Abstract
Heterochromatin protein 1α (HP1α), a key player in the establishment and maintenance of higher-order chromatin regulates key cellular processes, including metaphase chromatid cohesion and centromere organization. However, how HP1α controls these processes is not well understood. Here we demonstrate that post-translational modifications of HP1α dictate its mitotic functions. HP1α is constitutively phosphorylated within its N-terminus whereas phosphorylation within the hinge domain occurs preferentially at G2/M phase of the cell cycle. The hinge-phosphorylated form of HP1α specifically localizes to kinetochores during early mitosis and this phosphorylation mediated by NDR1 kinase is required for mitotic progression and for Sgo1 binding to mitotic centromeres. Cells lacking NDR kinase show loss of mitosis-specific phosphorylation of HP1α leading to prometaphase arrest. Our results reveal that NDR kinase catalyzes the hinge-specific phosphorylation of human HP1α during G2/M in vivo and this orchestrates accurate chromosome alignment and mitotic progression.
Keywords: cell cycle, HP1α, mitosis, NDR kinases, phosphorylation
Introduction
Posttranslational modifications of proteins regulate epigenetic control of gene expression. Modifications in histone and non-histone proteins dictate higher order chromatin structure that in turn modulates gene silencing and chromosome segregation1,2. Heterochromatin is a specialized state of higher order chromatin that typically represents gene poor, highly condensed and transcriptionally silent regions and is characterized by the presence of histone H3K9 trimethylation marks 3,4. Heterochromatin protein 1 (HP1) is the key reader of this histone mark and is responsible for recruiting other effector molecules that together, establish the heterochromatin domain 5,6.
HP1 consists of an N-terminal chromodomain (CD) that recognizes and binds to methylated H3K9 4, a C-terminal chromoshadow domain (CSD) responsible for HP1 dimerization and protein-protein interaction 7–9 and a flexible hinge region (H) of variable length that separates the chromo and chromo shadow domains and is also thought to mediate chromatin binding 10,11. The interactions of several histone and non-histone proteins with the isoforms of HP1 (α, β and γ) in mammalian cells have already been implicated in essential cellular process, including the establishment and maintenance of higher-order chromatin, metaphase chromatid cohesion and centromere architecture 5,12,13. Post-translational modifications to all isoforms of HP1 at both CD and CSD modulate its interaction with other proteins or histone marks or regulate its function 14–16.
Protein phosphorylation is a key regulatory post-translational event that controls protein function and localization 17. The interplay between protein kinases and their specific substrates dictate crucial cellular processes ranging from cell cycle progression to chromatin organization. HP1, originally identified from a silenced region of Drosophila polytene chromosome is hyperphosphorylated by casein kinase II (CKII) 18–22. In Drosophila, the hyperphosphorylation of HP1 is implicated with heterochromatin assembly 18, whereas hypo-phosphorylated HP1 remains in a complex with an HMG-like HP1/ORC-Associated Protein (HOAP) and ORC 23. Since the discovery of HP1 in Drosophila, orthologs have been identified in various species including fission yeast, chicken, mouse and human with remarkable evolutionary conservation 7,24–26. Similarly, the phosphorylation of the fission yeast homolog of HP1, Swi6 regulates the binding of transcription regulators to chromatin and affects the transcriptional status at the heterochromatin 27. Swi6 phosphorylation by CKII is required for gene silencing but not for chromosome segregation and sister chromatid cohesion suggesting that independent mechanisms regulate each of these events 28. Hsk1, another protein kinase, also interacts with Swi6 and is required for heterochromatic gene silencing, suggesting that multiple kinases may be involved in phosphorylating Swi6 29.
Mammalian HP1 is also highly phosphorylated and phosphorylation of HP1α at the N-terminal serine residues imparts stronger binding affinity to methylated H3K9 resulting in more robust chromatin association 14,30,31. CKII phosphorylates HP1β at the chromo shadow domain following DNA damage that in contrast results in disruption of chromatin binding of HP1β 32. Though in vitro data suggest that CKII can phosphorylate HP1α, there is no in vivo evidence that supports the role of CKII in HP1α phosphorylation. The elucidation of the biological function of phosphorylation of HP1α and the identification of an in vivo kinase that phosphorylates HP1α in mammals remain elusive.
NDR ((Nuclear-Dbf2-related) kinases are highly conserved kinases that control vital cellular processes in various organisms, including mitotic exit, cytokinesis, cell growth and proliferation and differentiation 33. The NDR kinase orthologs have been shown to be required for the MEN (mitosis exit network) in budding yeast and for SIN (septation initiation network) in fission yeast 34–36. Dbf2 orthologs in close association with upstream Ste-20-like kinases and MOB (Mps-one-binding) co-activators together constitute the Hippo pathway and coordinate key cellular processes like cell growth, proliferation and tumorigenesis 37–39. In humans, NDR kinases have been shown to be required for G1/S transition, centrosome duplication and for mitotic chromosome alignment 40. To date, the cell cycle protein p21 is the only known in vivo substrate identified for NDR kinase in human cells 40. Recent work demonstrated that NDR1 kinase is required for accurate chromosome alignment 41 but the relevant substrates remain to be identified.
In this study, we have identified that NDR kinase phosphorylates HP1α within its hinge domain predominantly during G2/M phase of the cell cycle. During early mitosis, hinge-phosphorylated HP1α localizes to kinetochores. Depletion of NDR kinase results in chromosomal alignment defects associated with defects in phosphorylation of HP1α at the hinge region and disruption of Sgo1 binding to centromeres. Our results demonstrate that NDR1 kinase-mediated phosphorylation of HP1α is required for accurate chromosome alignment and mitotic progression in mammalian cells.
Results
NDR kinase associates with HP1α
In a screen to identify the substrates for NDR kinases, we have detected HP1α, a protein that regulates heterochromatin organization and cell cycle progression, as an NDR kinase interacting protein. To verify the interaction between NDR kinase and HP1α, we co-transfected HA-NDR1 and YFP-HP1α, followed by HA immunoprecipitations to demonstrate the interaction of NDR1 with HP1α (Fig. 1a and Supplementary Fig. 1a). Similarly, transient transfection of HA-HP1α and T7-NDR1 followed by immunoprecipitation using HA antibody confirmed the interaction of NDR1 and HP1α (Fig. 1b and Supplementary Fig. 1b).
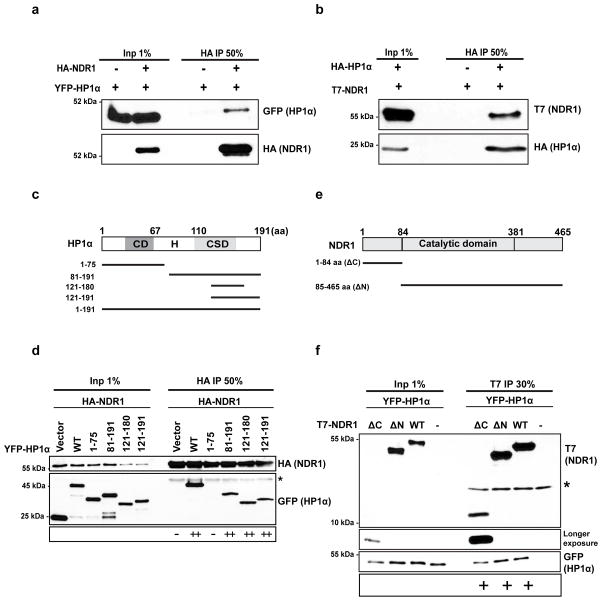
Figure 1. NDR1 associates with HP1α.
(a) Immunoprecipitation using HA antibody in cells expressing YFP-HP1α with (+) or without (−) HA-NDR1. Note the interaction between HA-NDR1 and YFP-HP1α (detected by GFP immunoblot). (b) Reciprocal immunoprecipitation using HA antibody in cells expressing T7-NDR1 with (+) or without (−) HA-HP1α. (c) Schematic representation of HP1α truncation mutants. (d). Immunoprecipitation using HA antibodies from cells expressing HA-NDR1 along with HP1α truncation mutants. Immunoblots using GFP antibody demonstrate robust interaction of NDR1 kinase with the mutants containing the chromoshadow domain (121-180/191) or with hinge region (81-191) but not with the chromo domain (1-75aa). YFP vector transfected with HA-NDR1 has been used as control. Extent of interaction is depicted below the immunoblots. (e) Schematic representation of truncation mutants of NDR1 kinase spanning hydrophobic N-terminal as well as the central catalytic/kinase domain. (f) Immunoprecipitation using T7 antibody from cells expressing YFP-HP1α and various truncation mutants of T7-NDR1 (ΔC and ΔN). Note that HP1α interacts with both the N- and C-terminus of the NDR1 kinase. Extent of interaction is depicted below the immunoblots.
To map the interacting domains between HP1α and NDR1, various truncation mutants of HP1α, 1-75aa (spanning the chromo domain); 81-191 (hinge and chromoshadow domain); 121-180 and 121-191 (chromoshadow domain) were generated (Fig. 1c). Co-transfection of HA-NDR1 along with YFP vector or YFP-HP1α full length or truncation mutants, followed by immunoprecipitation using HA antibody revealed the association of NDR1 predominantly with the chromoshadow domain of HP1α (Fig. 1d). This is further confirmed by the fact that a chromoshadow domain mutant of HP1α that has lost its ability to bind PXVXL/I ligands (HP1α-W174A) failed to interact with NDR1 (Supplementary Fig. 1b). To identify the NDR1 domain that associates with HP1α, we generated NDR1 truncation mutants, 1-84 aa (spanning N-terminal regulatory sequence, ΔC) and 85-465 aa, (containing a C-terminal hydrophobic motif and the catalytic domain, ΔN) (Fig. 1e). Co-IP of these mutants with HP1α demonstrated that multiple regions on NDR1 interacted with HP1α (Fig. 1f).
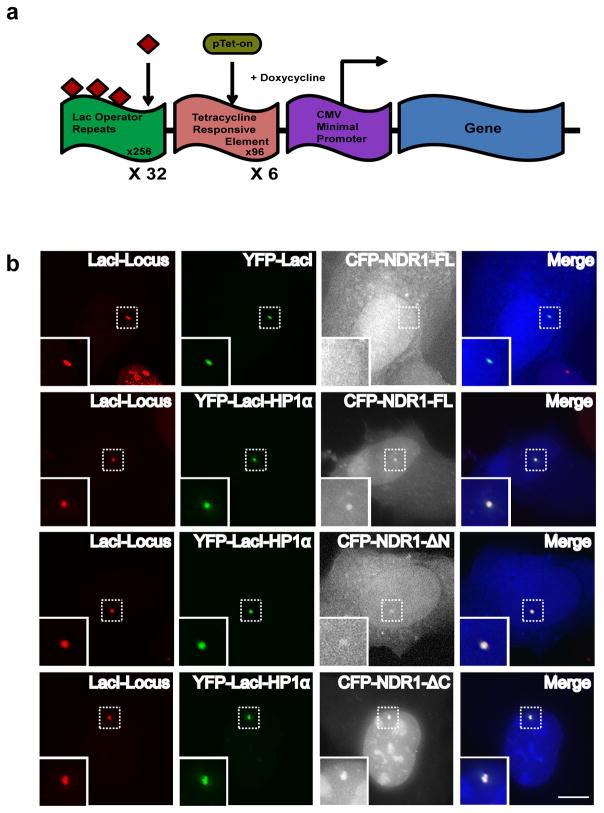
To study the association of NDR kinases with HP1α at an in vivo locus, we utilized a cell line originally developed by Spector and colleagues 42. The locus has several hundred copies of the lac operator repeats that can be easily visualized by the presence of a Cherry-lac repressor (LacI) 43,44 (Fig. 2a and 2b). We generated a triple fusion protein consisting of YFP-LacI-HP1α and examined if the accumulation of HP1α at the site was sufficient to recruit NDR1 to that site. In addition to the tethered chromatin locus, a fraction of the YFP-LacI-HP1α was also present at heterochromatic regions within the nucleus that is apparent upon overexposure (Supplementary Fig. 1c). CFP-NDR1 is predominantly a cytoplasmic protein with low amounts of it being detected in the nucleus. However upon tethering of HP1α, the CFP-NDR1 was present in the nuclear chromatin site in 18% of the cells (Fig. 2b). We also co-expressed CFP-NDR1-1-84aa (CFP-NDR1-ΔC, Fig. 2b) or CFP-NDR1-85-465aa (CFP-NDR1-ΔN, Fig. 2b) in YFP-LacI-HP1α expressing cells. CFP-NDR1-85-465 (ΔN) was recruited to the chromatin locus in similar percentage of the cells (19%; Fig. 2b). CFP-NDR1 expressing only the N-terminus (1-84aa), CFP-NDR1-ΔC, was predominantly localized in the nucleus, suggesting that the C-terminus of this kinase contains a nuclear export signal. The nuclear enriched CFP-NDR1-ΔC accumulated at the YFP-LacI-HP1α tethered locus in 37% of cells (Fig. 2b). Similar to the in vitro results, both the ΔC- and ΔN- NDR1 mutants were found to associate with the HP1α-tethered chromatin locus. We observed a weak signal of NDR1 kinase at the CLTon locus (Fig. 2b) since the endogenous HP1α is known to localize to these sites42. However the signal intensity of NDR1 when HP1α is tethered to the locus was much higher than when HP1α is not tethered.
Figure 2. Association of NDR1 with HP1α at an in vivo reporter heterochromatic locus.
(a) Schematic representation of the U2OS 2-6-3 CLTon array that has been stably integrated in human cells [adapted and modified, 42–44]. (b) The chromatin locus is visualized by Cherry-LacI staining. Cells were co-transfected with YFP-LacI and CFP-NDR-FL or with YFP-LacI-HP1α and CFP-NDR1 or its truncation mutants. YFP-LacI does not recruit CFP-NDR1 to the locus, whereas YFP-LacI-HP1α can efficiently recruit CFP-NDR1 to the chromatin locus. Cells co-transfected with YFP-LacI-HP1α and CFP-NDR1ΔN or CFP-NDR1ΔC shows accumulation at the HP1α containing locus. Scale bar represents 10μm.
NDR1 kinase phosphorylates HP1α in vitro
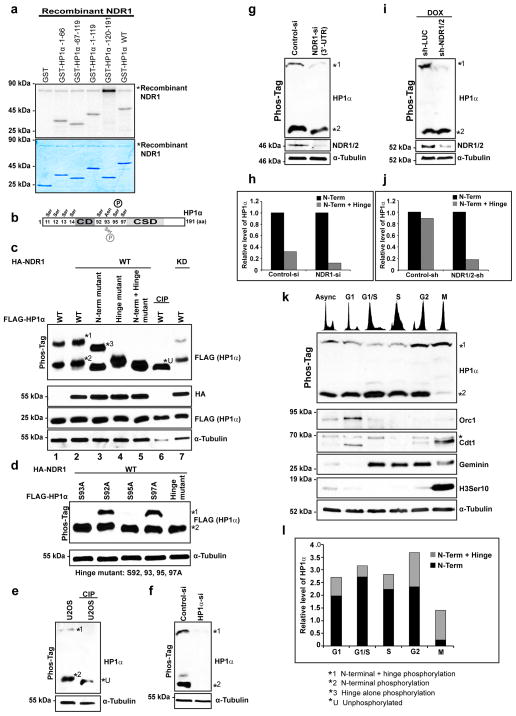
We next addressed the role of NDR kinase in HP1α phosphorylation employing in vitro kinase assays. Recombinant NDR1 kinase was used in the kinase assay with GST, GST- HP1α-WT, GST-HP1α-1-66aa, GST-HP1α-67-119aa, GST-HP1α-1-119aa and GST-HP1α-120-191aa as substrates (Fig. 3a). HP1α constructs with the chromodomain and/or the hinge domain (region spanning 1-119aa), but not the chromoshadow domain (120-191aa), could be efficiently phosphorylated by NDR1 kinase (Fig. 3a). Autophosphorylation of recombinant NDR1 was also observed, this was especially prominent in the absence of relevant substrates (120-191aa). Similarly, NDR kinase purified from HEK 293 cells exhibited similar ability to modify HP1α (Supplementary Fig. 2a).
Figure 3. NDR kinase phosphorylates HP1α.
(a) In vitro kinase assay using recombinant NDR1 kinase and GST or GST-HP1α WT or truncation mutants 1-66, 67-119, 1-119 and 120-191 as substrates. Coomassie stained gel is shown below for the substrates. (b) Schematic representation of the various phosphorylation sites on human HP1α. Note that the serine 93 of mouse HP1α is substituted with asparagine in human. (c) Phos-tag PAGE analysis of lysates from cells transfected with FLAG-mHP1α with HA-NDR1 kinase or HA-NDR1.K118A or without either kinase. Two forms of HP1α, faster migrating form (*2) and slower migrating form (*1) are discernible in wild-type HP1α. CIP treatment shows the mobility of dephosphorylated form. Phos-tag PAGE analysis of lysates expressing HA-NDR1 with FLAG-mHP1α- N-terminal mutant (S11-14A) represents a slower migrating hinge-alone phosphorylated form (*3) and a faster migrating unphosphorylated form (*U). The hinge mutant only shows faster migrating form (*2). α-Tubulin is shown as loading control in regular SDS-PAGE. (d) Phos-tag PAGE analysis of lysates expressing HA-NDR1 along with single point mutants in the hinge region (S92A; S93A; S95A; S97A) of mouse HP1α and the combination of all (hinge mutant). (e) PhosTag PAGE analysis of U2OS lysates treated with phosphatase followed by immunoblot analysis using HP1α antibody. (f) PhosTag PAGE analysis of lysates from HP1α siRNA treated cells followed by immunoblot analysis using HP1α antibody. (g, h) PhosTag PAGE analysis of lysates from NDR depleted cells using siRNA or shRNA (i, j) approach. Note the slower migrating mitotic-specific phosphorylated form of HP1α is reduced upon NDR1/2 depletion. (k) PhosTag PAGE analysis of lysates from U2OS cells synchronized at different stages of the cell cycle and immunoblot analysis with HP1α antibodies to detect endogenous HP1α. Note the slower migrating form is predominantly present during G2 and mitosis. Flow cytometry profile is provided at the top of the panel. Orc1 (present predominantly during G1); Cdt1 (present during G1, phosphorylated during M); Geminin (present in post-G1 cells) and Histone 3 phosphorylated at serine 10 (mitosis-specific) are shown in regular SDS-PAGE. (l) Quantitation of the relative levels of N-terminal and N-terminal + hinge phosphorylation of HP1α during different stages of the cell cycle based on the immunoblot in Fig. k.
NDR kinase phosphorylates HP1α in vivo
HP1α has several predicted phosphorylation sites in the N-terminus and within the hinge region. Recent studies have demonstrated that mammalian HP1α is phosphorylated at the N-terminal and hinge region 14,30,45. Here we selected several putative serine residues of mammalian HP1α based on the phosphosite predictions and evaluated the in vivo phosphorylation status of these residues46 (Fig. 3b). To demonstrate in vivo phosphorylation as well as to map the sites of phosphorylation within HP1α by NDR1 kinase, we co-transfected FLAG-tagged wild-type or several different point mutants (substitutions of serine to alanine of predicted phosphorylation site at N-terminal and hinge region) of mouse HP1α and human HA-NDR1 in U2OS cells. The lysates were analyzed by phos-tag-PAGE. Phos-tag forms a complex with Mn+2 and traps R-OPO3−2, this enhances the mobility shifts of phosphorylated proteins47. Wild-type HP1α was highly phosphorylated in cells co-transfected with NDR kinase in vivo with two distinct phosphorylation patterns (Fig. 3c, lane 2, see *1 and *2). Both the forms were sensitive to phosphatase treatment (CIP) and produce a faster migrating non-phosphorylated form of HP1α (Fig. 3c, lane 6, *U). In the absence of NDR1 kinase, the slower migrating phosphorylated band (*1) of HP1α was somewhat reduced (Fig. 3c, lane 1). The co-transfection with a kinase-deficient mutant form of NDR1 (K118A), also showed reduced phosphorylation of HP1α (Fig. 3c, lane 7). To further map the residues on HP1α that are phosphorylated by NDR kinases, we utilized full-length HP1α mutants containing amino acids substitutions that should result in a non-phosphorylatable N-terminus (N-terminal mutant, Serine at positions 11, 12, 13, 14 were substituted with alanine) or non-phosphorylatable hinge region (Hinge mutant, Serine at positions 92, 93, 95 and 97 were substituted with alanine) or a combination of both as well as the single point mutants of N-terminal (S11A, S12A, S13A, S14A) and hinge region (S92A, S93A, S95A, S97A). An HP1α mutant where all the serines at the N-terminal (S11, 12, 13, 14A) and at the hinge (S92, 93, 95, 97A) were substituted with alanines, a complete loss of phosphorylation was observed (Fig. 3c, lane 5). Further, mobility of this mutant HP1α was equivalent to that of the WT-HP1α treated with CIP (Fig. 3c, compare lanes 5 & 6). The faster migrating band of N-terminal mutant (Fig. 3c, lane 3) showed mobility similar to the unphosphorylated form (*U, Fig. 3c, lane 6). Also, the slower migrating form in the N-terminal mutant-transfected cells (*3) migrated faster compared to the slower-migrating form in wild type-transfected cells (*2), (Fig. 3c, compare lane 2 and 3). Further, dissection of the N-terminal end revealed that the point mutant, S14A the predominant phosphorylation site (Supplementary Fig. 2b, lane 4) showed similar profile to that of the N-terminal mutant (Supplementary Fig. 2b, lane 5). Further analysis using the hinge region mutant (S92A, S93A, S95A, S97A) co-transfected with NDR kinase revealed a loss of the slower migrating form altogether (Fig. 3c, lane 4). However, these mutants showed no change in the mobility of the faster migrating form suggesting that changes in the phosphorylation within the hinge region does not affect the N-terminal phosphorylation (Fig. 3d). Both S93A and S95A mutations of the hinge region resulted in disappearance of slower migrating form suggesting that these two sites represent the primary phosphorylation sites of NDR1 in the hinge region (Fig. 3d and Supplementary Fig. 2c). Interestingly, even though human and mouse HP1α are highly conserved (~99%), serine 93 of mouse HP1α is substituted with asparagine in human (Fig. 3b). We therefore suggest that the predominant site of phosphorylation within the hinge region of human HP1α is Serine 95 (Fig. 3d). A fainter slower migrating form can also be observed upon overexposure in S95A mutant perhaps due to the presence of intact serine 93 in this mutant (Supplementary Fig. 2c). These above results revealed that the fast migrating form (*2 in Fig. 3c and 3d) represented HP1α phosphorylated at the N-terminal region alone whereas the slower migrating form (*1) denoted HP1α phosphorylated in the hinge region in addition to the N-terminus.
Immunoblots using HP1α antibody to detect endogenous HP1α on Phos-tag gels revealed two forms of HP1α in human cells similar to our observations when mouse HP1α was transiently expressed (Fig. 3e, U2OS, asynchronous). Furthermore, phosphatase treatment showed that both the forms collapsed to an unphosphorylated form (Fig. 3e). Further, it was evident that the N-terminal domain was constitutively phosphorylated, since unphosphorylated form of HP1α was never detectable. Analysis of HP1α siRNA treated extracts further confirmed the specificity of both the phosphorylated forms (Fig. 3f).
To address if NDR1 kinase phosphorylates HP1α in vivo, we next examined the status of phosphorylation of HP1α in cells lacking NDR kinase. Phos-tag electrophoresis analysis revealed that in NDR kinase-depleted cells (using both siRNA as well as shRNA), there was a significant reduction in the slower-migrating band of HP1α (*1) without any significant alteration in the faster migrating band (*2; Fig. 3g–j). The decrease in the phosphorylation at the hinge region (slower migrating band) of HP1α upon NDR depletion further confirmed that NDR kinase is responsible for phosphorylating HP1α within the hinge region in vivo (Fig. 3g–j). While the in vitro data demonstrated that NDR kinase could also phosphorylate HP1α at the N-terminal domain, no significant and consistent changes in the N-terminal phosphorylation of HP1α were observed in NDR1/2-depleted cells. These results suggest that other kinases (in addition to NDR kinase) may phosphorylate HP1α at its N-terminus domain in vivo.
We next examined if phosphorylation of HP1α at specific serine residues in the N-terminus as well as the hinge region are cell cycle regulated. During G1, G1/S, S and G2 phase, the faster migrating form representing the N-terminal phosphorylation of endogenous HP1α was predominant, whereas cells at G2 also showed increased levels of the slower migrating form of HP1α representing phosphorylation at the N-terminus+hinge regions (Fig. 3k and 3l). By mitosis, the slower migrating form represented the prominent form of HP1α in the cells (Fig. 3k and 3l). Interestingly, there was an increase in the levels of slower migrating form (N-terminal+hinge) of HP1α in G2 and M phase with a concomitant decrease in the faster migrating form specifically during mitosis (Fig. 3k and 3l). In addition, we have also observed other migrating forms of HP1α in between form *1 and *2, especially in mitotic extracts (Fig. 3k). We believe that these represent other phosphorylated forms of HP1α that are present at lower levels.
Similar observations, where we detect slower migrating forms during mitosis were made in cells co-transfected with FLAG-HP1α and HA-NDR1 and synchronized at mitosis. In nocodazole-arrested cells expressing only the FLAG-HP1α the slower migrating forms were apparent in addition to the faster migrating one (Supplementary Fig. 2d). Contrary to the data obtained with endogenous HP1α (see Fig. 3k and 3l), FLAG-HP1α-expressing prometaphase-arrested cells continued to show higher levels of N-terminal-alone phosphorylated faster migrating form of HP1α (the ratio of faster migrating [lower] versus slower migrating [upper] band in the absence NDR1 is 1.85/1) (Supplementary Fig. 2d). We speculated that the cellular pool of NDR1 might be limiting for the cells to phosphorylate the hinge domain of most of the exogenously expressed HP1α. To test this, we co-expressed both HP1α and NDR1 and in prometaphase cells the ratio of the slower migrating form to the faster migrating form was found altered with increased levels of the slower migrating form being apparent (the ratio changed to 1/1.29) (Supplementary Fig. 2d). Based on all the data we conclude that during mitosis most of the cellular HP1α undergoes hinge-specific phosphorylation by NDR kinase.
S95 phosphoHP1α localizes to centromeres during mitosis
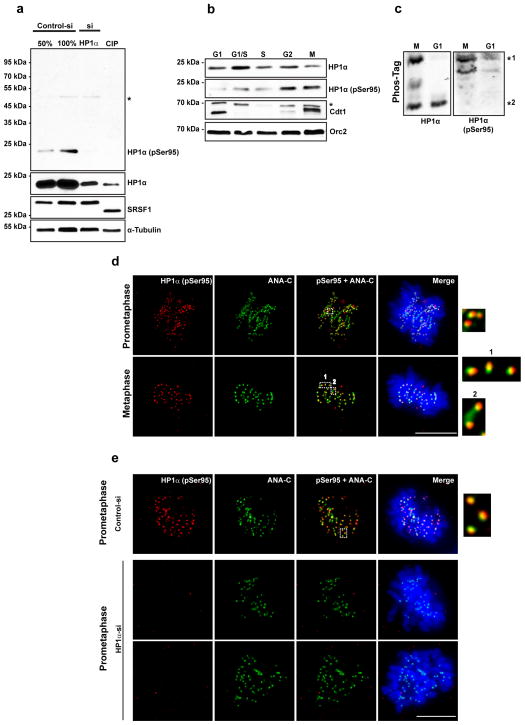
To address the functional relevance of the cell cycle-regulated hinge-specific phosphorylation of HP1α in human cells, we have generated HP1α-S95-specific phospho-antibody. Depletion of HP1α using HP1α-specific siRNA oligonucleotides followed by immunoblot analysis in HeLa whole cell extracts using HP1α antibody as well as S95-specific phospho-antibody revealed that the S95 antibody detected HP1α only in control cells but not in HP1α-depleted cells (Fig. 4a). Further, immunoblot analysis in cell lysates treated with CIP using the S95 antibody showed significantly reduced signal further confirming the specificity of this antibody (Fig. 4a). We have shown that HP1α is constitutively phosphorylated in vivo. The HP1α antibody that can recognize both phosphorylated and unphosphorylated forms recognizes the phosphorylated HP1α more prominently than the unphosphorylated form (Fig. 4a). We next examined the cellular abundance of hinge-phosphorylated form of HP1α during different stages of the cell cycle using S95 specific-antibody. Hinge-phosphorylated form of HP1α was most prominent during G2 and mitosis (Fig. 4b) with reduced levels during G1 phase, corroborating our previous results (Fig. 3k). In contrast, the HP1α antibody showed reduced signals during mitosis, because the overall levels of HP1α during G1/S seem to be much higher than during mitosis. Furthermore, phos-tag immunoblot analysis in M-phase and G1 synchronized cell extracts revealed that S95 antibody detected the N-terminal + hinge phosphorylated form of HP1α (slower migrating form *1) (Fig. 4c). S95 antibody occasionally detected a band that has the mobility in between the two phosphorylated forms. This may represent the hinge-only phosphorylated form. These results further confirm that the slower migrating form of HP1α present during G2 and mitosis is phosphorylated predominantly at serine 95.
Figure 4. Hinge-specific phosphorylated form of HP1α localizes at kinetochores during early mitosis.
(a). Immunoblot analysis of lysates using HP1α-S95 phospho-antibody from HeLa cells that are depleted of HP1α using siRNA and treated with CIP on regular SDS-PAGE. SRSF1 is shown as control. (b) Immunoblot analysis of lysates from different stages of the cell cycle using HP1α-S95 phospho-antibody on regular SDS-PAGE. Note the increased signal with S95 antibody during G2 and Mitosis. (c) PhosTag PAGE analysis of lysates from cells synchronized at Mitosis (M) or G1 phase and immunoblot analysis with HP1α and HP1α-S95 phospho-antibodies. Note that S95 phospho-Ab does not detect the faster form of HP1α. (d) Immunofluorescence localization of hinge-specific phosphorylation of HP1α using S95 phospho-Ab (red) along with centromere marker ANA-C (green) in human HeLa cells. Note the association of phosphorylated form of HP1α in the vicinity of centromeres in prophase and metaphase. Inset (5X) shows that the red signals are exterior to green, suggesting the presence of this modified form of HP1α at outer kinetochores. Scale bar represents 10μm. (e) Immunofluorescence using S95 and ANA-C antibodies in U2OS cells treated with control or HP1α siRNA. Note the loss of centromeric signal of S95 in HP1α siRNA-treated cells. Inset (5X) shows that the red signals are exterior to green. Scale bar represents 10μm.
We next examined the localization of the hinge-specific phosphorylation of HP1α using affinity purified S95 phospho-antibody. Co-immunolocalization using S95 antibody and centromere specific antibody (ANA-C) in human cell lines, including HeLa cells (Fig. 4d) and U2OS cells (Supplementary Fig. 3a) demonstrated that the hinge-specific form of HP1α was present in the vicinity of the centromeres prominently during prophase. In most of the occasions, the S95 signal was located on the outer edge of the ANA-C (recognizing CENP-A) antibody-stained centromeres, reminiscent of kinetochore distribution (please see the insets in Fig. 4d–e). Co-immunolocalization using S95 antibody and outer kinetochore specific-Mad1 in human HeLa cells further confirmed the kinetochore association of phosphorylated HP1α (Supplementary Fig. 3e and 3f). The kinetochore association of hinge-specific phosphorylated form of HP1α reduced further as the cells progressed through mitosis and was no longer evident during anaphase and telophase (Supplementary Fig. 3a). However, an antibody against HP1α that recognizes all the forms of HP1α detected HP1α at centric and pericentromeric heterochromatin throughout mitosis (Supplementary Fig. 3d). These results demonstrate that the S95-specific phosphorylated form of HP1α preferentially localizes at kinetochores during early mitosis and is lost from these sites as the cells progress through mitosis. Use of pre-immune serum as well as peptide block experiments confirmed the specificity of the antibody (Supplementary Fig. 3b and 3c). To further confirm the specificity of the S95 antibody, we carried out immunostaining using this antibody in U2OS cells treated with HP1α siRNA. Specific signals at the centromeres using S95 antibody were almost completely diminished in HP1α siRNA-treated cells but not in the control cells (Fig. 4e).
Depletion of NDR kinase results in mitotic abnormalities
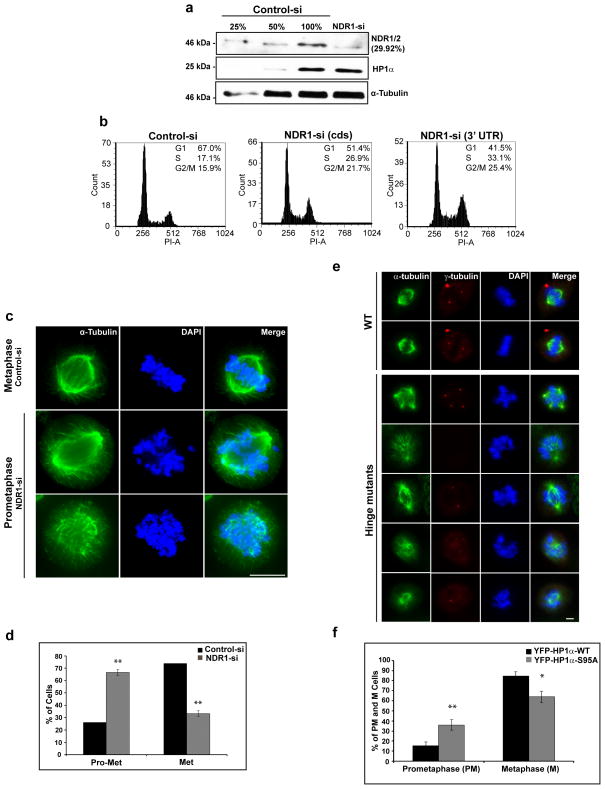
In order to address the physiological relevance of NDR kinase-mediated phosphorylation of HP1α we examined how NDR1 influences HP1α function in vivo. We conducted RNAi-mediated knockdown of NDR using two small interfering RNA (siRNA against 3′ UTR and coding region of NDR1) oligonucleotides as well as short hairpin RNA (shRNA) in human U2OS cell lines 48. We achieved >70% knockdown of NDR (both by siRNA and shRNA treatment) and found that the cells lacking these kinases did not show significant alteration in the total cellular levels of HP1α and other HP1 isoforms (Fig. 5a and Supplementary Fig. 4a). Immunoblot with an antibody that detected both NDR1 and 2 in control and NDR1 si-treated cells indicated that depletion of NDR1 also reduces the levels of NDR2. Human genome contains two NDR genes, NDR1 and NDR2. We have observed that both NDR1 and NDR2 kinases interact with one another (Supplementary Fig. 4b) and therefore there seems to be co-operativity between these two kinases. We have also observed that NDR1 homodimerizes/homomultimerizes (Supplementary Fig. 4c). We are currently investigating the homo-di/multimerization of NDR1 and its heteromerization with NDR2 and its impact on the functionality of these kinases.
Figure 5. Hinge-specific phosphorylation of HP1α is required for accurate chromosome alignment and mitotic progression.
(a) Immunoblot analysis of lysates from cells that are depleted of NDR kinase using siRNA. The upper panel denotes the % of whole cell extracts (WCE) loaded in control; the knockdown represents 100% WCE. Note that >70% knock-down of NDR1 was achieved. (b) Flow cytometry in control and NDR depleted cells using two different siRNA targeting the coding region (cds) and 3′ UTR of NDR1. (c) Immunofluorescence analysis using α-tubulin antibodies to study mitotic phenotype. (d) Note an increase in prometaphase population upon NDR depletion. Error bars represent SD of three independent experiments. Statistical significance was determined by Student’s t-test. Mean ± SD, *p<0.05, **p<0.01 and ***p<0.001. Scale bar represents 10μm. (e) Overexpression of FLAG-S95A and hinge mutant (S92, S93, 95, 97A) of mouse HP1α in human U2OS cells and immunofluorescence analysis using α- and γ-tubulin. Note mitotic abnormalities in cells expressing mutants. Scale bar represents 10μm. (f) Transient transfection of YFP-HP1α-WT and YFP-HP1α-S95A mutant, followed by statistical analysis of mitotic population. Note the accumulation of prometaphase stage in cells expressing YFP-HP1α-S95A mutants. Error bars represent SD of three independent experiments. Statistical significance was determined by Student’s t-test. Mean ± SD, *p<0.05, **p<0.01 and ***p<0.001. Scale bar represents 10μm.
Flow cytometry analysis of NDR1-depleted cells demonstrated a decrease in G1-phase with a concomitant increase in G2/M population (Fig. 5b and Supplementary Fig. 4d), suggesting that NDR kinase is required for cell cycle progression. We have observed significant loss of hinge-specific mitotic phosphorylation of HP1α in NDR1-depleted cells (Figure 3g–j), suggesting that this post-translational modification may have a crucial function in mitotic progression.
Phenotypic analysis of mitotic cells demonstrated that in control cells, a total of 26±0.02% (for control siRNA-treated cells) and 25±11.6% (shLUC control cells) of mitotic cells were present in prometaphase stage (± denotes standard deviation of the mean from three independent experiments). We observed an increase in prometaphase population (67±2% for NDR1 siRNA- and 60±15.7% for NDR shRNA-treated cells; ± denotes standard deviation of the mean from three independent experiments) with a concomitant decrease in metaphase populations in the NDR kinase-depleted cells (Fig. 5c, 5d & Supplementary Fig. 4e and 4f). These results indicate that NDR1-depleted cells show defects in chromosome alignment and therefore display defects in mitotic progression.
Since we observed mitotic defects with cells arrested during prometaphase and loss of hinge-specific phosphorylation of HP1α upon NDR-depletion, we examined if phosphorylation of HP1α is required for mitotic progression. To address the role of phosphorylation within the hinge region of HP1α, we conducted overexpression of the non-phosphorylatable mutants (FLAG tagged mouse HP1α mutated at S95A or S92, 93, 95, 97A) and examined its effect on mitosis. We observed variable chromosome alignment defects in cells expressing hinge mutants in comparison to controls (Fig. 5e). Further, we have also observed aberrant spindle morphology. Defective spindles could also cause delay in the alignment of chromosomes and the extent of these defects could be attributed to the penetrance of the phenotype. Moreover, transient over-expression of YFP-HP1α-S95A mutant significantly increased the prometaphase population over cells expressing wild type HP1α (Fig. 5f). In control cells we observed 16±4% of prometaphase, 84±4% of metaphase, in contrast, S95A expressing cells showed 36±5% of cells in prometaphase and 64±5% in metaphase (± denotes standard deviation of the mean from three independent experiments). Therefore, the HP1α-S95A hinge mutants of HP1α phenocopy ‘NDR depletion, further supporting the role of NDR1-mediated phosphorylation of HP1α in proper chromosome alignment and hence mitotic progression perhaps mediating the prometaphase to metaphase transition. We also depleted endogenous NDR1 from HeLa cells followed by the immunolocalization of hinge phosphorylated HP1α at the mitotic centromere using S95-specific phospho-antibody. We observed a reduction of phospho-HP1α at mitotic centromeres upon NDR1 depletion (Supplementary Fig. 4g), indicating the NDR1 mediated phosphorylation of HP1α is required for the localization of HP1α to centromeres.
We next evaluated the localization of YFP- HP1α-S95A mutant as well as YFP-HP1α-S95E mutant in cells where the endogenous HP1α (using 3′UTR specific siRNAs) was depleted. Immunoblots demonstrated that 3′UTR specific siRNA treatment resulted in the reduction of endogenous HP1α but did not alter the overall levels of YFP-tagged HP1α mutants (Supplementary Fig. 5a). YFP-HP1α-S95A localization to the mitotic centromeres was reduced in the absence of endogenous HP1α (Supplementary Fig. 5b). YFP-HP1α-S95A associated with centromeres only in cells that contained intact endogenous HP1α. This is explained by our observation that the mutants can efficiently interact with the WT-HP1α and therefore can be recruited to the centromeres by virtue of their association with the WT-HP1α (Supplementary Fig. 5c). On the other hand, the phospho-mimetic mutant of HP1α (YFP-HP1α-S95E) bound to mitotic centromeres in the presence or absence of endogenous HP1α (Supplementary Fig. 5b). These results suggest that the mitotic defects in NDR-depleted cells may be due to changes in the phosphorylation status of HP1α, since reduced phosphorylation in the hinge-domain of HP1α reduced its ability to localize to centromeric region.
Phospho S95 facilitates Sgo1 binding to mitotic centromeres
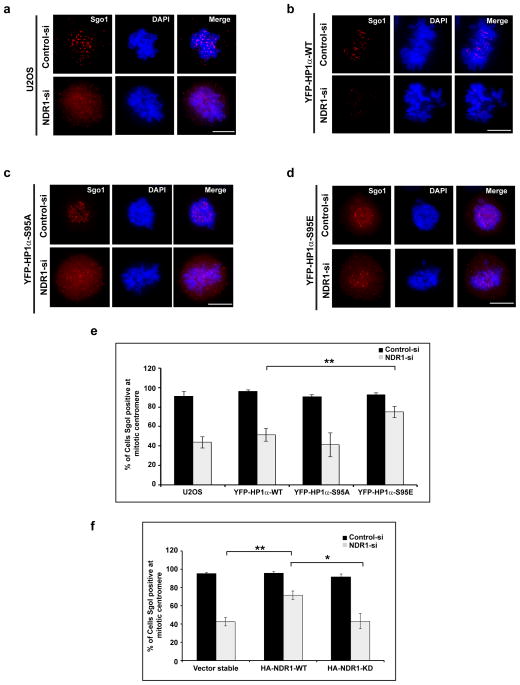
Next we were interested to understand the role of hinge-phosphorylation of HP1α in mitotic progression. HP1-mediated targeting of the human Shugoshin (Sgo1) to mitotic centromeres is required for centromeric sister-chromatid cohesion 28. We examined the localization of Sgo1 to centromeres during mitosis in NDR1-siRNA and HP1α-siRNA treated cells. We have systematically counted (>150 cells) and looked for centromeric staining of Sgo1 in prometaphase cells. Presence of clear puncta was categorized as positive, whereas diffuse signal with no enrichment at the centromeres were categorized as negative. We observed a significant decrease in the localization of Sgo1 to mitotic centromeres in cells lacking NDR1 as well as HP1α (around 91±5% cells showed Sgo1 staining on mitotic centromere in control siRNA treated cells, whereas 44±5.6% for NDR1 siRNA- and 16±1.1% for HP1α siRNA-treated cells. (± denotes standard deviation of the mean from three independent experiments) (Fig. 6a & 6e; Supplementary Fig. 5d). In order to address if the hinge-specific phosphorylation of HP1α facilitates the Sgo1 localization to mitotic centromeres, we individually depleted either NDR1 kinase or HP1α (utilizing siRNA oligonucleotides targeting the 3′UTR of NDR1 and HP1α, respectively) in cells stably expressing YFP-HP1α-WT, YFP-HP1α-S95A or YFP-HP1α-S95E. Depletion of NDR1 or HP1α in YFP-HP1α-S95A background continued to show lack of Sgo1 association to mitotic centromeres (91±5% in controls, 41±12.3% for NDR1 siRNA- and 21±4.7% for HP1α siRNA-treated cells; ± denotes standard deviation of the mean. Data were calculated from triplicates) (Fig. 6c & 6e; Supplementary Fig. 5d). We have observed similar loss of Sgo1 staining from centromeres in YFP-HP1α-WT cells upon NDR1 depletion (96±1.5% in controls, 51±6.5% for NDR1 siRNA- treated cells; ± denotes standard deviation of the mean. Data were calculated from triplicates) (Fig. 6b & 6e) Interestingly, in NDR1 or HP1α-depleted cells expressing YFP-HP1α-S95E, the number of mitotic cells that decorated Sgo1 at centromeres increased significantly (75±5.8% for NDR1 siRNA- and 69±8% for HP1α siRNA-treated cells; ± denotes standard deviation of the mean. Data were calculated from triplicates) (Fig. 6d & 6e; Supplementary Fig. 5d). Similarly, localization of Sgo1 at mitotic centromeres was partially rescued in endogenous NDR1-depleted cells (using 3′UTR specific-siRNA) that were stably expressing HA-NDR1, but not in cells expressing vector alone or the kinase-deficient NDR1 (Figure 6f). These results demonstrate that NDR1 kinase (and perhaps NDR1 kinase-mediated phosphorylation of HP1α at the hinge region) directly/indirectly facilitates the localization of Sgo1 to mitotic centromeres. It is well established that the recruitment of Sgo1 to mitotic centromeres is crucial for chromosome segregation 28. We demonstrate that NDR-depletion causes significant reduction in HP1α phosphorylation, defects in Sgo1 localization to centromeres and mitotic abnormalities. Based on these results, we propose that the hinge phosphorylation of HP1α during mitosis by NDR1 facilitates the proper loading of Sgo1 to centromeres (Fig. 7).
Figure 6. Hinge-specific phosphorylation of HP1α facilitates Sgo1 binding to mitotic centromeres.
(a). Immunofluorescence using Sgo1 antibody in NDR1-depleted cells in U2OS cells; (b) YFP-HP1α-WT; (c) YFP-HP1α-S95A and (d) YFP-HP1α-S95E expressing cells. DNA is counterstained with DAPI. The bar represents 10 μm. (e) Quantification of the presence of Sgo1 at mitotic centromeres (prometaphase stage). Note the increased binding of Sgo1 to centromeres in NDR1-depleted YFP-HP1α-S95E expressing cells. Error bars represent SD of three independent experiments. Statistical significance was determined by Student’s t-test. Mean ± SD, *p<0.05 and **p<0.01. (f) Quantification of Sgo1 presence (Sgo1+) at mitotic centromeres in cells expressing empty vector, HA-NDR1-WT or HA-NDR1-KD (K118A) and lacking NDR1 kinase. Error bars represent SD of three independent experiments. Statistical significance was determined by Student’s t-test. Mean ± SD, *p<0.05 and **p<0.01. Note the rescue of Sgo1 signal at centromeres in HA-NDR1 expressing cells.
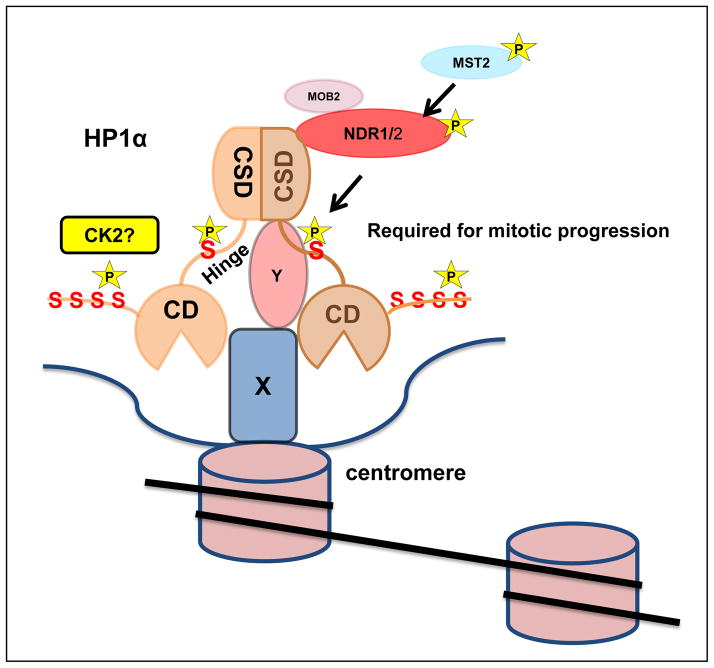
Figure 7. Phosphorylation of HP1α is required for accurate mitotic progression.
Model depicting the association of phosphorylated HP1α within the N-terminus associating with the chromatin mark H3trimethyl-K9. NDR kinase binds to the chromoshadow domain of HP1α and phosphorylates the hinge domain during G2/M stages of cell cycle that in turn may facilitate its association with centromeric proteins (X and Y represent unknown proteins at centromere; Sgo1 is a good candidate for Y) and accurate chromosome alignment and mitotic progression. Stars represent phosphorylation sites.
Discussion
HP1α, a heterochromatin-associated protein, facilitates the assembly of higher order chromatin structure and plays key roles in various chromosome functions including sister chromatid cohesion and transcriptional gene silencing 27. Similar to histones, HP1 isoforms are decorated with extensive post-translational modifications that are often linked with their biological functions 14. In this study, we demonstrate that mammalian HP1α is constitutively phosphorylated within its N-terminus and additional phosphorylation within the hinge domain becomes apparent as the cells progress from G2 to mitosis and this modification is required for mitotic progression.
HP1 has been shown to be required for accurate chromosome segregation in Drosophila embryos 49, in fission yeast 24,50 as well as in mammalian cells 51,52. HP1/Swi6 in fission yeast is required for the recruitment of cohesins to centromeres and the establishment of the centromeric histone H3 variant CENP-ACnp1, which is crucial to regulate chromosome alignment and segregation 1,24,53–55. The targeting of HP1α to centromeric heterochromatin occurs through two independent mechanisms. In interphase nuclei, the tri- methylated lysine 9 of histone H3 is important for targeting HP1α to centromeric heterochromatin 3,4 and the phosphorylation at N-terminal serine residues of HP1α enhances the interaction of HP1α with tri-methylated H3K9 30. However, during mitosis phosphorylation by Aurora B kinase at Serine 10 of H3 (H3S10) disrupts this interaction and releases most of the HP1α from chromatin except at centromeres 56. Therefore, a chromodomain (CD) independent event operates to recruit HP1α to mitotic centromeres, involving an interaction between chromosomal passenger protein INCENP and HP1 chromoshadow domain (CSD) through the PXVXL/I motif 57,58. Characterization of centromeric proteins that specifically associate with the hinge-phosphorylated HP1α at kinetochores will be a goal for future studies.
A recent study by Hiragami-Hamada and colleagues described a role for the N-terminal phosphorylation of HP1α in enhanced chromatin binding 30. Even though it is largely believed that CKII is the kinase that phosphorylates HP1α, in vivo data obtained upon CKII depletions show no effect on HP1α phosphorylation 30. We demonstrate that mammalian HP1α is a mitotic substrate for NDR (Nuclear Dbf2 related) kinase. Using immunoprecipitation experiments we have demonstrated the interaction of NDR1 kinase and HP1α. We were however unable to detect a direct interaction between HP1α and NDR1. This is not surprising since the interaction between kinase and its substrates are known to be highly transient in nature and have strong dependency on the cellular context 59,60. However, HP1α-W174A, a chromo-shadow domain mutant that has lost its ability to bind PXVXL/I ligands showed almost complete loss of interaction with NDR1 kinase.
The NDR family of serine threonine kinases, exist as four related kinases in human cells: NDR1, NDR2, LATS1 (large tumor suppressor-1) and LATS2 and all of them play key roles in many vital cellular processes, including cell growth, proliferation and differentiation, G1/S transition, centrosome duplication, mitotic chromosome alignment, mitotic exit and cytokinesis33. The cell cycle protein p21 is the only known in vivo substrate identified for NDR kinase in human cells 40. Recent work demonstrated that NDR1 kinase is required for accurate chromosome alignment 41. Further, it was demonstrated that the activation of NDR by the upstream pathway comprising of MST2 kinase, Furry and MOB, determine the accuracy of chromosome alignment on the metaphase plates 41. We have observed that in the NDR1-depleted cells, chromosomes failed to align at the metaphase plate with cells accumulating in prometaphase. Furthermore a small population of cells showed spindle abnormalities consistent with the previously described role of NDR1 in maintaining spindle integrity and centrosome structure 61. Work by Mizuno and coworkers have demonstrated that the kinase activity of NDR1 is required for chromosome alignment 41. However, the substrates that are phosphorylated by NDR1 kinase that may drive accurate chromosome alignment had yet to be identified.
We have now demonstrated that HP1α is a mitotic substrate for NDR1 kinase and that the phosphorylation of HP1α is crucial for chromosome alignment and mitotic progression. Our present data indicate that NDR1 kinase is responsible for the G2/M phosphorylation within the hinge region of HP1α, as depletion of NDR1 results in significantly reduced phosphorylation of HP1α within the hinge region. This is consistent with the fact that the kinase activity of NDR1 was reported to be enhanced about 4.4 fold in mitotic cells as compared to S-phase cells 41. We observe that the hinge-specific phosphorylated form of HP1α localizes at kinetochores predominantly during prophase and prometaphase and is subsequently released from these sites during later stages of mitosis. Further, we observed pro-metaphase arrest in cells that were either depleted of NDR1 or in which the HP1α-S95A hinge mutant of HP1α was over-expressed. These results suggest that the hinge-specific phosphorylation of HP1α is crucial for mitotic progression. Furthermore, NDR1 or HP1α depletion caused reduced association of Sgo1 to centromeres. Sgo1 has been shown to protect centromeric sister chromatid cohesion during prophase 62. Our results demonstrate that the phosphorylated form of HP1α facilitates the localization of Sgo1 to the mitotic centromeres. We also observed that the HP1α-S95A mutant failed to localize to the centromeres suggesting that the phosphorylated form of HP1α may tether Sgo1 directly or indirectly to the mitotic centromeres. HP1α has previously been implicated in promoting Sgo1 centromeric localization 28. However, a recent study has suggested that a Sgo1 mutant deficient in HP1 binding is functional in sister chromatid cohesion and localizes to centromeres 58. Our data supports a role for NDR1-mediated hinge-specific phosphorylation of HP1α in facilitating Sgo1 binding to centromeres directly or indirectly. In addition, several redundant mechanisms are known to enable the Sgo1 targeting to mitotic centromeres 58.
Earlier studies reported defects in mitotic chromosome alignment and in the activation of Aurora B upon NDR1 depletion 41,63. It is a possibility that the hinge-specific phosphorylation of HP1α protein, in addition to governing Sgo1 binding to centromeres, also modulates the Aurora B activity in order to promote accurate and faithful mitotic chromosome alignment and segregation. Activation of Aurora B, as well as its positioning at heterochromatin has been shown to require INCENP 64,65. Furthermore, Aurora B associates with C-terminus of INCENP, while the hinge region of HP1α binds to the N-terminus of INCENP 65,66. Phosphorylation of HP1α at the hinge region might modulate the interaction of HP1α with Sgo1 and perhaps INCENP at centromeres (Fig. 7). It is likely that the lack of NDR1 kinase might affect multiple pathways that prevent Sgo1 association to mitotic centromeres.
Our data supports a role for NDR-mediated hinge-specific phosphorylated HP1α in recruiting Sgo1 directly or indirectly to the mitotic centromeres. However, a possibility of HP1α independent role of NDR1 in regulating Sgo1 can also be considered. The role of Sgo1 in cohesion protection has been shown to be antagonized by the mitotic kinase Plk1 and aurora B 67. On the other hand the kinase activity of Aurora B was increased in cells depleted of NDR1 or MST1, an upstream kinase of NDR1 63. Moreover, similar to NDR1 depletion, depletion of MST1 also resulted into mitotic arrest with unaligned mitotic chromosomes and the activation of the Mad2- and BubR1-dependent spindle checkpoint response 63. Therefore, it is possible that depletion of NDR1 hyperactivates Aurora B, which in turn destabilizes the Sgo1 on mitotic chromosome. Similarly, depletion of either Mad2 or Aurora B has been shown to abrogate the mitotic-arrest phenotype induced by Bub1 and Sgo1 depletion spindle checkpoint response 63. Therefore, it is possible that depletion of NDR1 hyperactivates Aurora B, which in turn destabilizes the Sgo1 on mitotic chromosome. Similarly, depletion of either Mad2 or Aurora B has been shown to abrogate the mitotic-arrest phenotype induced by Bub1 and Sgo1 depletion 68. Future studies will focus on the role of NDR1 kinase and phosphorylated HP1α in the loading of Sgo1 to the centromeres.
Materials and Methods
Cell Culture and Generation of Stable Cell Lines
Cell lines used in this study were obtained from the American Type Culture Collection grown in Dulbecco’s modified Eagle’s medium (DMEM) containing high glucose, supplemented either with 10% fetal bovine serum (FBS; Hyclone for U2OS) or reduced tetracyclin fetal bovine serum (Tet system approved FBS; Clontech for 2-6-3 CLTon). Cells were transfected with Lipofectamine 2000 (Invitrogen) as per the manufacturers’ protocol. For generation of stable tetracycline-inducible cell lines expressing tetracyclin-regulated shRNA against NDR1 and 2, U2OS cells-TetON were transfected with pTER-NDR1/2 and pTER-Luciferase (control shRNA) [kind gift from Brian A Hemmings]. Cell clones were selected and maintained by growing them in the presence of 200 μg/ml zeocin and 200 μg/ml hygromycin. YFP- HP1α-WT, mS95A and mS95E cell lines were generated by transfecting the corresponding constructs in U2OS cell lines followed by selecting and maintaining them in the media containing G418 (Invitrogen). Lenti-viral expression system was utilized for stable integration of HA-NDR1-WT or HA-NDR1-KD or empty vector in U2OS cells. Cell clones were selected by growing them in the presence of puromycin (2μg/ml).
Plasmids and antibodies
For different experiments, wild type human NDR1 cDNA was cloned in pCGN (XbaI/BamHI) or pCGT (XbaI/BamHI).. The C-terminal (amino acids 1-84, 1-100, 1-175 and 1-225) and N-terminal (amino acids 85-435, 142-465 and 188-465) deletion mutants were cloned in pCGT or pECFP-C1 vectors. The coding region of human NDR1 was also cloned into pECFP-C1 vector. The coding region of human HP1α was cloned into pEYFP–LacI vector (modified from pEGFP-LacI; kindly provided by Miroslav Dundr) 69. The YFP tagged wild type (1-191aa) and truncated mutants (1-75, 81-191, 121-180 and 121-191aa) of human HP1α were kindly provided by Yasushi Hiraoka 57. GST tagged wild type and truncated mutants (1-66, 67-119, 1-119 and 120-191aa) and FLAG tagged wild type mouse HP1α and its serine to alanine point mutants were the kind gifts from Pierre Chambon and Jun-ichi Nakayama, respectively. The antibodies used in this study were as follows: anti-T7 (Novagen 69522, 1:5000), anti-HA (12CA5, 1:500) anti-HP1α (for WB, Chemicon MAB3446, 1:500 and immunostaining for WB, Chemicon MAB3584, 1:100), anti-HP1β (Chemicon MAB3448, 1:500), anti-HP1γ (Chemicon MAB3450, 1:500), ANA-C (Sigma ANA-C, detecting CENP-A, 1:15), Anti-NDR1/2 (Millipore MABS56, 1:500) Anti-NDR-N-terminal (gift from Brian A Hemmings), anti-GFP (Covance 118R, 1:500), anti-HA, anti-FLAG (M2 FLAG Sigma F3165, 1:500), anti-Mad1 (Sigma M8069, 1:100) and anti-α-tubulin (Sigma T5168, WB-1:5000, IF- 1:2000), γ-tubulin (Sigma, T5792, 1:2000), H3K9-me3 (Upstate 07-523, 1:500) anti-phospho H3Ser 10 (Upstate 06-570, 1:600), anti-Geminin (Santa cruz sc13015, 1:300) and anti-Sgo1 (gift from Hongtao Yu). Uncropped scans of the western blots are provided in Supplementary Fig. 6.
Generation of phospho-specific HP1α.antibody
Abgent custom services were used to generate and purify the phospho-specific antibody against the modified peptide SNKRKSNFBNSADDIKSKKKRC. The antibodies are purified using peptide affinity purification by Abgent custom services. The peptides are conjugated to solid phase packed in columns for purification purposes. Filtered sera are passed through non-phospho peptide (SNKRKSNFSNSADDIKSKKKRC) column and bound antibody is eluted. The pass through is then loaded to phosho peptide column. Bound antibody is eluted as the phospho-antibody.
Cell Synchronization
U2OS cells were synchronized at different stages of the cell cycle either by double thymidine block and release or nocodazole block and release. Briefly, cells were grown in the presence of 2 mM thymidine for 24 hr. After the first block, thymidine was removed by repeatedly washing the cells in PBS and cells were released for 12 hr in fresh medium. Finally, cells were synchronized at G1/S by further growing them in the presence of 2 mM thymidine for 24 hr. The cells were released in fresh medium for 4 hr to collect them in S phase; 8 hr for G2 phase and monitored for appearance of rounded cells for mitosis. Alternately, cells were synchronized at prometaphase by treating them with 50 ng/ml Nocodazole for 12–16 hr. G1 population was collected by washing the nocodazole-arrested cells in PBS and releasing them in the fresh medium for 5–6 hr. Synchronized samples were evaluated by flow cytometry and immunoblot analysis.
Immunoprecipitation and flow cytometry
For co-IP, T7/HA tagged wild type and various mutants of NDR1 were co-transfected with YFP-tagged HP1α (both wild type and mutants) in U2OS cells. Cells were lysed, 24 hours post-transfection, in IP buffer [50 mM HEPES (pH 7.9), 5% glycerol, 200 mM KCl, 0.1% Triton X and 1 mM CaCl2 supplemented with protease and phosphatase inhibitors and 0.5 uM okadaic acid] followed by treatment with 2U of micrococcal nuclease (Sigma) for 20 min at room temperature. Digestion was stopped by the addition of 0.5 M EDTA. After pre-clearing with Gammabind Sepharose beads for 1 hour, the lysates were incubated with appropriate antibody- conjugated agarose overnight. The beads were washed in the same IP buffer and finally denatured by the addition of Laemmli buffer. The complex was analyzed by Western. Uncropped western blots have been supplied as supplementary Fig. 6.
For flow cytometry, cells were fixed in chilled ethanol overnight after resuspending in PBS + 1% NGS. After two rounds of washing, cells were resuspended in PBS + 1% NGS with 120μg/ml propidium iodide (PI) and 10μg/ml RNase A followed by 30 min incubation at 37°C. DNA content was measured by flow cytometry.
Immunofluorescence analysis
The 2-6-3 CLTon and U2OS/HeLa cells were either fixed in in ice-cold methanol at −20°C or 2% PFA for 15 minutes at room temperature. The cells were permeabilized in 0.5% Triton X-100 in PBS for 7 minutes, stained with DAPI followed by washing in PBS and finally were mounted in either PPD or Vectashield (Vector Laboratories Inc.). For Ser 95 specific phosphor-antibody, cells were first pre-extracted in 0.3% CSK buffer for 3 min followed by 2% PFA fixation for 15 minutes. Antibody staining was carried out using standard procedures. A Zeiss Axioimager z1 fluorescence microscope (Carl Zeiss Inc.) equipped with Chroma filters (Chroma Technology) was used for cellular examination. Axiovision software (Zeiss) was used to collect digital images from a Hamamatsu cooled CCD camera. Images were also acquired using a Delta Vision optical sectioning deconvolution instrument (Applied Precision) on an Olympus microscope.
For figure 4 (and supplementary Figure 3), images were acquired using a deltavision optical sectioning microscope. For individual images, 6 sections of 0.3μm were used for processing by deconvolution and quick projection.
Depletion of human NDR1/2
Two small interfering RNA (siRNAs) targeting human NDR1 (IDT, USA) against 3′ UTR (sense:5′ CCAAUAUGUCAUAGUAAAGUCUCCT3′, anti-sense: 3′GUGGUUAUACAGUAUCAUUUCAGAGGA5′) and cds (sense:5′ AGAAAUAGACGUCAGCUAGCCUUCT3′, anti-sense: 3′UUUCUUUAUCUGCAGUCGAUCGGAAGA5′) were delivered into cells twice at a gap of 24 hr, at a final concentration of 10μM, mediated by Lipofectamine RNAimax (Invitrogen). siRNA against HP1α and control luciferase were described elsewhere 70. Sequence of siRNA targeting human HP1α against 3′ UTR are as follows: sense:5′ GACAUGUUGAGAUGGAAAGGAUGTT 3′, anti-sense: 3′GACUGUACAACUCUACCUUUCCUACAA5′.
pTER-NDR1/2 constructs were used to express tetracyclin inducible shRNA against NDR1 and 2 to allow stable inducible knockdown of targeted proteins (a kind gift from Dr. B. Hemmings) 46. Doxycyclin (2 μg/ml) was added to induce the shRNA expression for 5 days, followed by lysis and immunoblot analysis. shRNA against firefly luciferase was used as control for these studies.
Kinase assay and phos tag analysis
In vitro kinase assay was performed either using GST-tagged human NDR1 purified from baculovirus-infected Sf9 cells (Sigma) or from HEK293 cells that were transfected with HA tagged version of NDR kinase as described 40 with minor modifications. Briefly, HEK293 cells were transfected with HA-NDR1 wild type and were stimulated with 1μM okadaic acid (Sigma) for 1 hr. The kinase was immunoprecipitated using anti-HA agarose from transfected HEK293. The HA-NDR1 was eluted by peptide elution. In vitro kinase assays was performed by incubating purified glutathione S-transferase (GST)-tagged HP1α, its truncated mutants and GST-tagged/the eluted kinase for 30 min at 30°C in the presence of [γ-32P] ATP in kinase buffer (25mM Tris pH 7.5, 10mM MgCl2,100μM ATP, 1mM DTT). The reaction was stopped by boiling the samples in Laemmli buffer for 5 min at 95°C. Samples were resolved on SDS-PAGE, stained with Coomassie blue, and exposed to a phosphorimager (Amersham Biosciences). To detect the phosphorylation based on band shift, phos-Tag SDS-PAGE was employed according to the manufacturer’s protocol (AAL-107; NARD Institute, Amagasaki, Japan). Protein samples treated with CIP (NEB, USA) was used as dephosphorylated control.
Supplementary Material
Acknowledgments
We thank members of the Prasanth laboratory for discussions and suggestions. We thank Drs. P. Chambon, J. Chen, W. Dai, M. Dundr, B. Hemmings, Y. Hiraoka, J. Nakayama, H. Yu, P. Silver, D. Spector and B. Stillman for providing reagents and suggestions. We would like to thank Drs. D. Rivier and S. Ceman for critical reading of the paper. This work was supported by NIH (1RO1GM088252) award to KVP and NSF (0843604), NSF career (1243372) and NIH (1RO1GM099669) awards to SGP.
Footnotes
Author Contributions
Conceived and designed the experiments- AC, SGP; performed the experiments- AC, SGP; analyzed the data- AC, SGP, KVP; contributed reagents/materials/analysis tools- AC, SGP, KVP; prepared the figures- AC.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 2.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 4.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 5.Kwon SH, Workman JL. The heterochromatin protein 1 (HP1) family: put away a bias toward HP1. Mol Cells. 2008;26:217–227. [PubMed] [Google Scholar]
- 6.Zeng W, Ball AR, Jr, Yokomori K. HP1: heterochromatin binding proteins working the genome. Epigenetics. 2010;5:287–292. doi: 10.4161/epi.5.4.11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aasland R, Stewart AF. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 1995;23:3168–3173. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brasher SV, et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. Embo J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiru A, et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. Embo J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meehan RR, Kao CF, Pennings S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. Embo J. 2003;22:3164–3174. doi: 10.1093/emboj/cdg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen AL, et al. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 12.Kwon SH, Workman JL. HP1c casts light on dark matter. Cell Cycle. 2011;10:625–630. doi: 10.4161/cc.10.4.14796. [DOI] [PubMed] [Google Scholar]
- 13.Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- 14.LeRoy G, et al. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol Cell Proteomics. 2009;8:2432–2442. doi: 10.1074/mcp.M900160-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Kirschmann DA, Wallrath LL. Does heterochromatin protein 1 always follow code? Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16462–16469. doi: 10.1073/pnas.162371699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat Cell Biol. 2006;8:407–415. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- 17.Krebs EG. Historical perspectives on protein phosphorylation and a classification system for protein kinases. Philos Trans R Soc Lond B Biol Sci. 1983;302:3–11. doi: 10.1098/rstb.1983.0033. [DOI] [PubMed] [Google Scholar]
- 18.Eissenberg JC, Ge YW, Hartnett T. Increased phosphorylation of HP1, a heterochromatin-associated protein of Drosophila, is correlated with heterochromatin assembly. J Biol Chem. 1994;269:21315–21321. [PubMed] [Google Scholar]
- 19.James TC, et al. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- 20.James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao T, Eissenberg JC. Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J Biol Chem. 1999;274:15095–15100. doi: 10.1074/jbc.274.21.15095. [DOI] [PubMed] [Google Scholar]
- 22.Zhao T, Heyduk T, Eissenberg JC. Phosphorylation site mutations in heterochromatin protein 1 (HP1) reduce or eliminate silencing activity. J Biol Chem. 2001;276:9512–9518. doi: 10.1074/jbc.M010098200. [DOI] [PubMed] [Google Scholar]
- 23.Shareef MM, et al. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol Biol Cell. 2001;12:1671–1685. doi: 10.1091/mbc.12.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekwall K, et al. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- 25.Koonin EV, Zhou S, Lucchesi JC. The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 1995;23:4229–4233. doi: 10.1093/nar/23.21.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaguchi K, Hidema S, Mizuno S. Chicken chromobox proteins: cDNA cloning of CHCB1, -2, -3 and their relation to W-heterochromatin. Exp Cell Res. 1998;242:303–314. doi: 10.1006/excr.1997.4082. [DOI] [PubMed] [Google Scholar]
- 27.Shimada A, Murakami Y. Dynamic regulation of heterochromatin function via phosphorylation of HP1-family proteins. Epigenetics. 2010;5:30–33. doi: 10.4161/epi.5.1.10605. [DOI] [PubMed] [Google Scholar]
- 28.Yamagishi Y, Sakuno T, Shimura M, Watanabe Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455:251–255. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- 29.Bailis JM, Bernard P, Antonelli R, Allshire RC, Forsburg SL. Hsk1-Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat Cell Biol. 2003;5:1111–1116. doi: 10.1038/ncb1069. [DOI] [PubMed] [Google Scholar]
- 30.Hiragami-Hamada K, et al. N-terminal phosphorylation of HP1{alpha} promotes its chromatin binding. Mol Cell Biol. 2011;31:1186–1200. doi: 10.1128/MCB.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 32.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 33.Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- 34.Grallert A, Connolly Y, Smith DL, Simanis V, Hagan IM. The S. pombe cytokinesis NDR kinase Sid2 activates Fin1 NIMA kinase to control mitotic commitment through Pom1/Wee1. Nat Cell Biol. 2012 doi: 10.1038/ncb2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr Biol. 2001;11:784–788. doi: 10.1016/s0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- 36.Toyn JH, Johnston LH. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. Embo J. 1994;13:1103–1113. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 39.Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 40.Cornils H, Kohler RS, Hergovich A, Hemmings BA. Human NDR kinases control G(1)/S cell cycle transition by directly regulating p21 stability. Mol Cell Biol. 2011;31:1382–1395. doi: 10.1128/MCB.01216-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiba S, Ikeda M, Katsunuma K, Ohashi K, Mizuno K. MST2- and Furry-mediated activation of NDR1 kinase is critical for precise alignment of mitotic chromosomes. Curr Biol. 2009;19:675–681. doi: 10.1016/j.cub.2009.02.054. [DOI] [PubMed] [Google Scholar]
- 42.Janicki SM, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sathyan KM, Shen Z, Tripathi V, Prasanth KV, Prasanth SG. A BEN-domain-containing protein associates with heterochromatin and represses transcription. J Cell Sci. 2011;124:3149–3163. doi: 10.1242/jcs.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen Z, et al. A WD-repeat protein stabilizes ORC binding to chromatin. Mol Cell. 2010;40:99–111. doi: 10.1016/j.molcel.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 46.Hornbeck PV, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. ( http://www.phosphosite.org/) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kosako H, et al. Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat Struct Mol Biol. 2009;16:1026–1035. doi: 10.1038/nsmb.1656. [DOI] [PubMed] [Google Scholar]
- 48.Vichalkovski A, et al. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr Biol. 2008;18:1889–1895. doi: 10.1016/j.cub.2008.10.060. [DOI] [PubMed] [Google Scholar]
- 49.Kellum R, Alberts BM. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci. 1995;108(Pt 4):1419–1431. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- 50.Nakayama J, Klar AJ, Grewal SI. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- 51.Quivy JP, Gerard A, Cook AJ, Roche D, Almouzni G. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol. 2008;15:972–979. doi: 10.1038/nsmb.1470. [DOI] [PubMed] [Google Scholar]
- 52.Kiyomitsu T, Iwasaki O, Obuse C, Yanagida M. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J Cell Biol. 2010;188:791–807. doi: 10.1083/jcb.200908096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernard P, et al. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 54.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nozawa RS, et al. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol. 2010;12:719–727. doi: 10.1038/ncb2075. [DOI] [PubMed] [Google Scholar]
- 56.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 57.Hayakawa T, Haraguchi T, Masumoto H, Hiraoka Y. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J Cell Sci. 2003;116:3327–3338. doi: 10.1242/jcs.00635. [DOI] [PubMed] [Google Scholar]
- 58.Kang J, et al. Mitotic centromeric targeting of HP1 and its binding to Sgo1 are dispensable for sister-chromatid cohesion in human cells. Mol Biol Cell. 2011;22:1181–1190. doi: 10.1091/mbc.E11-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan CS, Linding R. Experimental and computational tools useful for (re)construction of dynamic kinase-substrate networks. Proteomics. 2009;9:5233–5242. doi: 10.1002/pmic.200900266. [DOI] [PubMed] [Google Scholar]
- 60.Pusch S, Dissmeyer N, Schnittger A. Bimolecular-fluorescence complementation assay to monitor kinase-substrate interactions in vivo. Methods Mol Biol. 2011;779:245–257. doi: 10.1007/978-1-61779-264-9_14. [DOI] [PubMed] [Google Scholar]
- 61.Hergovich A, Lamla S, Nigg EA, Hemmings BA. Centrosome-associated NDR kinase regulates centrosome duplication. Mol Cell. 2007;25:625–634. doi: 10.1016/j.molcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 62.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 63.Oh HJ, et al. MST1 limits the kinase activity of aurora B to promote stable kinetochore-microtubule attachment. Curr Biol. 2010;20:416–422. doi: 10.1016/j.cub.2009.12.054. [DOI] [PubMed] [Google Scholar]
- 64.Sessa F, et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 65.Terada Y. Aurora-B/AIM-1 regulates the dynamic behavior of HP1alpha at the G2-M transition. Mol Biol Cell. 2006;17:3232–3241. doi: 10.1091/mbc.E05-09-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ainsztein AM, Kandels-Lewis SE, Mackay AM, Earnshaw WC. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J Cell Biol. 1998;143:1763–1774. doi: 10.1083/jcb.143.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shintomi K, Hirano T. Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes Dev. 2009;23:2224–2236. doi: 10.1101/gad.1844309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang Z, Sun Y, Harley SE, Zou H, Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci U S A. 2004;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 70.Prasanth SG, Shen Z, Prasanth KV, Stillman B. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc Natl Acad Sci U S A. 2010;107:15093–15098. doi: 10.1073/pnas.1009945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.