Abstract
Tissue homeostasis and regenerative capacity rely on rare populations of somatic stem cells endowed with the potential to self-renew and differentiate. During aging, many tissues show a decline in regenerative potential coupled with a loss of stem cell function. Cells including somatic stem cells have evolved a series of checks and balances to sense and repair cellular damage to maximize tissue function. However, during aging the mechanisms that protect normal cell function begin to fail. In this review, we will discuss how common cellular mechanisms that maintain tissue fidelity and organismal lifespan impact somatic stem cell function. We will highlight context-dependent changes and commonalities that define aging, by focusing on three age-sensitive stem cell compartments: blood, neural, and muscle. Understanding the interaction between extrinsic regulators and intrinsic effectors that operate within different stem cell compartments is likely to have important implications for identifying strategies to improve health span and treat age-related degenerative diseases.
1. INTRODUCTION
Aging leads to profound effects on many, if not all tissues of the body, including muscle weakness (Lang et al., 2010), graying and loss of hair (Nishimura, Granter, & Fisher, 2005), a decline in cognition (Bishop, Lu, & Yankner, 2010), and impaired immune function (Geiger, de Haan, & Florian, 2013). The regenerative response of tissues after injury is often delayed leading to slower repair of parenchyma that is commonly replaced by accumulation of adipogenic or fibrogenic accumulation (Kapetanaki, Mora, & Rojas, 2013).
Maintenance and repair of many adult tissues rely on stem cells. These cells reside at the top of a cellular hierarchy endowed with the ability to self-renew and differentiate, whereas their downstream progeny is restricted to replenishing the differentiated tissue (Orford & Scadden, 2008; Simons & Clevers, 2011). Stem cells spend relatively long periods of time in a quiescent state compared to their progeny, which proliferate to produce numerous differentiated cells that replace or repair the tissue throughout the lifespan of the organism (Li & Clevers, 2010; Orford & Scadden, 2008). In response to increased demand such as growth or regeneration after injury, stem cells break from quiescence, enter the cell cycle, and divide either symmetrically or asymmetrically to replace the stem cell pool and the committed progenitor pool. To avoid abnormal growth or loss of tissues, the balance between production of stem cells and differentiated progeny needs to be tightly regulated. Multiple levels of cell autonomous and extrinsic factors tightly control fate decisions of stem cells. For example, a specialized microenvironment, also known as the stem cell niche, provides extrinsic signals in the form of paracrine or juxtacrine signaling that is essential for maintenance of stem cell function and restricting stem cell numbers (Li & Clevers, 2010; Morrison & Spradling, 2008). It is possible that extrinsic signals derived from the local niche and systemic environment shape the epigenetic landscape of the stem cell, which influences gene expression to dictate stem cell fate (Pollina & Brunet, 2011).
Recent technological advances in genetic reporters and cell surface marker detection have revealed a greater complexity in stem cell populations than previously anticipated (Grompe, 2012; Simons & Clevers, 2011). Across different niches, stem cells with a restricted proliferative history, termed slow dividing stem cells, are endowed with high self-renewing potential compared with stem cells from the same tissue that have undergone more divisions during their history (Chakkalakal, Jones, Basson, & Brack, 2012; Foudi et al., 2009; Wilson et al., 2008; Zhang, Cheong, Ciapurin, McDermitt, & Tumbar, 2009). That slow dividing cell give rise to frequently dividing cells, but not vice versa, demonstrates a hierarchical relationship that is controlled by or correlated with proliferative output. As the markers to define stem cells increase, the degree of heterogeneity within a population is becoming appreciated. Within the same tissue, subsets of stem cells can be indiscriminately identified that are biased to differentiate into distinct cell types, albeit restricted in the same developmental lineage. Due to this level of complexity, it is possible that changes in function between two points (i.e., adult and aged) are a feature of extrinsic and intrinsic changes in all stem cells or the expansion of biased subsets over others.
Studies on stem cell aging and the molecular regulation of lifespan were pioneered in nonmammalian systems (Jones & Rando, 2011; Kenyon, 2010). In Drosophila, the number of stem cells in the testis and ovary declines during aging, due in part to age-dependent changes in the niche (Boyle, Wong, Rocha, & Jones, 2007; Pan et al., 2007). Moreover, deregulation of Notch/JNK (Jun-activated kinase) signaling during aging causes loss of intestinal tissue homeostasis through overproliferation and inappropriate differentiation of intestinal stem cells (ISCs) (Biteau, Hochmuth, & Jasper, 2008). Remarkably, reducing proliferation of ISCs through repression of insulin-like growth factor (IGF) and JNK stress pathways demonstrates an inverse correlation between lifespan and ISC proliferation. Maximal lifespan was achieved when ISC proliferation was reduced, which correlated with improved metabolic homeostasis of aged Drosophila (Biteau et al., 2010). This demonstrates a direct link between lifespan and stem cell activity, at least in the intestine. Moreover, stem cell function and lifespan are affected by metabolic and epigenetic factors that change with age (Bratic & Larsson, 2013; Eijkelenboom & Burgering, 2013; Laplante & Sabatini, 2012; Pollina & Brunet, 2011).
At the organismal level, aging is based on a chronological clock. At the cellular level, age can be broken down into two components, replicative and chronological age. Replicative age relates to the proliferative output of stem cells during their history. Chronological aging is linked to the age of the intracellular constituents of stem cells. Hierarchically upstream stem cells are widely considered restricted in their proliferative output compared to downstream progenitors. Therefore, aging of quiescent stem cells occurs primarily on a chronological clock, whereas the age of downstream progeny is based on a replicative clock.
During each round of division a cell has to faithfully copy its DNA, repair any errors, and transcribe and translate proteins necessary for ensuring the appropriate fate and functionality of the cell. Each of these processes is error prone and demands high-fidelity repair processes and functional checkpoints. In the absence of replication, the cellular constituents may be the same age as the organism itself. In a nonreplicating cell, the cellular constituents must repair faulty DNA and remove damaged and misfolded proteins that are normally cleared through cell division to avoid protein toxicity. Whether a stem cell ages on a chronological or replicative clock will affect the cellular damage and repair processes that are invoked, which ultimately may impact the aging phenotype of the stem cell.
In the mammalian organism, it is unquestionable that tissue homeostasis and regenerative capacity decline during aging. It is clear that age-dependent stem cell dysfunction can manifest in many forms, such as depletion of available stem cells, deregulation of cell fate, that is, loss of self-renewal and/or differentiation, increased apoptosis, and senescence. However age-dependent changes across different niches are not conserved. For example, as we discuss later, the number of skeletal muscle stem cells and neural stem cells (NSCs) decreases, whereas the number of blood forming stem cells is maintained during aging. These differences may reflect the extent of accrued damage, the capacity to repair, or the ability to persist in spite of damage. It is noteworthy that the defects that impact stem cell function with age are unique to stem cells, such as impaired self-renewal potential; however, some changes such as apoptosis or senescence may be generalizable to all types of somatic cells during aging and organismal lifespan.
In this review, we discuss the mechanisms and consequences of cellular and organismal aging that are used reiteratively in the regulation of mammalian somatic stem cells. We also highlight the common and context-specific age-dependent changes of stem cell function by focusing on three paradigmatic stem cell populations: blood, neural, and skeletal muscle, due to their previously characterized aged phenotypes.
2. MOLECULAR PLAYERS OF CELLULAR AGING
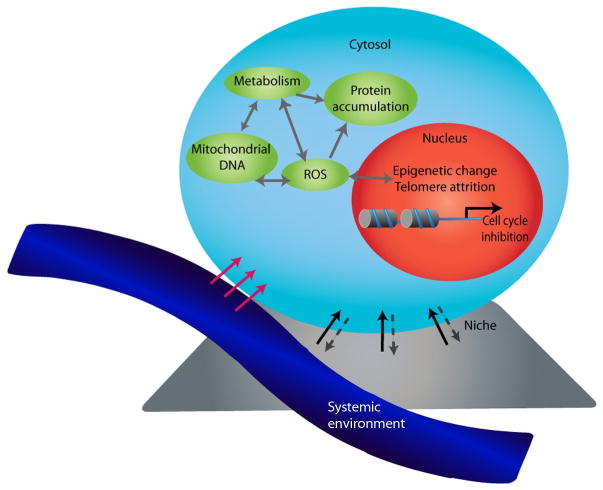
In this section, we outline the cell-intrinsic mechanisms and extrinsic modifiers that have been demonstrated to control cellular aging (Fig. 14.1).
Figure 41.1.
Multiple components driving stem cell aging. Stem cell aging occurs through changes in both cell-extrinsic regulators and intrinsic effectors. Cell-external influences such as changes in systemic factors from the bloodstream (blue) or factors from the stem cell niche (gray) can alter stem cell function by inducing signaling pathways and modifying the epigenetic and genetic signature of stem cells. The aged stem cell may also directly alter the niche. Aging is also driven by deregulation of cell-intrinsic effectors that control proteostasis, mitochondrial function, and metabolic control. Many of these modifications are interrelated suggesting one can impact another, leading to stem cell failure at multiple levels. ROS, reactive oxygen species; mtDNA, mitochondrial DNA.
2.1. Genome
DNA is relatively unstable and prone to DNA mutations through by-products of cellular metabolism such as reactive oxygen species (ROS) and environmentally induced lesions such as exposure to ultraviolet (UV) or irradiation (IR). Accumulation of irreversible genomic DNA damage has been implicated as a prominent cause of aging (Eijkelenboom & Burgering, 2013; Kenyon, 2010; Sperka, Wang, & Rudolph, 2012). The WRN (Werner syndrome ATP-dependent) helicase and ATM (Ataxia Telangiectasia Mutated) kinase are essential for DNA repair. Deletion of either leads to a premature aging phenotype in mice (Lombard et al., 2000; Wong et al., 2003).
Maintenance of genomic integrity and fidelity is dependent on protective DNA repair mechanisms. ROS-induced damage is repaired by base excision repair, whereas repair of double strand breaks (DSBs) after IR is achieved by nucleotide excision repair (Blanpain, Mohrin, Sotiropoulou, & Passegue, 2011). Cells invoke distinct methods to repair DSBs depending on the cell cycle status; quiescent cells are repaired by non-homologous end joining (NHEJ), proliferating cells are repaired by homologous recombination (HR). Therefore, due to their quiescent nature, somatic stem cells may rely on NHEJ for repair. NHEJ is a more error-prone repair mechanism, as it does not rely on the other intact DNA strand as a template. Therefore, it is possible that the quiescent state of stem cells may increase their likelihood of accumulating DNA damage and impacting stem cell function; however, it is also possible that when the quiescent stem cell enters the cell cycle any damage will be repaired using HR.
Telomere integrity is cited as a major regulator of lifespan longevity (Flores et al., 2008; Vaziri & Benchimol, 1996). Age-associated telomere reduction has been shown to threaten chromosome integrity of highly proliferative aging tissues (Vaziri & Benchimol, 1996). A gradual decline of telo-mere length with age has been observed in mouse (Flores et al., 2008) and human tissues (Harley, Futcher, & Greider, 1990). Further evidence supports the notion that age-related decrease in telomere length could occur through loss of telomerase, which maintains telomere length. Telomerase mutations, which are found in patients with dyskeratosis congenita, who have shorter telomeres, show a premature aging phenotype (Mitchell, Wood, & Collins, 1999). In a mouse model, loss of telomerase promotes lineage skewedness, which is one of the aging phenotypes in HSCs (hematopoietic stem cells) (Ju et al., 2007). Overexpression of telomerase delays the appearance of age-dependent phenotypes and shows cancer resistance (Tomas-Loba et al., 2008). These studies show that shortened telomeres or a deficit in telomerase function can cause a functional decline of tissues. Therefore, the proliferative output and levels of telomerase will influence the contribution of telomere biology to stem cell decline during aging.
Accumulation of DNA damage results in a cell checkpoint response, involving the upregulation of cell cycle inhibitors, such as p16Ink4a, p19Arf, and p53 that leads to cell cycle arrest, senescence, apoptosis, or differentiation (Signer & Morrison, 2013; Sperka et al., 2012). p16Ink4a, p19Arf, and p53 are increased with age (Sahin & DePinho, 2012; Sperka et al., 2012) and their loss is associated with tumor incidence (Kemp, Donehower, Bradley, & Balmain, 1993; Matheu et al., 2007). Intuitively, higher expression of tumor suppressors may be associated with increased incidences of cancers with age as a compensatory mechanism to prevent the prevalence of tumors (Matheu et al., 2007). Insufficiency of BubR1, which is one of the major components of the mitotic checkpoint for spindle assembly, increases expression of both p16Ink4a and p19Arf that causes premature aging by progressively increased aneuploidy and senescence (Baker et al., 2004). As expected, loss of p16Ink4a attenuates age-dependent decline in proliferation and function of stem cells (Janzen et al., 2006; Krishnamurthy et al., 2006; Molofsky et al., 2006). In contrast to differentiated tissue, there is a scarcity of data showing that senescence occurs to stem cells in vivo. Interestingly, the influence of senescent cells on tissue homeostasis may be through their influence on neighboring cells via secretion of paracrine factors (Burtner & Kennedy, 2010; Campisi, 2005; Krishnamurthy et al., 2004).
2.2. Mitochondrial DNA mutation
Oxidative phosphorylation in the mitochondrial electron transport chain generates ROS, which is highly reactive and toxic to mitochondrial (mt) DNA, leads to a decline in mitochondrial functions (Kujoth et al., 2005; Trifunovic et al., 2004). With age, increased levels of ROS have been observed along with dysfunctional mitochondria and considered as a cause of aging (Bratic & Larsson, 2013; Trifunovic et al., 2004).
Can mtDNA mutations affect lifespan and aging? Homozygous knock-in mice engineered to express mtDNA polymerase that have defective proof-reading, show increased mtDNA point mutations and deletions, which is associated with reduced lifespan and premature aging phenotypes (Kujoth et al., 2005; Trifunovic et al., 2004). In addition, mice that lack mtDNA polymerase exonuclease display neural and hematopoietic progenitor dysfunction and a progeria phenotype (Ahlqvist et al., 2012). In contrast, introduction of random point mutations in mtDNA to a mouse model was not sufficient to reduce lifespan (Edgar et al., 2009). Therefore, the extent of mtDNA damage or the specific mutation in the mitochondrial genome will influence the impact to the stem cell.
2.3. Epigenome
The epigenetic code enables cells to receive and remember environmentally induced signals to create a more stable state (Cavalli & Paro, 1998; Grewal & Klar, 1996). DNA methylation and histone modifications provide transient regulation of gene expression, but not through the DNA sequence itself. The identification of specific enzymes that balance these modifications suggests that modulation of the epigenome may influence stem cell fate and the aging process.
Histone acetylases and deacetylases add and remove acetyl groups on histones, respectively. Sirtuins (Sirt) are a family of histone deacetylases that have been shown to regulate organismal lifespan as well as oxidative stress and DNA damages (Kaeberlein, McVey, & Guarente, 1999; Kennedy, Austriaco, Zhang, & Guarente, 1995; Mostoslavsky et al., 2006; Rodgers et al., 2005). Sirt1 and Sirt2 have been implicated in life extension in different model organisms (Kaeberlein et al., 1999; Kennedy et al., 1995). However, moderate overexpression of Sirt1 in mice was not sufficient to increase lifespan (Herranz et al., 2010). In contrast, overexpression of Sirt6 extends lifespan of male mice by regulating the IGF signaling pathway (Kanfi et al., 2012), which has a key role in aging (Kenyon, 2010).
Histone methylation also plays a role in aging. The ASH-2 trithorax complex 9, which trimethylates H3K4, is a lifespan regulator in Caenorhabditis elegans. Deficiencies in the ASH-2 complex and the H3K4 methyltransferase SET-2 are shown to extend worm lifespan (Greer et al., 2010). Moreover, deficiencies in the ASH-2 complex only in parents can be inherited to descendants spanning several generations (Greer et al., 2011). Therefore, ancestral chromatin states may be incompletely reprogrammed and influence gene expression of descendants during future generations.
Chromatin modifiers have been shown to control cellular proliferation, metabolism, and even longevity (Florian et al., 2012; Greer et al., 2010; Jacobs, Kieboom, Marino, DePinho, & van Lohuizen, 1999). The polycomb repressive complex (PRC), which consists of PRC1 (core subunit Bmi1, Cbx, Ring1, and Phc) and PRC2 (structural members including Ezh2, Eed, and Suz12) directly methylate specific lysines on histones to control levels of gene expression (Margueron & Reinberg, 2011; Sauvageau & Sauvageau, 2010). Bmi1 has been related to organismal longevity (Greer et al., 2010), self-renewal and differentiation of HSCs (Hidalgo et al., 2012; Lessard & Sauvageau, 2003; Park et al., 2003), NSC (Fasano et al., 2007; Molofsky et al., 2003), lung (Zacharek et al., 2011), prostate (Lukacs, Memarzadeh, Wu, & Witte, 2010), and epidermal stem cells (Ezhkova et al., 2009). Expression of p16Ink4a and p19Arf, which are encoded from the Cdkn2a locus, increases with age (Krishnamurthy et al., 2004). The Ink4a/Arf locus is regulated by PRC1 including Bmi1 (Lessard & Sauvageau, 2003; Lukacs et al., 2010; Molofsky et al., 2003; Park et al., 2003; Zacharek et al., 2011). Indeed, attenuation of p16Ink4a partially rescues self-renewal functions of stem cells lacking Bmi1 (Lessard & Sauvageau, 2003; Molofsky et al., 2003; Park et al., 2003; Sauvageau & Sauvageau, 2010). A recent report shows association of PRC2 with aging phenotypes in HSCs (Beerman et al., 2013). Given the roles of PRC-mediated transcriptional regulation of specific genes critical for self-renewal, differentiation, and DNA damage repair in adult stem cells their contribution would not be surprising.
The addition of methyl groups to specific regions of the DNA sequence suppresses gene transcription. DNA methylation patterns are controlled by at least three DNA methyltransferases, DNMT1, DNMT3A, and DNMT3B that catalyze the transfer of a methyl group to DNA (Smith & Meissner, 2013). Aged tissues show a decrease in DNA methylation (Maegawa et al., 2010; Pollina & Brunet, 2011). However, specific loci tend to become hypermethylated (Maegawa et al., 2010). A conditional loss-of-function approach to delete Dnmt1 from hair follicle stem cells led to stem cell failure and an exaggerated aged phenotype to the hair follicles and skin (Li et al., 2012). In addition, inhibition of DNMT1 or 3A in a mesenchymal stem cell population increased cell senescence in vitro, through the upregulation of cell cycle inhibitors P16Ink4a and p21Cip1/Waf1 (So, Jung, Lee, Kim, & Kang, 2011). Understanding the function of the methylated target genes will be important to decipher whether global or specific methylation drives stem cell aging.
2.4. Protein homeostasis
Tight regulation of the proteome is required for normal cell function. The accumulation of misfolded or aggregated proteins can disrupt intracellular signaling cascades and induce toxicity and apoptosis. In response to stress, cellular homeostasis is achieved by removal of the cell via apoptosis or removal of damaged macromolecules via autophagy. Autophagy is a potent repressor of unwanted apoptosis or necrosis possibly through liberation of Bcl-2 (Lee et al., 2009; Pattingre et al., 2005). Removal of unwanted proteins relies on autophagasomes, chaperones, lysosomes, and ubiquitin-proteasome system (UPS) (Ellis & Pinheiro, 2002; Hartl, Bracher, & Hayer-Hartl, 2011). Increased protein accumulation in aged tissues suggests that homeostasis of the proteome is deregulated during aging (Chiti, Stefani, Taddei, Ramponi, & Dobson, 2003). UPS is one of the main proteolytic mechanisms to ensure degradation of damaged proteins (Hartl et al., 2011; Rubinsztein, Marino, & Kroemer, 2011). UPS selectively tags poly-ubiquitin chains on damaged proteins and degrades the tagged proteins by proteasome complex machinery. During aging, proteasome activity declines (Conconi, Szweda, Levine, Stadtman, & Friguet, 1996; Shibatani, Nazir, & Ward, 1996).
Genetic inhibition of autophagy induces premature aging phenotypes. In C. elegans, loss of members of the autophagy pathway, Atg3 (autophagy-related 3), Atg9, Atg18, and Beclin1, decrease the lifespan (Toth et al., 2008). In addition, the deletion of Sestrin1 (activator the AMP-responsive protein kinase and repressor of mTOR pathway) (Lee, Budanov, et al., 2010), possibly through decreased autophagy, reduces the longevity of Drosophila. In contrast, overexpression of Atg8 increased Drosophila lifespan (Simonsen et al., 2008). In mice liver, expression levels of Lamp2a decline with age. Overexpression of hepatocyte-specific Lamp2a, a key protein in the chaperone-mediated autophagy (CMA) pathway, prevents the aging-associated defect in CMA, which leads to a reduction in toxic proteins and aggregates, and apoptotic cells within the liver (Zhang & Cuervo, 2008). These studies indicate that aging is correlated with a decline in autophagy function.
2.5. Energy metabolism
Mitochondrial and metabolic activity during development and normal aging may influence life span and rate of aging phenotypes (Sahin & DePinho, 2012; Signer & Morrison, 2013). The FoxO family of transcription factors, as a part of IGF/mTOR (target of rapamycin) pathway has been shown to regulate metabolism and oxidative stress by promoting antioxidant enzymes (Murphy et al., 2003; Wang, Bohmann, & Jasper, 2005). Stimulation of the IGF pathway induces the PI3K/AKT/mTORC2 pathway, which leads to inactivation of FoxO due to phosphorylation (Kenyon, 2010). Caloric restriction (CR) has been shown to increase lifespan in various species from yeast to mammals (Kenyon, 2010; Mair & Dillin, 2008). Although the underlying mechanisms behind lifespan extension are not fully resolved, recent papers show that CR preserves numbers and functions of stem cells (Cerletti, Jang, Finley, Haigis, & Wagers, 2012; Yilmaz et al., 2012). Genetic analysis in rodents shows modulation of TOR pathway is a major effector of CR response. mTOR exists as two complexes, mTORC1 and mTORC2. As mentioned before, mTORC2 is a downstream target of IGF-PI3K pathway that activates the Akt-FoxO signaling pathway to promote cell proliferation and regulate oxidative stress. In contrast, mTORC1 promotes protein translation, ribosome biogenesis, and regulates autophagy (Laplante & Sabatini, 2012). Therefore, mTOR influences many critical cellular processes and will likely be involved in age-associated stem cell decline.
3. EXTRINSIC REGULATION OF AGED CELLULAR AND TISSUE HOMEOSTASIS
In Section 2, the intrinsic effectors of aged cell function were considered. However, it is known that tissue-specific stem cells reside in niches. Across different mammalian stem cell compartments niches have been operationally defined (Morrison & Spradling, 2008). Local signaling from the niche regulates tissue maintenance by preserving the function of stem cells. Studies from drosophila have provided direct evidence how local signaling factors from the niche are required for maintenance of stem cell number and function throughout life (Boyle et al., 2007; Pan et al., 2007).
In addition to the niche, tissue-specific stem cell niches lie near blood vessels, containing numerous soluble growth factors and cytokines that can potentially influence stem cell function. Direct evidence of a systemic influence on stem cell function during aging was provided using heterochronic parabiosis, the surgical pairing of two mice to achieve a shared circulation between young and aged mice (Conboy, Conboy, & Rando, 2013). This system has been used to determine whether the decline in tissue-specific stem cell function with age was due to cell-intrinsic irreversible, age-related changes or cell-extrinsic influence by the environment in muscle (Brack et al., 2007; Conboy et al., 2005), liver (Conboy et al., 2005), heart (Loffredo et al., 2013), and the central nervous system (Ruckh et al., 2012; Villeda et al., 2011). The data from these diverse tissue types indicate that the aged systemic environment markedly contributes to aged stem cell phenotypes and nonstem cell phenotypes.
Another example of tissue rejuvenation was provided by aged skin. It is known that Nf-κb increases with age in multiple tissues (Chambers et al., 2007; Helenius, Hanninen, Lehtinen, & Salminen, 1996; Korhonen, Helenius, & Salminen, 1997). Nf-κb is activated by different cell stressors that accelerate aging such as DNA damage and oxidative stress (Pasparakis, 2009). Moreover, signals such as Sirt1 and FoxO3 that positively regulate longevity are found to repress Nf-κb (Lin, Hron, & Peng, 2004; Yeung et al., 2004). Remarkably, blocking Nf-κb activity for 2 weeks in aged skin led to partial rejuvenation of the gene profile, reverting half of the age-dependent genes back to youthful levels (Adler, Kawahara, Segal, & Chang, 2008). Moreover, many of the phenotypes associated with aged skin, such as increased epidermal thickness, increased senescence, and decreased proliferation were all partially restored back to levels seen in younger skin. Together, these results demonstrate the potential of multiple aged tissues to undergo molecular and phenotypic rejuvenation after modulation of the extrinsic environment.
4. AGING IN TISSUE-SPECIFIC STEM CELLS
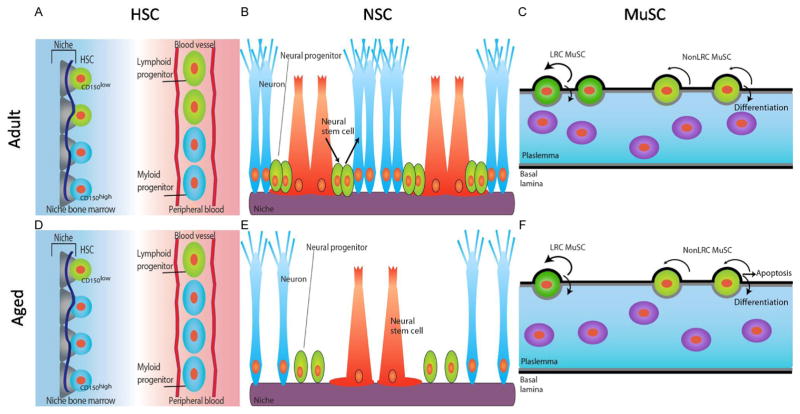
The general cellular processes that breakdown during aging appear to impact many different cell types including somatic stem cells. The unique functions, intrinsic regulators, and specialized environments of tissue-specific stem cells are likely to impose distinct responses and phenotypic consequences to aging. In this section, we will discuss age-related phenotypes and the mechanisms that lead to stem cell demise in three distinct stem cell niches (Fig. 14.2).
Figure 14.2.
Schematic diagrams of adult stem cells and niche in bone marrow, brain, and muscle tissue. (A, D) Quiescent adult hematopoietic stem cells (HSCs) are located in the endosteal region of adult (A) and aged (D) bone marrow (left) residing in a niche composed of endothelial cells, osteoblasts (gray), and perivascular cells (dark blue line). The adult HSC pool consists of lymphoid-biased (CD150low) HSCs (green) and myeloid-biased (CD150high) HSCs (blue), with high self-renewal potential and balanced differentiation toward lymphoid (green, elongated) and myeloid progenitor cells (blue, elongated) in the peripheral blood (right, A and D). In aged HSC niche, selective expansion/retention of myeloid-biased CD150high HSCs skewing differentiation toward the myeloid lineage due to the robust self-renewal potential of aged CD150high HSCs compared to CD150low HSCs. (B, E) Neural stem cells (NSCs) reside at subgranular zone (SGZ) of the dentate gyrus of the hippocampus in adult (B) and aged (E) brain. NSCs (red) can give rise to neural progenitors (green), which progressively differentiate into neurons (blue). Systemic factors from the blood and local niche factors (purple) control NSC number and function. With age, number of NSCs and adult neurogenesis (making new neurons) declines. (C, F) Muscle satellite stem cells (MuSCs) in adult (C) and aged (F) muscle fibers. MuSCs (green) reside between the plasmalemma (gray line) and the basal lamina (black line) of muscle fibers. Long-term label retaining (LRC) MuSCs (dark green) have a high capacity for self-renewal and differentiation into muscle. nonLRC MuSCs (light green) are capable of differentiation but have limited self-renewal capacity. With age, the number of LRC MuSCs and fusion competent progenitors declines leading to a reduced number of functional stem cells and defective muscle repair.
4.1. Hematopoietic stem cells
HSCs reside in the bone marrow, have long-term self-renewal potential, and can differentiate into committed progenitors that are critical for generating downstream progeny of the blood system (Orkin & Zon, 2008).
Aging in the blood system drives changes in HSC numbers, decreases regenerative potential, and skews differentiation potential toward myeloid lineages (Beerman, Maloney, Weissmann, & Rossi, 2010; Geiger et al., 2013). These phenotypes are contributed to by both cell-autonomous and cell-extrinsic factors. Contrary to other stem cell niches, the number of HSCs in the bone marrow increases with age (Beerman, Bhattacharya, et al., 2010; Beerman et al., 2013; Challen, Boles, Chambers, & Goodell, 2010). Intuitively, increased HSCs in aged bone marrow may seem beneficial. However, analyzed on a per-cell basis or in a competitive transplant setting, the gold standard for assessing stem cell activity, aged HSCs show defects in self-renewal potential and long-term reconstitution of the blood (Chambers et al., 2007; Janzen et al., 2006; Rossi et al., 2005). Therefore, HSCs are functionally impaired due in part to cell-autonomous defects. It remains to be answered why the number of HSCs in aged population increases.
One of the hallmarks of aging in HSCs is a skewed differentiation potential toward the myeloid lineage at the expense of the lymphoid and erythroid lineages (Beerman, Bhattacharya, et al., 2010; Benz et al., 2012; Challen et al., 2010; Dykstra, Olthof, Schreuder, Ritsema, & de Haan, 2011; Morita, Ema, & Nakauchi, 2010; Rossi et al., 2005). Such biased lineage potentials in aged HSCs are not from changes in differentiation potential of individual HSCs but rather by changes in composition of the HSC pool (Beerman, Bhattacharya, et al., 2010; Beerman, Maloney, et al., 2010; Challen et al., 2010; Morita et al., 2010). Prevalence of myeloid-biased (CD150high) HSCs may be a result of more robust self-renewal potential than of lymphoid-biased (CD150low) HSCs (Beerman, Bhattacharya, et al., 2010; Challen et al., 2010). Both aged CD150high and CD150low HSCs show reduced proliferative capacity and homing to the bone marrow. This indicates that the functional heterogeneity within the HSC pool is cell autonomously maintained and has the potential to skew lineage phenotypes over time.
Genomic integrity of any cell is dependent on protective DNA repair mechanisms. With age, DNA damage accumulates in HSCs, indicating a decrease in the function of appropriate repair mechanisms (Rossi, Seita, et al., 2007). The mode of DNA repair differs between quiescent and cycling stem cells. Quiescent HSCs rely on NHEJ for DNA repair, which is a more error-prone mechanism and will impact the accrual of DNA damage throughout life, leading to genomic rearrangements (Mohrin et al., 2010). Whether the accrued damage is cleared when aged quiescent HSCs enter into the cell cycle and take up a more effective mode of DNA repair will determine the extent that DNA damage accumulation impacts aged stem cell function.
Mutation or deletion in DNA repair components such as Msh2−/− (mismatch repair) (Reese, Liu, & Gerson, 2003), Brca2−/− (HR) (Navarro et al., 2006), XpdTTD (nucleotide excision repair) (Rossi, Bryder, et al., 2007), Ku80−/− (NHEJ) (Rossi, Bryder, et al., 2007), and Lig4Y288C (NHEJ) (Nijnik et al., 2007) all show reduced HSC function, leading to depletion of HSCs in mice. Deletion of DNA damage sensor Atm causes elevation of ROS, and loss of HSC quiescence, defects in repopulating capacity, and ultimately a depletion of the HSC pool (Ito et al., 2006). Antioxidant treatment on Atm−/− HSCs rescues the reduced repopulating potential when challenged by serial transplantation. This implies that elevated ROS can cause aging phenotypes in HSCs.
ROS can also mutate mtDNA, which is irreversible and deleterious to mitochondrial function (Kujoth et al., 2005; Trifunovic et al., 2004). In HSCs, the skewed differentiation toward myeloid lineage was observed in mutant mtDNA polymerase γ mice, and this phenotype was rescued by antioxidant-mediated ROS inhibition (Trifunovic et al., 2004), which implies mtDNA mutagenesis modulates adult stem cell function, thus leading to aging phenotypes. However, Bryder and colleagues show stark transcriptional differences between the aged mutant and wild-type mice. Moreover, unlike physiological aging that has epigenetic and genetic signature (Chambers et al., 2007), the transcriptional signature of HSCs is not impacted by the mitochondrial mutation (Norddahl et al., 2011). To date, it remains to be investigated whether mtDNA mutation directly contributes to physiological stem cell aging.
Other major players of cell-intrinsic regulation of HSC aging are oxidative damage, metabolic stress, and autophagy. Autophagy is a stress-response mechanism to clear damaged proteins and a major contributor in aging (Rubinsztein et al., 2011). Loss of autophagy by deletion of Atg7 (Mortensen et al., 2011) and Fip200 (Liu et al., 2010), members of autophagosome, increases ROS levels that lead to HSC depletion in mice, which indicates that autophagy is essential for HSC homeostasis. Is aging of the stem cell caused by a decline in autophagy? Recently, Passegue and colleagues challenged this notion (Warr et al., 2013). The authors revealed that freshly isolated aged HSCs have basal levels of autophagy, unlike adult HSCs that only when activated and stressed, mount an autophagic response. Prevention of the autophagic response led to HSC apoptosis, suggesting that steady-state autophagy is essential for aged HSC maintenance.
Oxidative stress and its regulation via FoxO are important for aging in HSCs. Conditional deletion of FoxO1, 3, and 4 leads to a loss in HSC quiescence, increased apoptosis, myeloid-biased differentiation, and decreased long-term HSC maintenance (Miyamoto et al., 2007; Tothova et al., 2007). A partial rescue of these phenotypes was observed using the ROS inhibitor N-acetyl-L-cysteine (Tothova et al., 2007), indicating that FoxO family members act as an important modulator of oxidative stress and HSC function throughout life.
The Sirtuin (Sirt) family is another candidate regulator of stem cell aging. Deletion of Sirt1 in adult HSCs causes increased proliferation and elevated DNA damage, which leads to loss of long-term HSC populations (Singh et al., 2013). Sirt3, which controls the activity of mitochondrial enzyme acetyl coenzyme A synthetase 2 (AceCS2) and electron transport chain complex 1 in mitochondria (Finkel, Deng, & Mostoslavsky, 2009), is critical for adult and aged HSC function under transplantation stress conditions (Brown et al., 2013). Interestingly, overexpression of Sirt3 can compensate for the ROS-mediated aged HSC phenotype (Bratic & Larsson, 2013). It remains unclear whether other members of Sirtuin family can also reverse aging-related phenotypes in aged stem cells.
mTORC1 functions as an energy sensor and attenuator of autophagy (Nicklin et al., 2009). Calorie restriction attenuates the mTORC1 signaling pathway, which increases stem cell proliferation and stem cells in the intestine through mTORC1 expressed in the niche (Yilmaz et al., 2012). With age, expression of mTORC1 increases in HSCs and its repression by rapamycin restores self-renewal of aged HSCs (Chen, Liu, Liu, & Zheng, 2009). In contrast, indirect activation of mTOR via conditional deletion of Pten in young HSCs induces a progeria phenotype and depletion of HSCs (Kalaitzidis et al., 2012; Lee, Nakada, et al., 2010; Magee et al., 2012). Therefore, HSCs and other stem cells are sensitive to metabolic output via mTORC1 levels. It is unlikely the requirement of mTORC1 is age specific or stem cell specific; however, any age-dependent change in metabolic regulation may increase the sensitivity of aged stem cells to mTORC1 levels.
Using genome-wide DNA methylation analysis of human and mouse HSCs, it was demonstrated that genes associated with myeloid lineage become hypomethylated with age. In contrast, site-specific hypermethylation was observed at genes associated with the PRC2 complex (Beerman et al., 2013; Bocker et al., 2011). Interestingly, age-dependent myeloid-skewedness and DNA methylation are largely dependent on the proliferative history of HSCs. Using proliferative demand as a driver of HSC aging, it was demonstrated that replicative aging is distinct from chronological aging at the DNA methylation level (Beerman et al., 2013). Both aged HSCs and adult HSCs under moderate proliferative demand underwent site-specific hypermethylation at the PRC2 complex, which was accompanied by transcriptional repression, as suggested previously (Chambers et al., 2007). In contrast, adult HSCs with a high level of experimentally enforced proliferation had a distinct methylome. Therefore, site-specific methylome changes appear to correlate with the functional decline of aged HSCs. Moreover, these data confirm at the epigenome level that stem cells with limited turnover throughout life experience the effects of chronological rather than proliferative aging.
The influence of the systemic environment and the local niche on HSC aging is becoming apparent. Recent data show that both systemic and niche-derived factors contribute to lineage skewedness of HSCs. Rante/Ccl5 cytokine, which is highly expressed in the local niche and aged blood, has been shown to increase myeloid progenitor proliferation, whereas knockout of Ccl5 increases lymphoid lineages during transplantation (Ergen, Boles, & Goodell, 2012). In addition, Challen et al. show lineage skewedness can be enhanced by differential responsiveness of TGFβ (Challen et al., 2010). The authors demonstrate that TGFβ provokes differential responses to myeloid-biased (CD150high) HSCs on activation. G-CSF is a well-known cytokine that promotes HSC proliferation, mobilization into the bloodstream, and eventual stem cell depletion (Ju et al., 2007; Song, Zhang, Ju, & Rudolph, 2012). Normal HSCs transplanted into telomerase-deficient mice that possess a hostile microenvironment show limited engraftment that was abrogated by G-CSF blocking antibody (Ju et al., 2007). Moreover, pharmacological strategies can partially reprogram the aged epigenetic HSC signature to that of adult HSCs (Alberts, Geneste, & Treisman, 1998), thus providing another example of molecular and functional reversion to a youthful state (Nebbioso, Carafa, Benedetti, & Altucci, 2012).
4.2. Neural stem cell
Neurons and glial cells are generated from NSCs, which reside in the sub-ventricular zone (SVZ) of the lateral ventricle and the subgranular zone (SGZ) of the dentate gyrus of the hippocampus (Kempermann, Jessberger, Steiner, & Kronenberg, 2004). Astroglial NSCs in the SVZ give rise to transit-amplifying cells, which are responsible for generating neurons that maintain the olfactory bulb (Imayoshi et al., 2008). NSCs from the SGZ generate neural progenitors that generate postmitotic neurons, which are critical for learning, memory, and behavior (Shors, Townsend, Zhao, Kozorovitskiy, & Gould, 2002). A progressive and dramatic decline in neurogenesis has been observed across different species during aging (Kempermann, Kuhn, & Gage, 1998; Kuhn, Dickinson-Anson, & Gage, 1996). This has been correlated with a decline in both the number of NSCs and proliferative expansion of transit-amplifying progenitors (Encinas et al., 2011; Lugert et al., 2010, 2012). Using markers to define distinct NSC subsets, Lugert et al. showed a reduction in the number of active NSCs due to their transition to a quiescent state in aged mice and therefore, preventing their contribution to neurogenesis (Lugert et al., 2010). In addition, a subset of aged cycling NSCs forms astrocytes instead of neurons (Encinas et al., 2011). Together, these changes are consistent with a failure to produce newborn neurons in the aged brain. Moreover, these studies highlight the heterogeneity within the NSC pool and the divergent responses of distinct subsets to respond to aging.
Several cell-intrinsic factors including the cell cycle inhibitors p16Ink4a and p57Kip2, mTOR signaling pathway, and FoxO family members have been correlated with aged phenotypes in NSCs (Furutachi, Matsumoto, Nakayama, & Gotoh, 2013; Molofsky et al., 2006; Nishino, Kim, Chada, & Morrison, 2008; Paik et al., 2009; Paliouras et al., 2012; Renault et al., 2009). Expression of p16Ink4a increases with age in neural progenitors (Molofsky et al., 2006). Deletion of the Ink4a locus reveals increased proliferation and neurogenesis from neural progenitors in SVZ compared to young counterpart, but not in the SGZ (Molofsky et al., 2006). In adult neural progenitors, p16Ink4a repression occurs through Hmga2, which is a member of the high-motility group A family that has no transcriptional activity but influences chromatin structure (Nishino et al., 2008). In aged neural progenitors, Hmga2 is repressed through upregulation of let-7b microRNA, leading to an increase in p16Ink4a expression. Overexpression of let-7b is sufficient to upregulate p16Ink4a expression by targeting Hmga2 expression. In contrast, deletion of Hmga2 leads to an increased expression of p16Ink4a, and a loss in the number and self-renewal potential of NSCs in fetal and young mice but not aged mice. This decline was partially rescued by p16Ink4a deletion. Together, these results indicate that changes in let-7b and Hmga2 may contribute to NSC aging phenotypes. It will be interesting to uncover why aged NSCs in SGZ of the aged brain were not sensitive to P16Ink4a levels.
Quiescence is critical for stem cell maintenance, in part through regulation of cell cycle inhibitors such as p57Kip2 (Matsumoto et al., 2011; Zou et al., 2011). Recently, Furutachi et al. showed a role of p57Kip2on SGZ NSC maintenance during aging (Furutachi et al., 2013). Short-term deletion of p57Kip2 specifically in Nestin-positive NSCs led to a transient amplification of radial NSCs and neurogenesis. In contrast, chronic deletion of p57Kip2 leads to a reduction in number of NSCs and neurogenesis, indicating that p57Kip2 is important for aged NSC maintenance. Moreover, these findings suggest that a short-term increase in NSC output is beneficial for neurogenesis, however, long-term proliferation leads to NSC exhaustion.
mTOR has been identified as a regulator of NSC quiescence (Paliouras et al., 2012). In the SVZ of adult mice, mTORC1 expression is absent in quiescent NSCs and present in transit-amplifying progenitors. In the aged SVZ, a decline in mTORC1 activity was observed along with reduced NSC proliferation. Reduction of mTORC1 by administration of rapamycin promotes adult NSC quiescence. Administration of EGF, strong inducer of mTOR, activated quiescent NSCs in aged mice, an effect abolished upon rapamycin treatment. This report indicates that mTOR activity determines the ratio between quiescent and activated NSCs.
Another cell-intrinsic player of aging in NSCs is the FoxO family. Recent studies show the role of FoxO proteins in regulating stem cell homeostasis in NSCs (Paik et al., 2009; Renault et al., 2009). One group utilized FoxO3−/− (Renault et al., 2009), and the other studied conditional deletion of FoxO1/3/4 (Paik et al., 2009). Both studies show that deletion of FoxO leads to a transient increase in the neonatal NSC pool in vivo but eventual depletion of NSCs in the adult. Reduced self-renewal potential assessed by in vitro neurosphere formation assay and differentiation potential was also observed in FoxO deleted mice. In the absence of FoxO, NSCs undergo increased oxidative stress, altered glucose metabolism (Renault et al., 2009), and elevated Wnt signaling (Paik et al., 2009). This suggests that the FoxO family regulates homeostasis of NSC through regulation of diverse genes and pathways.
The decline in neurogenesis during aging may also be regulated by changes in systemic factors in the blood circulation. Wyss-Coray and colleagues analyzed the systemic factors that influence neurogenesis using heterochronic parabiosis and identified that elevated levels of chemokine CC-chemokine ligand 11 (Ccl11) in plasma from aged mice and adult mice paired with aged mice (Villeda et al., 2011). After systemic delivery of Ccl11, adult mice developed learning and memory deficits, which were abrogated with coinjection of antibodies neutralizing Ccl11 along with Ccl11. Therefore, aging-related increase in Ccl11 is at least partly responsible for the reduction in aged neurogenesis. How Ccl11 affects neurogenesis remains unclear.
In addition to the systemic environment, changes in the NSC niche may impose age-dependent changes to NSC function. Components of the Wnt signaling pathway regulate adult neurogenesis (Seib et al., 2013). A recent study shows that expression of the soluble Wnt inhibitor, Dkk1 (Seib et al., 2013) increased with age in SGZ NSCs, which decreased adult neurogenesis. Conditional deletion of Dkk1 in Nestin-positive NSCs upregulated Wnt signaling, which led to increased self-renewal and differentiation into new neurons (Seib et al., 2013). Deletion of Dkk1 in aged mice showed evidence of enhanced behavioral performance levels, similar to young counterparts. This report emphasizes that changes in niche factors during aging affect NSCs function. Interestingly, exercise provides a positive stimulus for hippocampal neurogenesis in adult and aged mice (van Praag, Christie, Sejnowski, & Gage, 1999; van Praag, Shubert, Zhao, & Gage, 2005).
Another factor enriched in aged niches is TGFβ, which has been shown to be increased in blood (Challen et al., 2010) and muscle (Carlson et al., 2009) and leads to aging phenotypes. Levels of TGFβ increase in endothelial cells of the NSC niche during aging and after exposure to IR (Pineda et al., 2013). Inhibition of TGFβ signaling by delivery of a neutralizing antibody or administration of a pharmacological TGFβ inhibitor restored neurogenesis of irradiated adult mice and aged mice compared to young mice. This study demonstrates that TGFβ from the local niche is one driver of stem cell aging and these changes can be reversed when inhibited by pharmacological intervention.
4.3. Skeletal muscle stem cells
The differentiated muscle fiber functions as a niche cell that provides cues to retain muscle stem cells or satellite cells (MuSCs) in a quiescent and non-differentiated state (Bischoff, 1986). In aged muscle, the number of MuSCs declines by approximately 50% compared to adult muscle (Brack, Bildsoe, & Hughes, 2005; Cerletti et al., 2012; Chakkalakal et al., 2012). It was recently demonstrated that aged MuSCs break out of their stable quiescent state due to a redeployment of the developmental mitogen, fibroblast growth factor-2 (FGF2) in aged muscle fibers but not by the MuSCs themselves (Chakkalakal et al., 2012). FGF2-mediated proliferation was associated with increased apoptosis, myogenic commitment, and stem cell decline. Therefore, the aged niche is a modulator of stem cell number, similar to that observed in the aged niche of Drosophila (Boyle et al., 2007; Pan et al., 2007). Interestingly, although proliferative output increased, a subset of MuSCs that had undergone fewer divisions throughout adult life were able to retain long-term self-renewal potential when transplanted into adult muscle. These data demonstrate the importance of the quiescent state for maintenance of stem cell potential. Using a genetic strategy to increase FGF signaling in MuSCs only via deletion of Sprouty1, an intracellular feedback inhibitor of FGF signaling (Shea et al., 2010), elevated FGF signaling in aged MuSCs exacerbated the age-dependent satellite cell decline. Importantly, MuSC loss negatively impacted regenerative capacity (Chakkalakal et al., 2012). In contrast, short-term deletion of Sprouty1, to acutely activate FGF signaling, was met with an enhanced regenerative outcome (Chakkalakal et al., 2012), confirming that FGF signaling acts as a potent myogenic factor to prime MuSCs for functional myogenic contribution (Shefer, Van de Mark, Richardson, & Yablonka-Reuveni, 2006). This demonstrates that cell fate decisions depend on the level and duration of signaling. Other ligands including TGFβ2 and Delta1 that increase and decrease, respectively, in aged muscle fibers may also participate in deregulation of aged satellite cell homeostasis (Carlson et al., 2009; Conboy, Conboy, Smythe, & Rando, 2003). It can be concluded that niche-derived changes occurring during homeostasis can impact the success of subsequent regenerative insults. In the future, it will be important to determine what drives induction of growth factors in the aged niche.
Transplantation assays to compare engraftment potential between adult and aged MuSCs have given conflicting results, depending on whether the adult recipients were preirradiated (Chakkalakal et al., 2012; Collins, Zammit, Ruiz, Morgan, & Partridge, 2007). Engraftment potential of aged MuSCs was comparable to adult MuSCs if the adult host was irradiated (Chakkalakal et al., 2012; Collins et al., 2007). It is known that a fraction of MuSCs are depleted in response to IR (Heslop, Morgan, & Partridge, 2000). Therefore, IR decreases the number and function of adult stem cell “competitors” in the recipient muscle and provides a less competitive environment for the engrafted aged MuSCs to reoccupy the muscle.
It is generally accepted that MuSCs lose their regenerative potential during aging, due to intrinsic defects and cell-extrinsic changes in the aged environment (Brack & Rando, 2012). As we discuss next, the mechanisms that cause those changes are beginning to be identified.
To date, age-dependent DNA damage has not been extensively studied in aged MuSCs. However, dysfunction of aged MuSCs and muscle regeneration is exacerbated in the absence of Ku80, a component of the nonhomologous end-joining complex, that participates in DNA repair (Didier, Hourde, Amthor, Marazzi, & Sassoon, 2012). This demonstrates that DNA repair processes facilitate MuSC maintenance; however, determining whether DNA damage is causative to MuSC aging will require the formal demonstration that augmentation of DNA repair pathways in aged MuSCs rescues their function.
In addition, consistent with increased DNA damage response and cell cycle arrest, it has been suggested that aged MuSCs display increased abundance of the Ink4a/Arf family of cell cycle inhibitors (Carlson, Hsu, & Conboy, 2008). This may partially explain the delay in MuSC activation in response to injury. Whether Ink4a/Arf-mediated senescence occurs in aged MuSCs remains unexplored.
Metabolic factors control the level of mitochondrial and nuclear DNA damage induced by ROS, a by-product of oxidative phosphorylation. CR is a known modulator of metabolism and lifespan (Mair & Dillin, 2008). Recently, CR was demonstrated to increase MuSC number and function in adult and aged mice (Cerletti et al., 2012). The reduction in caloric load in adult and aged mice was correlated with increased number of mitochondria and elevated expression of well-known metabolic regulators Sirtuin and FoxO in FACS sorted MuSCs. Dissecting the complex relationship between DNA repair, metabolism, and autophagy and how they contribute to stem cell aging will require sophisticated genetic approaches.
To date, the contribution of the extrinsic environment to MuSC decline during aging is more clearly defined. It has been demonstrated that extrinsic modifications can rejuvenate MuSC function and muscle repair in the context of heterochronic tissue transplants (Carlson & Faulkner, 1989) and heterochronic parabiosis (Brack et al., 2007; Conboy et al., 2005). The extent of rejuvenation will likely depend on a multitude of factors including the age of the organism, the type and severity of injury, and the inflammation invoked by the injury (Shavlakadze, McGeachie, & Grounds, 2010; Smythe et al., 2008).
Using parabiotic pairings, Conboy et al. showed that unidentified factors in the serum of young mice could reverse the proliferative decline of MuSCs and that proliferation of adult MuSCs was impaired when exposed to aged systemic environment (Conboy et al., 2005). This seminal work provided a novel paradigm demonstrating that at least a subset of tissue-specific stem cells in aged organisms are functionally competent if exposed to a favorable milieu.
Early activation of adult MuSCs requires a high Notch/low Wnt state, followed by a low Notch/High Wnt state for activated MuSC to commit to differentiation (Brack, Conboy, Conboy, Shen, & Rando, 2008; Conboy & Rando, 2002). In aged muscle, both signaling cascades are deregulated; early Notch signaling is repressed and Wnt signaling is derepressed, leading to delayed satellite cell entry and a myogenic-to-fibrogenic fate conversion of a subset of aged MuSCs, respectively (Brack et al., 2007; Conboy et al., 2003). Serum factors have been identified that inhibit Notch and upregulate Wnt signaling. Elevated TGFβ2 in aged serum is able to repress Notch signaling and to increase cell cycle inhibitors of the Ink4a/Arf family to repress cell cycle entry of aged MuSCs. The identification of a Frizzled-binding protein that was upregulated in aged serum and when depleted, abrogated the fate change induced by aged serum, suggested a component of the Wnt pathway was involved (Brack et al., 2007). This was later identified as a complement family member (C1q) that activated Wnt signaling in an age-dependent manner (Naito et al., 2012). This series of papers shows that the systemic aged environment modulates satellite cell fate changes, but whether the subset of aged MuSCs that loses myogenic fate is intrinsically distinct from those that retain a MuSC identity remains unknown. Overall, the changes that occur to the aged niche are likely multifactorial and unlikely controlled through a single factor.
5. DISCUSSION AND CONCLUDING COMMENTS
Our understanding of the molecular control of stem cell function and the mechanisms that go awry during aging is rapidly expanding due to technological advances. For example, the identification of markers that allow unambiguous isolation of specialized stem cells, advances in sequencing technology that enable molecular analysis of rare stem cells, and appropriate assays to interrogate stem cell function in vivo. Through these advances, a picture emerges of many overlapping mechanisms that impact stem cell function in different niches during the aging process. The contribution of age-dependent changes to DNA fidelity, the epigenetic landscape, metabolic stress, and extrinsic factors all impact stem cell function during aging. Therefore, it is clear that the major regulators of general cellular fitness and lifespan also govern stem cell aging. This begs the question, what is unique about stem cell aging? Stem cells are unique in their ability to self-renew, a function that is lost during aging. Moreover, stem cells are relatively scarce compared to other cell types; therefore, the consequences of stem cell decline during aging are severe. Hence, identifying the mechanisms responsible for stem cell decline whether specific to stem cells or general regulators of cell aging and lifespan will illustrate the careful balance between tissue homeostasis, aging, and cancer.
In this review, we focused on three well-defined stem cell systems that have been characterized during aging: blood, neural, and muscle. Unfortunately, the specter of aging does not spare a single body part and therefore most stem cell compartments will undergo some form of age-related decline. As we begin to uncover age-dependent changes in other stem cell niches, context-specific differences and commonalities that define aging will become crystallized for the interests of human health.
A common theme across stem cells niches is the observed hierarchical relationship between stem cells of different potential, with upstream stem cells endowed with long-term self-renewal potential and a propensity to maintain a dormant quiescent state, relative to their more committed daughters. Can the loss of self-renewal potential during aging be explained by an increase in downstream stem cells at expense of the hierarchically upstream subset? Both aged HSCs and MuSCs retain a small fraction of label retaining cells endowed with self-renewal potential, however, the relative fraction of the more committed subset increases in the aged MuSC pool. It remains unknown whether this hierarchical skewing occurs in the HSC or NSC compartment during aging. If so, this could explain how the HSC pool is maintained in aged bone marrow at the same time that functionality declines.
It is interesting that unlike the NSC and MuSC compartment, the number of HSCs during aging does not decline. One interpretation is that the HSC system has compensatory mechanisms to prevent stem cell loss. Preservation of the quiescent state is critical for stem cell maintenance. However, the turnover of HSCs, MuSCs, and NSCs is relatively infrequent and therefore unlikely to explain the stark differences. In the future, direct comparisons between stem cell compartments may reveal context-dependent differences in protective or stress pathways that lead to preservation or loss of one stem cell compartment relative to another.
The identification of functionally heterogeneous subsets of adult stem cells under different molecular control dictates that any changes in stem cell function of a whole population maybe through selective loss or expansion of subsets during stress, turnover, and aging. A clonal subset that is intrinsically superior or acquires increased fitness under selective pressure will dominate the aged stem cell pool. Therefore, age-dependent changes in the extrinsic milieu and epigenetic state of the stem cell may cause functional changes to the whole pool of stem cells or allow for selection of one subset at the expense of the other. Determining the consequence of clonal expansion or loss in specific subsets will require a more comprehensive study into stem cell heterogeneity and specific signals that govern the functional properties of such subsets.
The clinical applications of stem cell augmentation are obvious. Several methods of intervention have been discussed that functionally improve aged stem cells. In some instances, the exposure of aged cells to a young environment restores cellular function, suggesting that the aged environment induces a repressive state that can be reversed in a youthful setting. Other promising strategies include the restoration of extrinsic signaling pathways such as Notch, Wnt, and TGFβ to youthful levels; decreasing stress pathways through metabolic reprogramming; or the reduction in inflammatory pathways all provide favorable functional outcome to stem cells and regenerative response of aged organisms. Whether these stem cell modifiers represent a functional compensation by an aged stem cell or true molecular rejuvenation to a young stem cell state remains to be tested. In the absence of full molecular rejuvenation, any functional improvement to aged stem cells is likely to be transient. It is also exciting to consider that stem cells can be modified through lifestyle factors. Exercise and reduced caloric intake appear to enhance tissue-specific stem cell activity in both adult and aged organisms. Understanding how aging of stem cells can be prevented to enhance tissue homeostasis and regeneration potential will be critical for healthy living and the treatment of age-related diseases in the ever expanding human aged population.
Acknowledgments
We apologize to our colleagues whose work could not be cited due to the space limitation. We would like to thank Brack lab members for critical discussion and reading of the manuscript and Justine Zayhowski for artwork. This work was supported by NIH Grants (R01 AR060868, R01 AR061002) and Ellison Medical Foundation New Scholars Award to ASB.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare no competing financial interest.
References
- Adler AS, Kawahara TL, Segal E, Chang HY. Reversal of aging by NFkappaB blockade. Cell Cycle. 2008;7:556–559. doi: 10.4161/cc.7.5.5490. [DOI] [PubMed] [Google Scholar]
- Ahlqvist KJ, Hamalainen RH, Yatsuga S, Uutela M, Terzioglu M, Gotz A, et al. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metabolism. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Alberts AS, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyper-acetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nature Genetics. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- Beerman I, Bhattacharya D, Zandi S, Sigvardsson M, Weissman IL, Bryder D, et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5465–5470. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I, Bock C, Garrison BS, Smith ZD, Gu H, Meissner A, et al. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell. 2013;12:413–425. doi: 10.1016/j.stem.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Beerman I, Maloney WJ, Weissmann IL, Rossi DJ. Stem cells and the aging hematopoietic system. Current Opinion in Immunology. 2010;22:500–506. doi: 10.1016/j.coi.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz C, Copley MR, Kent DG, Wohrer S, Cortes A, Aghaeepour N, et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 2012;10:273–283. doi: 10.1016/j.stem.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Bischoff R. Proliferation of muscle satellite cells on intact myofibers in culture. Developmental Biology. 1986;115:129–139. doi: 10.1016/0012-1606(86)90234-4. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–535. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genetics. 2010;6:e1001159. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Mohrin M, Sotiropoulou PA, Passegue E. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell. 2011;8:16–29. doi: 10.1016/j.stem.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood. 2011;117:e182–e189. doi: 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- Boyle M, Wong C, Rocha M, Jones DL. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell. 2007;1:470–478. doi: 10.1016/j.stem.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Brack AS, Bildsoe H, Hughes SM. Evidence that satellite cell decrement contributes to preferential decline in nuclear number from large fibres during murine age-related muscle atrophy. Journal of Cell Science. 2005;118:4813–4821. doi: 10.1242/jcs.02602. [DOI] [PubMed] [Google Scholar]
- Brack AS, Conboy IM, Conboy MJ, Shen J, Rando TA. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell. 2008;2:50–59. doi: 10.1016/j.stem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- Brack AS, Rando TA. Tissue-specific stem cells: Lessons from the skeletal muscle satellite cell. Cell Stem Cell. 2012;10:504–514. doi: 10.1016/j.stem.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratic A, Larsson NG. The role of mitochondria in aging. Journal of Clinical Investigation. 2013;123:951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Xie S, Qiu X, Mohrin M, Shin J, Liu Y, et al. SIRT3 reverses aging-associated degeneration. Cell Reports. 2013;3:319–327. doi: 10.1016/j.celrep.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Kennedy BK. Progeria syndromes and ageing: What is the connection? Nature Reviews Molecular Cell Biology. 2010;11:567–578. doi: 10.1038/nrm2944. [DOI] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: Good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Carlson ME, Conboy MJ, Hsu M, Barchas L, Jeong J, Agrawal A, et al. Relative roles of TGF-beta1 and Wnt in the systemic regulation and aging of satellite cell responses. Aging Cell. 2009;8:676–689. doi: 10.1111/j.1474-9726.2009.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BM, Faulkner JA. Muscle transplantation between young and old rats: Age of host determines recovery. The American Journal of Physiology. 1989;256:C1262–C1266. doi: 10.1152/ajpcell.1989.256.6.C1262. [DOI] [PubMed] [Google Scholar]
- Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454:528–532. doi: 10.1038/nature07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Cerletti M, Jang YC, Finley LW, Haigis MC, Wagers AJ. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell. 2012;10:515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6:265–278. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dys-regulation. PLoS Biology. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Science Signaling. 2009;2:ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- Collins CA, Zammit PS, Ruiz AP, Morgan JE, Partridge TA. A population of myogenic stem cells that survives skeletal muscle aging. Stem Cells. 2007;25:885–894. doi: 10.1634/stemcells.2006-0372. [DOI] [PubMed] [Google Scholar]
- Conboy MJ, Conboy IM, Rando TA. Heterochronic parabiosis: Historical perspective and methodological considerations for studies of aging and longevity. Aging Cell. 2013;12:525–530. doi: 10.1111/acel.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Rando TA. The regulation of notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Developmental Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- Conconi M, Szweda LI, Levine RL, Stadtman ER, Friguet B. Age-related decline of rat liver multicatalytic proteinase activity and protection from oxidative inactivation by heat-shock protein 90. Archives of Biochemistry and Biophysics. 1996;331:232–240. doi: 10.1006/abbi.1996.0303. [DOI] [PubMed] [Google Scholar]
- Didier N, Hourde C, Amthor H, Marazzi G, Sassoon D. Loss of a single allele for Ku80 leads to progenitor dysfunction and accelerated aging in skeletal muscle. EMBO Molecular Medicine. 2012;4:910–923. doi: 10.1002/emmm.201101075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra B, Olthof S, Schreuder J, Ritsema M, de Haan G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. The Journal of Experimental Medicine. 2011;208:2691–2703. doi: 10.1084/jem.20111490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar D, Shabalina I, Camara Y, Wredenberg A, Calvaruso MA, Nijtmans L, et al. Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metabolism. 2009;10:131–138. doi: 10.1016/j.cmet.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Eijkelenboom A, Burgering BM. FOXOs: Signalling integrators for homeo-stasis maintenance. Nature Reviews Molecular Cell Biology. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- Ellis RJ, Pinheiro TJ. Medicine: Danger—Misfolding proteins. Nature. 2002;416:483–484. doi: 10.1038/416483a. [DOI] [PubMed] [Google Scholar]
- Encinas JM, Michurina TV, Peunova N, Park JH, Tordo J, Peterson DA, et al. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8:566–579. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergen AV, Boles NC, Goodell MA. Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood. 2012;119:2500–2509. doi: 10.1182/blood-2011-11-391730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores I, Canela A, Vera E, Tejera A, Cotsarelis G, Blasco MA. The longest telomeres: A general signature of adult stem cell compartments. Genes & Development. 2008;22:654–667. doi: 10.1101/gad.451008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian MC, Dorr K, Niebel A, Daria D, Schrezenmeier H, Rojewski M, et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell. 2012;10:520–530. doi: 10.1016/j.stem.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nature Biotechnology. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S, Matsumoto A, Nakayama KI, Gotoh Y. p57 controls adult neural stem cell quiescence and modulates the pace of lifelong neurogenesis. The EMBO Journal. 2013;32:970–981. doi: 10.1038/emboj.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger H, de Haan G, Florian MC. The ageing haematopoietic stem cell compartment. Nature Reviews Immunology. 2013;13:376–389. doi: 10.1038/nri3433. [DOI] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Klar AJ. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- Grompe M. Tissue stem cells: New tools and functional diversity. Cell Stem Cell. 2012;10:685–689. doi: 10.1016/j.stem.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Helenius M, Hanninen M, Lehtinen SK, Salminen A. Changes associated with aging and replicative senescence in the regulation of transcription factor nuclear factor-kappa B. The Biochemical Journal. 1996;318(Pt. 2):603–608. doi: 10.1042/bj3180603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, et al. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nature Communications. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop L, Morgan JE, Partridge TA. Evidence for a myogenic stem cell that is exhausted in dystrophic muscle. Journal of Cell Science. 2000;113(Pt. 12):2299–2308. doi: 10.1242/jcs.113.12.2299. [DOI] [PubMed] [Google Scholar]
- Hidalgo I, Herrera-Merchan A, Ligos JM, Carramolino L, Nunez J, Martinez F, et al. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;11:649–662. doi: 10.1016/j.stem.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Ohtsuka T, Takao K, Miyakawa T, Yamaguchi M, et al. Roles of continuous neurogenesis in the structural and functional integrity of the adult forebrain. Nature Neuroscience. 2008;11:1153–1161. doi: 10.1038/nn.2185. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nature Medicine. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- Jones DL, Rando TA. Emerging models and paradigms for stem cell ageing. Nature Cell Biology. 2011;13:506–512. doi: 10.1038/ncb0511-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Z, Jiang H, Jaworski M, Rathinam C, Gompf A, Klein C, et al. Telomere dysfunction induces environmental alterations limiting hematopoietic stem cell function and engraftment. Nature Medicine. 2007;13:742–747. doi: 10.1038/nm1578. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Development. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaitzidis D, Sykes SM, Wang Z, Punt N, Tang Y, Ragu C, et al. mTOR complex 1 plays critical roles in hematopoiesis and Pten-loss-evoked leukemogenesis. Cell Stem Cell. 2012;11:429–439. doi: 10.1016/j.stem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- Kapetanaki MG, Mora AL, Rojas M. Influence of age on wound healing and fibrosis. The Journal of Pathology. 2013;229:310–322. doi: 10.1002/path.4122. [DOI] [PubMed] [Google Scholar]
- Kemp CJ, Donehower LA, Bradley A, Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74:813–822. doi: 10.1016/0092-8674(93)90461-x. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Jessberger S, Steiner B, Kronenberg G. Milestones of neuronal development in the adult hippocampus. Trends in Neurosciences. 2004;27:447–452. doi: 10.1016/j.tins.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Kuhn HG, Gage FH. Experience-induced neurogenesis in the senescent dentate gyrus. The Journal of Neuroscience. 1998;18:3206–3212. doi: 10.1523/JNEUROSCI.18-09-03206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Korhonen P, Helenius M, Salminen A. Age-related changes in the regulation of transcription factor NF-kappa B in rat brain. Neuroscience Letters. 1997;225:61–64. doi: 10.1016/s0304-3940(97)00190-0. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, et al. Ink4a/Arf expression is a biomarker of aging. Journal of Clinical Investigation. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: Age-related decrease of neuronal progenitor proliferation. The Journal of Neuroscience. 1996;16:2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Lang T, Streeper T, Cawthon P, Baldwin K, Taaffe DR, Harris TB. Sarcopenia: Etiology, clinical consequences, intervention, and assessment. Osteoporosis International. 2010;21:543–559. doi: 10.1007/s00198-009-1059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, et al. FLIP-mediated autophagy regulation in cell death control. Nature Cell Biology. 2009;11:1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Nakada D, Yilmaz OH, Tothova Z, Joseph NM, Lim MS, et al. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 2010;7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jiang TX, Hughes MW, Wu P, Yu J, Widelitz RB, et al. Progressive alopecia reveals decreasing stem cell activation probability during aging of mice with epidermal deletion of DNA methyltransferase 1. The Journal of Investigative Dermatology. 2012;132:2681–2690. doi: 10.1038/jid.2012.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Hron JD, Peng SL. Regulation of NF-kappaB, Th activation, and autoinflammation by the fork head transcription factor Foxo3a. Immunity. 2004;21:203–213. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Liu F, Lee JY, Wei H, Tanabe O, Engel JD, Morrison SJ, et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–4814. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo FS, Steinhauser ML, Jay SM, Gannon J, Pancoast JR, Yalamanchi P, et al. Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell. 2013;153:828–839. doi: 10.1016/j.cell.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Beard C, Johnson B, Marciniak RA, Dausman J, Bronson R, et al. Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Molecular and Cellular Biology. 2000;20:3286–3291. doi: 10.1128/mcb.20.9.3286-3291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugert S, Basak O, Knuckles P, Haussler U, Fabel K, Gotz M, et al. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Lugert S, Vogt M, Tchorz JS, Muller M, Giachino C, Taylor V. Homeo-static neurogenesis in the adult hippocampus does not involve amplification of Ascl1 (high) intermediate progenitors. Nature Communications. 2012;3:670. doi: 10.1038/ncomms1670. [DOI] [PubMed] [Google Scholar]
- Lukacs RU, Memarzadeh S, Wu H, Witte ON. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell. 2010;7:682–693. doi: 10.1016/j.stem.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Research. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JA, Ikenoue T, Nakada D, Lee JY, Guan KL, Morrison SJ. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012;11:415–428. doi: 10.1016/j.stem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Dillin A. Aging and survival: The genetics of life span extension by dietary restriction. Annual Review of Biochemistry. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Takeishi S, Kanie T, Susaki E, Onoyama I, Tateishi Y, et al. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2011;9:262–271. doi: 10.1016/j.stem.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. The Journal of Experimental Medicine. 2010;207:1173–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. The Journal of Experimental Medicine. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Naito AT, Sumida T, Nomura S, Liu ML, Higo T, Nakagawa A, et al. Complement C1q activates canonical Wnt signaling and promotes aging-related phenotypes. Cell. 2012;149:1298–1313. doi: 10.1016/j.cell.2012.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro S, Meza NW, Quintana-Bustamante O, Casado JA, Jacome A, McAllister K, et al. Hematopoietic dysfunction in a mouse model for Fanconi anemia group D1. Molecular Therapy. 2006;14:525–535. doi: 10.1016/j.ymthe.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Nebbioso A, Carafa V, Benedetti R, Altucci L. Trials with ‘epigenetic’ drugs: An update. Molecular Oncology. 2012;6:657–682. doi: 10.1016/j.molonc.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Granter SR, Fisher DE. Mechanisms of hair graying: Incomplete melanocyte stem cell maintenance in the niche. Science. 2005;307:720–724. doi: 10.1126/science.1099593. [DOI] [PubMed] [Google Scholar]
- Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norddahl GL, Pronk CJ, Wahlestedt M, Sten G, Nygren JM, Ugale A, et al. Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell. 2011;8:499–510. doi: 10.1016/j.stem.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nature Reviews Genetics. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliouras GN, Hamilton LK, Aumont A, Joppe SE, Barnabe-Heider F, Fernandes KJ. Mammalian target of rapamycin signaling is a key regulator of the transit-amplifying progenitor pool in the adult and aging forebrain. The Journal of Neuroscience. 2012;32:15012–15026. doi: 10.1523/JNEUROSCI.2248-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Chen S, Weng C, Call G, Zhu D, Tang H, et al. Stem cell aging is controlled both intrinsically and extrinsically in the Drosophila ovary. Cell Stem Cell. 2007;1:458–469. doi: 10.1016/j.stem.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: Implications for inflammatory diseases. Nature Reviews Immunology. 2009;9:778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Pineda JR, Daynac M, Chicheportiche A, Cebrian-Silla A, Sii Felice K, Garcia-Verdugo JM, et al. Vascular-derived TGF-beta increases in the stem cell niche and perturbs neurogenesis during aging and following irradiation in the adult mouse brain. EMBO Molecular Medicine. 2013;5:548–562. doi: 10.1002/emmm.201202197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollina EA, Brunet A. Epigenetic regulation of aging stem cells. Oncogene. 2011;30:3105–3126. doi: 10.1038/onc.2011.45. [DOI] [PubMed] [Google Scholar]
- Reese JS, Liu L, Gerson SL. Repopulating defect of mismatch repair-deficient hematopoietic stem cells. Blood. 2003;102:1626–1633. doi: 10.1182/blood-2002-10-3035. [DOI] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi DJ, Seita J, Czechowicz A, Bhattacharya D, Bryder D, Weissman IL. Hematopoietic stem cell quiescence attenuates DNA damage response and permits DNA damage accumulation during aging. Cell Cycle. 2007;6:2371–2376. doi: 10.4161/cc.6.19.4759. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Ruckh JM, Zhao JW, Shadrach JL, van Wijngaarden P, Rao TN, Wagers AJ, et al. Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell. 2012;10:96–103. doi: 10.1016/j.stem.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, DePinho RA. Axis of ageing: Telomeres, p53 and mitochondria. Nature Reviews Molecular Cell Biology. 2012;13:397–404. doi: 10.1038/nrm3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib DR, Corsini NS, Ellwanger K, Plaas C, Mateos A, Pitzer C, et al. Loss of Dickkopf-1 restores neurogenesis in old age and counteracts cognitive decline. Cell Stem Cell. 2013;12:204–214. doi: 10.1016/j.stem.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Shavlakadze T, McGeachie J, Grounds MD. Delayed but excellent myogenic stem cell response of regenerating geriatric skeletal muscles in mice. Biogerontology. 2010;11:363–376. doi: 10.1007/s10522-009-9260-0. [DOI] [PubMed] [Google Scholar]
- Shea KL, Xiang W, LaPorta VS, Licht JD, Keller C, Basson MA, et al. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 2010;6:117–129. doi: 10.1016/j.stem.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer G, Van de Mark DP, Richardson JB, Yablonka-Reuveni Z. Satellite-cell pool size does matter: Defining the myogenic potency of aging skeletal muscle. Developmental Biology. 2006;294:50–66. doi: 10.1016/j.ydbio.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibatani T, Nazir M, Ward WF. Alteration of rat liver 20S proteasome activities by age and food restriction. The Journals of Gerontology Series A, Biological Sciences and Medical Sciences. 1996;51:B316–B322. doi: 10.1093/gerona/51a.5.b316. [DOI] [PubMed] [Google Scholar]
- Shors TJ, Townsend DA, Zhao M, Kozorovitskiy Y, Gould E. Neurogenesis may relate to some but not all types of hippocampal-dependent learning. Hippocampus. 2002;12:578–584. doi: 10.1002/hipo.10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Morrison SJ. Mechanisms that regulate stem cell aging and life span. Cell Stem Cell. 2013;12:152–165. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- Singh SK, Williams CA, Klarmann K, Burkett SS, Keller JR, Oberdoerffer P. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. The Journal of Experimental Medicine. 2013;210:987–1001. doi: 10.1084/jem.20121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: Roles in mammalian development. Nature Reviews Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Smythe GM, Shavlakadze T, Roberts P, Davies MJ, McGeachie JK, Grounds MD. Age influences the early events of skeletal muscle regeneration: Studies of whole muscle grafts transplanted between young (8 weeks) and old (13–21 months) mice. Experimental Gerontology. 2008;43:550–562. doi: 10.1016/j.exger.2008.02.005. [DOI] [PubMed] [Google Scholar]
- So AY, Jung JW, Lee S, Kim HS, Kang KS. DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs. PLoS One. 2011;6:e19503. doi: 10.1371/journal.pone.0019503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Zhang J, Ju Z, Rudolph KL. Telomere dysfunctional environment induces loss of quiescence and inherent impairments of hematopoietic stem cell function. Aging Cell. 2012;11:449–455. doi: 10.1111/j.1474-9726.2012.00802.x. [DOI] [PubMed] [Google Scholar]
- Sperka T, Wang J, Rudolph KL. DNA damage checkpoints in stem cells, age-ing and cancer. Nature Reviews Molecular Cell Biology. 2012;13:579–590. doi: 10.1038/nrm3420. [DOI] [PubMed] [Google Scholar]
- Tomas-Loba A, Flores I, Fernandez-Marcos PJ, Cayuela ML, Maraver A, Tejera A, et al. Telomerase reverse transcriptase delays aging in cancer-resistant mice. Cell. 2008;135:609–622. doi: 10.1016/j.cell.2008.09.034. [DOI] [PubMed] [Google Scholar]
- Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- van Praag H, Christie BR, Sejnowski TJ, Gage FH. Running enhances neurogenesis, learning, and long-term potentiation in mice. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13427–13431. doi: 10.1073/pnas.96.23.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praag H, Shubert T, Zhao C, Gage FH. Exercise enhances learning and hippocampal neurogenesis in aged mice. The Journal of Neuroscience. 2005;25:8680–8685. doi: 10.1523/JNEUROSCI.1731-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Benchimol S. From telomere loss to p53 induction and activation of a DNA-damage pathway at senescence: The telomere loss/DNA damage model of cell aging. Experimental Gerontology. 1996;31:295–301. doi: 10.1016/0531-5565(95)02025-x. [DOI] [PubMed] [Google Scholar]
- Villeda SA, Luo J, Mosher KI, Zou B, Britschgi M, Bieri G, et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477:90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- Wong KK, Maser RS, Bachoo RM, Menon J, Carrasco DR, Gu Y, et al. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature. 2003;421:643–648. doi: 10.1038/nature01385. [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. The EMBO Journal. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharek SJ, Fillmore CM, Lau AN, Gludish DW, Chou A, Ho JW, et al. Lung stem cell self-renewal relies on BMI1-dependent control of expression at imprinted loci. Cell Stem Cell. 2011;9:272–281. doi: 10.1016/j.stem.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YV, Cheong J, Ciapurin N, McDermitt DJ, Tumbar T. Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell. 2009;5:267–278. doi: 10.1016/j.stem.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nature Medicine. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P, Yoshihara H, Hosokawa K, Tai I, Shinmyozu K, Tsukahara F, et al. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell. 2011;9:247–261. doi: 10.1016/j.stem.2011.07.003. [DOI] [PubMed] [Google Scholar]