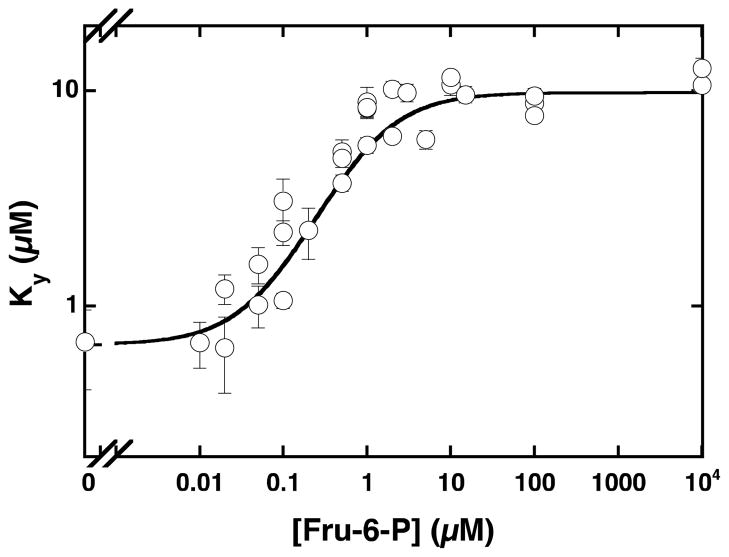
To establish the values for the binding of Fru-6-P and PEP, and coupling between PEP and Fru-6-P by tryptophan fluorescence in the absence of turnover, we introduced a tryptophan at position 313 (L313W). To ensure that this variant behaves similarly to wild type protein, we measured the parameters for the binding and coupling of PEP and Fru-6-P using the initial velocity experiments described in the materials and methods section. The resulting values are presented in Table 2 and correlate well with the values for the wild type enzyme. The change in the intrinsic tryptophan fluorescence as a function of PEP concentration was measured at increasing concentrations of Fru-6-P (Figure 4) and fit to equation 2. The Hill number for the binding of PEP to TtPFK is equal to 1, suggesting that there is no homotropic cooperativity in the PEP binding to free enzyme. The Hill number was not changed with the addition of Fru-6-P (data not shown). The values for the dissociation constants for PEP were plotted as a function of Fru-6-P concentration (Figure 5) and fit to equation 3b to obtain the values for the dissociation constant for Fru-6-P and the coupling constant (Table 2). The values for the dissociation constant for PEP and the coupling constant obtained from the tryptophan fluorescence experiments agree well with the values obtained from the initial velocity experiments. The dissociation constant for Fru-6-P binding obtained using tryptophan fluorescence experiments is over 200-fold lower compared to the value determined from the initial velocity experiments. This has also been previously seen for the Fru-6-P binding to E. coli PFK in the absence of MgATP (33).
Figure 4.
The plot of relative fluorescence intensity of the L313W variant at 25°C as a function of PEP concentration at Fru-6-P concentrations of 0 μM (solid circles), 0.2 μM (open squares), 1 μM (open triangles), 5 μM (open circles), 100 μM (solid squares). The sample was excited at 295 nm and the fluorescence intensity was detected using the 2-mm 335 nm Schott cut-on filter.
Figure 5.
The plot of apparent dissociation constants (Ky) for PEP as a function of Fru-6-P concentration. The data were fit to equation 3A to obtain the dissociation constants for Fru-6-P ( ) and the coupling constant Qay.
Discussion
The Michaelis constant for Fru-6-P of 27 μM and the specific activity of 41 Units/mg reported here agree relatively well with the values reported by Yoshida and Xu et al. (Km of 15 μM and specific activity of 57 Units/mg). The lower specific activity we report likely can be attributed to difference in buffer pH (EPPS pH8 vs Tris-HCl pH8.4), since the specific activity increases with pH (22). The specific activity of TtPFK at 25°C is significantly lower than that of EcPFK and BsPFK (Table 1). The low activity at room temperature is not surprising since it has been noted that typically the activities of the thermophilic enzymes at their native temperatures are comparable to those of the enzymes from mesophiles (34–36). As a consequence, at room temperature the activity of the enzyme from the thermophile is expected to be low.
The binding of Fru-6-P in the absence of PEP is weakly cooperative, resulting in a Hill number of 1.6. Binding of PEP to TtPFK results in a decrease in apparent binding affinity for Fru-6-P, however, it also induces further homotropic cooperativity in Fru-6-P binding, increasing the Hill number to 2.5. Thus the homotropic cooperativity of Fru-6-P binding is different for the free enzyme compared to the PEP-bound enzyme (Figure 2). The specific activity of the enzyme is not changed in the presence of PEP, confirming that PEP is a K-type effector. This finding is of importance because we were able to show that there is no change in kcat at saturating concentrations of the inhibitor. Previously, the lack of the kcat effect of PEP was only inferred from the experiments performed at subsaturating inhibitor concentrations (37). In order to understand the effect of an inhibitor on the turnover of an enzyme, it is necessary to measure the turnover of the enzyme species that is bound and remains bound to the inhibitor at all substrate concentrations. Given the antagonism that exists between the binding of inhibitor and the binding of substrate, both the substrate and the inhibitor will bind preferentially to the free enzyme, forming either YE or EA species. Consequently the population of the enzyme in the ternary complex, YEA, is very small at relatively low concentrations of A and Y. The turnover that is seen under these conditions comes mostly from the inhibitor-free form, EA, and thus does not report on the effect of the inhibitor on the catalytic activity of the enzyme. In contrast, when the inhibitor is fully saturating, the enzyme exists in either the YE or YEA forms depending on the concentration of substrate. The binding of substrate uniquely forms the ternary complex YEA from which turnover can potentially occur. In the present study we are able to demonstrate unambiguously that the turnover seen for the EA form of the enzyme is the same as that seen for the YEA form, indicating that the binding of PEP does not affect the catalytic activity of TtPFK. Consequently, one can conclude that, despite the substantial conformational perturbations introduced by the binding of inhibitor alone (5–9), the positioning of residues that define the transition state for the reaction must be unaffected by whether or not the inhibitor is bound as long as Fru-6-P is present.
It is interesting to note that TtPFK exhibits tighter PEP binding compared to the PFK’s from Bacillus stearothermophilus (20) and E. coli (14) while having a considerably weaker coupling between PEP and Fru-6-P binding (Table 1). This observation serves as another reminder that the binding affinity of the allosteric effector does not correlate with the magnitude of the allosteric effect it is able to produce, contrary to predictions often made by 2-state models. Similar observations have been made when comparing the binding and allosteric affects of PEP to those of phosphoglycolate in Bacillus stearothermophilus PFK. While the binding affinity of phosphoglycolate to BsPFK is 10-fold weaker than that of PEP, it has a 3.5-fold greater inhibitory effect on the binding of Fru-6-P (13). Another example of the independence of binding and coupling is the E187A substitution in EcPFK, which leads to the loss of activating effect of MgADP, while the binding of MgADP remains virtually unaffected in this variant (38). Similar behavior was noted for the R252E variant of EcPFK, which can bind both PEP and MgADP, but exhibits neither inhibition by PEP, nor activation by MgADP (39).
The coupling between PEP and Fru-6-P binding was also measured using intrinsic tryptophan fluorescence. The advantage of this method is that is it directly measures the dissociation constants for the substrate and the inhibitor in the absence of the second substrate, MgATP. Since the wild type TtPFK does not have a native tryptophan, a tryptophan was introduced at position 313, the position of the native tryptophan in E. coli PFK. With initial velocity experiments we verified that the binding and coupling parameters for PEP and Fru-6-P of the L313W variant are similar to those of the wild type enzyme and concluded that this variant is a suitable alternative to the wild type enzyme for the fluorescence studies (Table 2). The values of the coupling constant obtained by either method agree very well (Table 2) and suggest that MgATP binding has no effect on the coupling between Fru-6-P and PEP. The dissociation constants for PEP obtained from the tryptophan fluorescence and the initial velocity experiments are also in good agreement. The only large discrepancy between the fluorescence and initial velocity results is the dissociation constant for Fru-6-P, which is roughly 200-fold lower when determined in the absence of MgATP. This comes as no surprise, as the antagonistic effect of MgATP on Fru-6-P binding is also seen in PFK’s from E. coli and B. stearothermophilus (33, 40). The results of this experiment also demonstrate the concept of reciprocity inferred from the thermodynamic linkage analysis in that the binding of the allosteric effector elicits the same response on the binding of the substrate as the substrate does on the allosteric effector.
PEP coupling free energy in TtPFK, determined with initial velocity measurements, increases with increasing temperature (Figure 3) indicating that the inhibition by PEP is entropically-driven. It is intriguing that the change in the enthalpy of inhibition is negative. This may suggest that any rearrangements in the three-dimensional static structures of the species on the right side of the disproportionation equilibrium (Scheme I) compared to those on the left hand side, would in fact reflect an activating effect of PEP on the enzyme. It is the change in the entropy between the two sides of the equilibrium that determines the positive sign of the coupling free energy and hence the inhibitory nature of PEP. The linearity of the data in Figure 3 within the sampled temperature range indicates that there is little if any change in the heat capacity associated with the inter-conversion among the free and liganded enzyme species represented in the disproportionation equilibrium. This suggests that the entropy associated with the inhibition by PEP stems mostly from the differences in the dynamic properties of the individual enzyme species as opposed to the differences in solvation of the different liganded states which usually produce substantial changes in ΔCp (41).
It is interesting to note that entropically-driven inhibition is also seen in the PFK from the moderate thermophile B. stearothermophilus (20), while the PFK from mesophilic E. coli displays enthalpically-driven inhibition by PEP (14). Although an analysis of the coupling parameter as a function of temperature has not been performed in bacterial PFK’s from other mesophiles and thermophiles, it is tempting to consider the possibility that the entropically-driven allosteric regulation is a common feature of the thermophilic PFK’s. It is also worth noting that while the magnitude of the coupling free energy of inhibition in TtPFK is relatively small at 25°C, at the physiologically relevant temperatures it may be comparable to that of EcPFK.
The binding of MgADP to TtPFK also had little effect on the specific activity of the enzyme but, interestingly, diminished the positive homotropic cooperativity in Fru-6-P binding, reducing the Hill numbers from 1.6 to 1 (Figure 1). In contrast to the substantial inhibition by PEP, the magnitude of the activation of TtPFK by MgADP is relatively small (Figure 2, Table 1). These finding are especially interesting in light of the results obtained by Yoshizaki (42), who measured the changes in the levels of metabolites in T. thermophilus under conditions of glycolysis and gluconeogenesis and reported that while the concentration of hexose phosphates and PEP varied inversely, the concentrations of adenylates (ATP, ADP, AMP) stayed constant. It is possible that TtPFK has evolved the ability to maintain a slight basal level of activation due to the 30–40 μM ADP present in the cell. However, since the levels of ADP do not change appreciably, it may not play a central role in the regulation of this enzyme. In contrast, the PEP concentrations changed from negligible under the conditions of glycolysis to 0.17 mM under the conditions of gluconeogenesis, which, given the strong binding affinity of PEP and a substantial coupling free energy of inhibition, would afford PEP a central role in regulating the PFK in T. thermophilus.
The analysis of the coupling coefficient of activation as a function of temperature revealed several interesting findings. First is that the activation of TtPFK by MgADP is entropically driven at 25°C, as is the inhibition by PEP. Second is that, unlike in the case of inhibition by PEP where the dependence of the logarithm of the coupling coefficient on reciprocal temperature is described well by a straight line, the variation of lnQax for MgADP as a function of reciprocal temperature exhibits pronounced curvature within the same temperature range. This non-linearity implies that there is a change in the heat capacity associated with the activation of TtPFK by MgADP. The negative change in heat capacity is conventionally linked to the removal of the non-polar surfaces from water (41), which suggests differences in the solvation states of the four species of the enzyme (E, XE, EA, and XEA) participating in Scheme I. The third observation is that at temperatures below 20°C, MgADP loses its activating effects and becomes an inhibitor as indicated by the change in sign in the value of lnQax. While the existence of such a crossover temperature is implicit in the dependence of the coupling free energy on entropy, enthalpy, and temperature (when entropy and enthalpy have the same sign), this crossover phenomenon is only infrequently observed at experimentally attainable temperatures. The few examples that have been reported include the allosteric effect of MgADP on the binding of Fru-6-P in BsPFK (at pH 6) and of IMP on the binding of MgADP in E. coli carbamyl-phosphate synthetase, which switch from activation to inhibition at temperatures below 16°C and 37°C, respectively (43). Similarly, the effect of PEP on the binding of MgATP to EcPFK, which is slightly activating at room temperature, becomes inhibitory at temperatures above 37°C (14).
Conclusions
The available crystal structures of type 1 ATP-dependent PFK-1 from several bacterial sources reveal a high overall structure conservation, and while no three-dimensional structure of TtPFK is currently available, we have no reason to believe that it is dramatically different given its high degree of sequence homology. The sequence of TtPFK is 57% identical to that of BsPFK and 46% identical to the sequence of EcPFK. Sequence similarities are 70% and 62%, respectively. Considering the likely conservation of structure, it is particularly interesting to observe the extent of variation in the functional properties of these PFK’s. In particular, TtPFK displays a very high binding affinity for its allosteric inhibitor PEP relative to the other prokaryotic PFKs commonly studied. At the same time, the allosteric coupling between the substrate and the inhibitor is much weaker (Table 1). These observations underscore the independence of binding and allosteric action, and they suggest that those aspects of the protein’s structure that are important for the binding of the allosteric effectors and substrates may well be different from the aspects responsible for establishing the magnitude of the allosteric effect those ligands are able to produce. However an understanding of the precise structural basis for these properties in any bacterial PFK remains to be determined.
The tight binding of PEP and Fru-6-P and the weaker coupling between the substrate and inhibitor binding, mean that, in the case of TtPFK, the ternary complex with the inhibitor is more easily attainable than in the PFK’s from E. coli and Bacillus stearothermophilus. Given the importance of the role that the ternary complex plays in defining the allosteric coupling, as evident in Scheme I, this feature suggests that TtPFK should prove to be a useful system with which to further probe the structural basis for the allosteric responsiveness of prokaryotic PFK. In addition, these features have also allowed us to unambiguously establish that PEP (and MgADP) have very little effect on kcat, even under conditions where they remain fully bound at the allosteric site during turnover.
Acknowledgments
Funding
This work was supported by National Institutes of Health Grant GM033216 and Robert A. Welch grant A1543.
Footnotes
Abbreviations: PFK, phosphofructokinase; TtPFK, Thermus thermophilus phosphofructokinase; BsPFK, Bacillus stearothermophilus phosphofructokinase; EcPFK, E. coli phosphofructokinase; Fru-6-P, fructose-6-phosphate; PEP, phospho(enol)pyruvate
References
- 1.Kemp RG, Gunasekera D. Evolution of the allosteric ligand sites of mammalian phosphofructo-1-kinase. Biochemistry. 2002;41:9426–9430. doi: 10.1021/bi020110d. [DOI] [PubMed] [Google Scholar]
- 2.Kemp RG, Foe LG. Allosteric regulatory properties of muscle phosphofructokinase. Mol Cell Biochem. 1983;57:147–154. doi: 10.1007/BF00849191. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart GD, Lardy HA. Rat-Liver Phosphofructokinase - Use of Fluorescence Polarization to Study Aggregation at Low Protein-Concentration. Biochemistry. 1980;19:1484–1490. doi: 10.1021/bi00548a035. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart GD, Lardy HA. Rat-Liver Phosphofructokinase - Kinetic and Physiological Ramifications of the Aggregation Behavior. Biochemistry. 1980;19:1491–1495. doi: 10.1021/bi00548a036. [DOI] [PubMed] [Google Scholar]
- 5.Evans PR, Farrants GW, Hudson PJ. Phosphofructokinase: structure and control. Philos Trans R Soc London Ser B Biol Sci. 1981;293:53–62. doi: 10.1098/rstb.1981.0059. [DOI] [PubMed] [Google Scholar]
- 6.Evans PR, Hudson PJ. Structure and control of phosphofructokinase from Bacillus stearothermophilus. Nature. 1979;279:500–504. doi: 10.1038/279500a0. [DOI] [PubMed] [Google Scholar]
- 7.Shirakihara Y, Evans PR. Crystal structure of the complex of phosphofructokinase from Escherichia coli with its reaction products. J Mol Biol. 1988;204:973–994. doi: 10.1016/0022-2836(88)90056-3. [DOI] [PubMed] [Google Scholar]
- 8.Rypniewski WR, Evans PR. Crystal structure of unliganded phosphofructokinase from Escherichia coli. J Mol Biol. 1989;207:805–821. doi: 10.1016/0022-2836(89)90246-5. [DOI] [PubMed] [Google Scholar]
- 9.Mosser R, Reddy MCM, Bruning JB, Sacchettini JC, Reinhart GD. Structure of the Apo Form of Bacillus stearothermophilus Phosphofructokinase. Biochemistry. 2012;51:769–775. doi: 10.1021/bi201548p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson JL, Reinhart GD. MgATP and fructose 6-phosphate interactions with phosphofructokinase from Escherichia coli. Biochemistry. 1992;31:11510–11518. doi: 10.1021/bi00161a032. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JL, Reinhart GD. Influence of substrates and MgADP on the time-resolved intrinsic fluorescence of phosphofructokinase from Escherichia coli. Correlation of tryptophan dynamics to coupling entropy. Biochemistry. 1994;33:2644–2650. doi: 10.1021/bi00175a037. [DOI] [PubMed] [Google Scholar]
- 12.Johnson JL, Reinhart GD. Influence of MgADP on phosphofructokinase from Escherichia coli - elucidation of coupling interactions with both substrates. Biochemistry. 1994;33:2635–2643. doi: 10.1021/bi00175a036. [DOI] [PubMed] [Google Scholar]
- 13.Tlapak-Simmons VL, Reinhart GD. Comparison of the inhibition by phospho(enol)pyruvate and phosphoglycolate of phosphofructokinase from B. stearothermophilus. Arch Biochem Biophys. 1994;308:226–230. doi: 10.1006/abbi.1994.1032. [DOI] [PubMed] [Google Scholar]
- 14.Johnson JL, Reinhart GD. Failure of a two-state model to describe the influence of phospho(enol)pyruvate on phosphofructokinase from Escherichia coli. Biochemistry. 1997;36:12814–12822. doi: 10.1021/bi970942p. [DOI] [PubMed] [Google Scholar]
- 15.Kimmel JL, Reinhart GD. Reevaluation of the accepted allosteric mechanism of phosphofructokinase from Bacillus stearothermophilus. Proc Natl Acad Sci USA. 2000;97:3844–3849. doi: 10.1073/pnas.050588097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riley-Lovingshimer MR, Reinhart GD. Equilibrium binding studies of a tryptophan-shifted mutant of phosphofructokinase from Bacillus stearothermophilus. Biochemistry. 2001;40:3002–3008. doi: 10.1021/bi002448i. [DOI] [PubMed] [Google Scholar]
- 17.Fenton AW, Paricharttanakul NM, Reinhart GD. Disentangling the web of allosteric communication in a homotetramer: heterotropic activation in phosphofructokinase from Escherichia coli. Biochemistry. 2004;43:14104–14110. doi: 10.1021/bi048569q. [DOI] [PubMed] [Google Scholar]
- 18.Ortigosa AD, Kimmel JL, Reinhart GD. Disentangling the web of allosteric communication in a homotetramer: heterotropic inhibition of phosphofructokinase from Bacillus stearothermophilus. Biochemistry. 2004;43:577–586. doi: 10.1021/bi035077p. [DOI] [PubMed] [Google Scholar]
- 19.Paricharttanakul NM, Ye S, Menefee AL, Javid-Majd F, Sacchettini JC, Reinhart GD. Kinetic and structural characterization of phosphofructokinase from Lactobacillus bulgaricus. Biochemistry. 2005;44:15280–15286. doi: 10.1021/bi051283g. [DOI] [PubMed] [Google Scholar]
- 20.Tlapak-Simmons VL, Reinhart GD. Obfuscation of allosteric structure-function relationships by enthalpy-entropy compensation. Biophys J. 1998;75:1010–1015. doi: 10.1016/S0006-3495(98)77589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida M, Oshima T, Imahori K. The thermostable allosteric enzyme: phosphofructokinase from an extreme thermophile. Biochem Biophys Res Commun. 1971;43:36–39. doi: 10.1016/s0006-291x(71)80081-5. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida M. Allosteric nature of thermostable phosphofructokinase from an extreme thermophilic bacterium. Biochemistry. 1972;11:1087–1093. doi: 10.1021/bi00756a022. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Oshima T, Yoshida M. Tetramer-dimer conversion of phosphofructokinase from Thermus thermophilus induced by its allosteric effectors. J Mol Biol. 1990;215:597–606. doi: 10.1016/S0022-2836(05)80171-8. [DOI] [PubMed] [Google Scholar]
- 24.Lovingshimer MR, Siegele D, Reinhart GD. Construction of an inducible, pfkA and pfkB deficient strain of Escherichia coli for the expression and purification of phosphofructokinase from bacterial sources. Protein Exp Purif. 2006;46:475–482. doi: 10.1016/j.pep.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Xu J, Seki M, Denda K, Yoshida M. Molecular-cloning of phosphofructokinase-1 gene from a thermophilic bacterium, Thermus-thermophilus. Biochem Biophys Res Commun. 1991;176:1313–1318. doi: 10.1016/0006-291x(91)90429-b. [DOI] [PubMed] [Google Scholar]
- 26.Symcox MM, Reinhart GD. A steady-state kinetic method for the verification of the rapid-equilibrium assumption in allosteric enzymes. Anal Biochem. 1992;206:394–399. doi: 10.1016/0003-2697(92)90384-j. [DOI] [PubMed] [Google Scholar]
- 27.Reinhart GD. The determination of thermodynamic allosteric parameters of an enzyme undergoing steady-state turnover. Arch Biochem Biophys. 1983;224:389–401. doi: 10.1016/0003-9861(83)90225-4. [DOI] [PubMed] [Google Scholar]
- 28.Reinhart GD. Influence of pH on the regulatory kinetics of rat liver phosphofructokinase: a thermodynamic linked-function analysis. Biochemistry. 1985;24:7166–7172. doi: 10.1021/bi00346a022. [DOI] [PubMed] [Google Scholar]
- 29.Reinhart GD. Quantitative analysis and interpretation of allosteric behavior. Methods Enzymol. 2004;380:187–203. doi: 10.1016/S0076-6879(04)80009-0. [DOI] [PubMed] [Google Scholar]
- 30.Reinhart GD, Hartleip SB, Symcox MM. Role of coupling entropy in establishing the nature and magnitude of allosteric response. Proc Natl Acad Sci USA. 1989;86:4032–4036. doi: 10.1073/pnas.86.11.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prabhu NV, Sharp KA. Heat capacity in proteins. Annu Rev Phys Chem. 2005;56:521–548. doi: 10.1146/annurev.physchem.56.092503.141202. [DOI] [PubMed] [Google Scholar]
- 32.Reinhart GD. Linked-function origins of cooperativity in a symmetrical dimer. Biophys Chem. 1988;30:159–172. doi: 10.1016/0301-4622(88)85013-0. [DOI] [PubMed] [Google Scholar]
- 33.Johnson JL, Reinhart GD. Mgatp and Fructose 6-Phosphate Interactions with Phosphofructokinase from Escherichia-Coli. Biochemistry. 1992;31:11510–11518. doi: 10.1021/bi00161a032. [DOI] [PubMed] [Google Scholar]
- 34.Plou FJ, Ballesteros A. Stability and stabilization of biocatalysts. Trends Biotechnol. 1999;17:304–306. doi: 10.1016/s0167-7799(99)01343-8. [DOI] [PubMed] [Google Scholar]
- 35.Somero GN. Temperature adaptation of enzymes - biological optimization through structure-function compromises. Annu Rev Ecol Syst. 1978;9:1–29. [Google Scholar]
- 36.Jaenicke R. Protein stability and molecular adaptation to extreme conditions. Eur J Biochem/ FEBS. 1991;202:715–728. doi: 10.1111/j.1432-1033.1991.tb16426.x. [DOI] [PubMed] [Google Scholar]
- 37.Blangy D, Buc H, Monod J. Kinetics of the allosteric interactions of phosphofructokinase from Escherichia coli. Journal of molecular biology. 1968;31:13–35. doi: 10.1016/0022-2836(68)90051-x. [DOI] [PubMed] [Google Scholar]
- 38.Pham AS, Janiak-Spens F, Reinhart GD. Persistent binding of MgADP to the E187A mutant of Escherichia coli phosphofructokinase in the absence of allosteric effects. Biochemistry. 2001;40:4140–4149. doi: 10.1021/bi001768z. [DOI] [PubMed] [Google Scholar]
- 39.Fenton AW, Paricharttanakul NM, Reinhart GD. Identification of substrate contact residues important for the allosteric regulation of phosphofructokinase from Eschericia coli. Biochemistry. 2003;42:6453–6459. doi: 10.1021/bi034273t. [DOI] [PubMed] [Google Scholar]
- 40.Kimmel JL, Reinhart GD. Reevaluation of the accepted allosteric mechanism of phosphofructokinase from Bacillus stearothermophilus. P Natl Acad Sci USA. 2000;97:3844–3849. doi: 10.1073/pnas.050588097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturtevant JM. Heat-capacity and entropy changes in processes involving proteins. Proc Natl Acad Sci USA. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshizaki F, Imahori K. Key role of phosphoenolpyruvate in the regulation of glycolysis-gluconeogenesis in Thermus thermophilus HB-8. Agric Biol Chem. 1979;43:537–545. [Google Scholar]
- 43.Braxton BL, Tlapak-Simmons VL, Reinhart GD. Temperature-induced inversion of allosteric phenomena. J Biol Chem. 1994;269:47–50. [PubMed] [Google Scholar]