Abstract
The potency of a series of Hexamethylene bis-acetamide (HMBA) derivatives inducing Hexamethylene bis-acetamide inducible protein 1 (HEXIM1) was determined in LNCaP prostate cancer cells. Several compounds with unsymmetrical structures showed significantly improved activity. Distinct from HMBA, these analogs have increased hydrophobicity and can improve the short half-life of HMBA, which is one of the factors that have limited the application of HMBA in clinics. The unsymmetrical scaffolds of the new analogs provide the basis for further lead optimization of the compounds using combinatorial chemistry strategy.
Keywords: HMBA, HEXIM1, derivatives, drug development
Hexamethylene bis-acetamide (HMBA), a small molecule, was investigated by the National Cancer Institute due to its potent anti-cancer and cell differentiation activities.1,2 The molecule failed at the Phase II clinical trial because of the dose-dependent toxicity.1 HMBA achieves its biological activity via Hexamethylene bis-acetamide inducible protein 1 (HEXIM1).3 HMBA significantly induces HEXIM1 expression in various cell lines at millimolar concentrations.3 However, for the agent to reach the active concentration in patients, it has to be administered via infusion of a high dosage,1,4 which causes significant toxicity.
HEXIM1 binds to 7SK RNA, a highly abundant non-coding RNA. Together they act as potent inhibitors of positive transcription elongation b (P-TEFb) and lead to inhibition of transcription. 5–8 The regulation of the relative ratio of inactive to active P-TEFb in cells by HEXIM1/7SK RNA plays a critical role in a wide range of cellular gene expression such as estrogen and glucocorticoid receptor regulated genes.7,9 HEXIM1 also has an important role in heart development and remodeling.10,11 HEXIM1 expression is decreased in a model of pathological hypertrophy in the adult heart12 and decreased HEXIM1 expression augmented angiotensinogen induced pathological hypertrophy.13 Recent research suggests that HEXIM1 suppresses cancer metastasis and Human immunodeficiency virus (HIV) replication.11,14,15 Another group reported on the inhibitory role of HEXIM1 in prostate tumorigenesis.16 Thus, drug candidates that can enhance the expression of HEXIM1 will have a great application in the treatment of cancer, heart disease, and acquired immunodeficiency syndrome (AIDS).
HMBA is the most potent and specific inducer for HEXIM1 thus far. However, this occurs only in in vitro cell cultures at concentrations ranging from one to five millimolars.2,3 It is difficult to reach high concentrations of HMBA in blood circulation due to its toxicity effects. There is an urgent need to optimize HMBA and improve its ability to induce HEXIM1 expression. 11,17 In the previous studies numerous HMBA analogs have been developed to improve the activity for inducing cell differentiation, but their ability to induce HEXIM1 expression has never been evaluated. 18 To obtain better HEXIM1 inducers, we designed and synthesized HMBA symmetrical and unsymmetrical derivatives. The structure of HMBA is represented as three different moieties, A, B and C, (Figure 1), which was used as the basis for our combinatorial chemistry approach to generate the new analogs. Since the direct binding target of HMBA is still unknown, the derivatives can be explored only via the traditional medicinal chemistry strategy, i.e., ligand based modification. Moreover, HMBA is a water- soluble molecule, which contributes greatly to its short half-life and poor tissue distribution. In the compound design, we introduced functional groups that can increase the hydrophobicity of the molecule.
Figure 1.
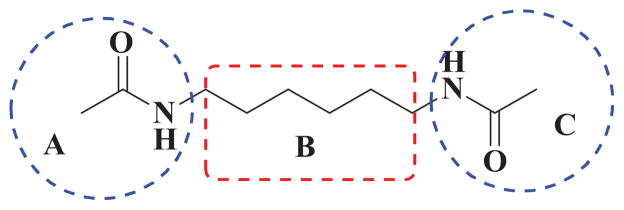
HMBA analog design
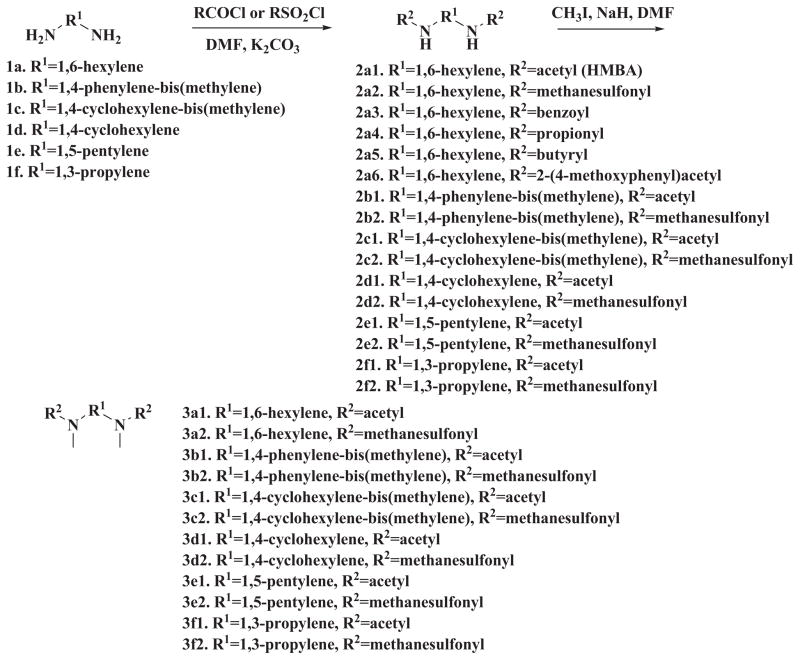
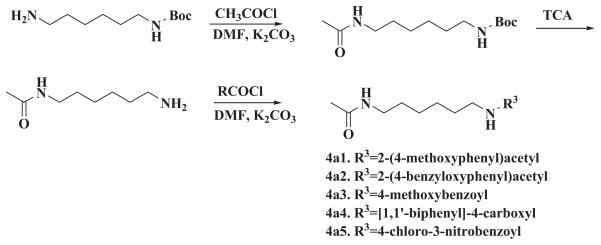
The synthesis of the symmetrical and unsymmetrical derivatives is described in Scheme 1 and 2, respectively. For the symmetrical analogs, moieties A and C were kept identical. Besides the acetyl group of HMBA, methanesulfonyl group, propionyl group or butyryl group were introduced. In addition, a bulky benzoyl group was introduced. The active proton in the amide moiety was replaced with methyl group to generate a series of analogs with lower solubility. For the moiety B, various linkers such as 1,3-propylene, 1,5-pentylene, 1,4-cyclohexylene, 1,4-phenylene-bis(methylene), 1,4-cyclohexylene-bis (methylene) were tested for optimization. Since the starting materials 1,4-di(aminomethyl)cyclohexane 1c and 1,4-diaminocyclohexane 1d are mixture of cis- and trans- isomers, the corresponding products 2c1, 2c2, 3c1, 3c2, 2d1, 2d2, 3d1, 3d2 are also mixture of cis-and trans-isomers. The further separation of cis-and trans-isomers was not carried out in this preliminary study. For the unsymmetrical analogs, the A and B moieties were kept the same as HMBA while the C moiety was modified with aryl or alkyl amide. A total of 32 compounds were synthesized, and these compounds were examined for their potency to induce HEXIM1 in LNCaP prostate cancer cells. HMBA (5 mM) was used as a positive control.
Scheme 1.
Synthesis of symmetrical HMBA analogs
Scheme 2.
Synthesis of unsymmetrical HMBA analogs
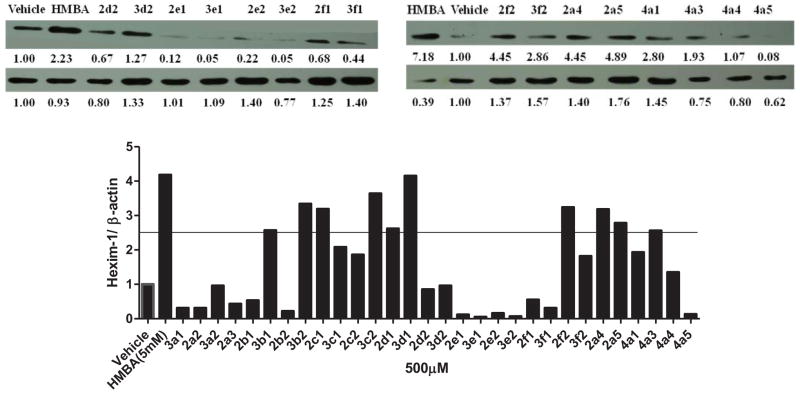
The biological activities of HMBA analogs were determined by assessing HEXIM1 expression using western blot analyses. The analogs were screened at 500 μM, a concentration that was 10 times lower than the concentration of HMBA used. Cell morphology was examined after the treatment to exclude compounds that caused cell toxicity. An increment of 2.5 fold in HEXIM1 expression compared to the control was set as a cutoff to select candidates for further analyses. In addition, solubility and structural characteristics were considered when selecting the candidates. Among the derivatives, 3b1, 3b2, 3c2, 2d1, 3d1, 2f2, 2a4 and 2a5 induced HEXIM1 expression above 2.5 fold compared to the vehicle treatment (Figure 2). Compounds 2c1 and 4a3 also reached this cutoff. However, cell morphology slightly changed after the treatment with these two compounds, which excluded them from further investigation. Compounds 3a2, 3d2, 3f2 did not reach the cutoff for HEXIM1. The reason could be that precipitation of these compounds during the treatment hindered entry to cells. Therefore, these three compounds were screened at lower concentrations. Compounds 4a1 and 4a4 have unsymmetrical novel structure, and are selected as candidates for further optimization. These two compounds were included for further analysis even if they did not reach the cutoff. Structurally, the aromatic and cyclohexylene moiety in compounds 3b1, 3b2, 2c1, 3c2, 2d1, and 3d1 resulted in enhanced activity. Compounds 2a4 and 2a5 with terminal propionyl and butyryl moieties exhibited better potency as well, suggesting that the bigger alkyl group at the terminal moiety allowed for enhanced activity. It is also possible that due to the higher hydrophobicity of these analogs when compared to HMBA, there is enhanced distribution of these compounds in the cells leading to improved biological activity (Figure 2).
Figure 2. Effect of HMBA analogs on HEXIM1 expression.
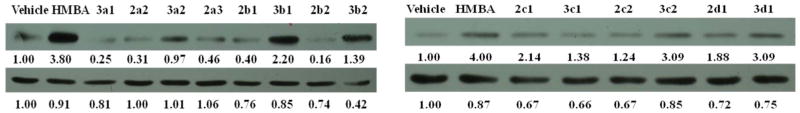
LNCaP cells were treated with vehicle, HMBA (5mM) and its analogs at 500 μM for 24h. Level of HEXIM1 (upper panel) and β-actin (lower panel) was analyzed by Western blot of cell extracts with HEXIM1 antibody and β-actin antibody respectively. The band intensities of HEXIM1 and β-actin were quantified using ImageJ (NIH) and listed below the bands. The results are the representative of three independent experiments.
Some compounds were further evaluated at a lower concentration of 100 μM (Figure 3). The compounds did not precipitate in cell culture, suggesting they were water soluble at 100 μM. Compounds 3c2, 2d1, 3d1, 2f2, 3f2, and 4a4 were not able to induce HEXIM1 expression, at 100 μM. However, compounds 3a2 and 3d2 showed significant activity at 100 μM that was not observed at 500 μM. The variation in potency could be due to the precipitation of the two compounds at 500 μM, which hampers cell penetration. Compounds 3b1 and 3b2 with the aromatic ring at B moiety showed good activity at 100 μM, and induced HEXIM1 expression by 3.8 and 2 fold, respectively. Compounds 2a4, 2a5, and 4a1 exhibited reduced activity at lower concentrations, suggesting dose dependent stimulation of HEXIM1 expression by these compounds. Due to the unsymmetrical structural characteristics of 4a1, a dose-dependent study was performed and the results are presented in Figure 4. The compound dose-dependently induced HEXIM1 expression suggesting that the unsymmetrical structure and increased hydrophobicity contribute to improved biological activity. Considering the aromatic moiety of compounds 3b1 and 3b2 and the bulky terminal moiety of compounds 2a4 and 2a5, more potent analogs could be developed if several of these structural units are combined in the new drug design.
Figure 3. Lower concentration of HMBA analogs on HEXIM1 expression.
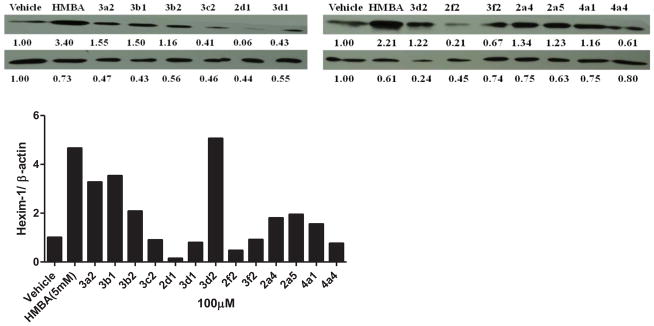
LNCaP cells were treated with vehicle, HMBA (5mM) and identified analogs at 100 μM for 24h. Level of HEXIM1 (upper panel) and β-actin (lower panel) was analyzed by Western blot of cell extracts with HEXIM1 antibody and β-actin antibody respectively. The band intensities of HEXIM1 and β-actin were quantified using ImageJ (NIH) and listed below the bands. The results are the representative of three independent experiments.
Figure 4. Dose-dependent study of compound 4a1 and HMBA on HEXIM1 expression.
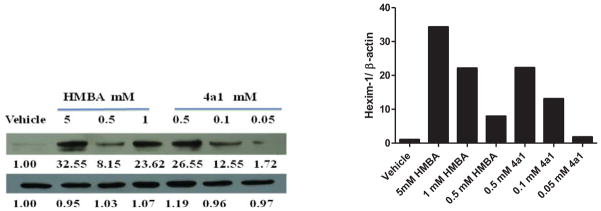
Level of HEXIM1 (upper panel) and β-actin (lower panel) was analyzed by Western blot of cell extracts with HEXIM1 antibody and β-actin antibody respectively.

In brief, our findings have provided unique molecular scaffolds that significantly induced HEXIM1 expression in prostate cancer cells, and have opened a new lead optimization direction of HMBA. To the best of our knowledge, this is the first study focusing on the lead optimization of HMBA to generate more potent HEXIM1 inducers. The results support the potential of unsymmetrical HMBA derivatives as lead compounds. More potent HEXIM1 inducers have potential application in cancer, AIDS, and heart disease treatment. Further lead optimization of HMBA to generate more potent HEXIM1 inducers is currently underway in our laboratory.
Acknowledgments
This work was supported by Center for Gene Regulation in Health and Disease (GRHD) of Cleveland State University and Ohio Department of Development (ODOD) to B.S. and NIH grant CA092440 to M.M.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Andreeff M, Stone R, Michaeli J, Young CW, Tong WP, Sogoloff H, Ervin T, Kufe D, Rifkind RA, Marks PA. Blood. 1992;80:2604. [PubMed] [Google Scholar]
- 2.Young CW, Fanucchi MP, Declan Walsh T, Baltzer L, Yaldaei S, Stevens YW, Gordon C, Tong W, Rifkind RA, Marks PA. Cancer Res. 1988;48:7304. [PubMed] [Google Scholar]
- 3.Turano M, Napolitano G, Dulac C, Majello B, Bensaude O, Lania L. J Cell Physiol. 2006;206:603. doi: 10.1002/jcp.20502. [DOI] [PubMed] [Google Scholar]
- 4.Conley BA, Egorin MJ, Sinibaldi V, Sewack G, Kloc C, Roberts L, Zuhowski EG, Forrest A, Van Echo DA. Cancer Chemother Pharmacol. 1992;31:37. doi: 10.1007/BF00695992. [DOI] [PubMed] [Google Scholar]
- 5.He N, Pezda AC, Zhou Q. Mol Cell Biol. 2006;26:7068. doi: 10.1128/MCB.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, Bensaude O. EMBO J. 2004;23:2608. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu N, Ouchida R, Yoshikawa N, Hisada T, Watanabe H, Okamoto K, Kusuhara M, Handa H, Morimoto C, Tanaka H. Proc Natl Acad Sci U S A. 2005;102:8555. doi: 10.1073/pnas.0409863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Mol Cell. 2003;12:971. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 9.Ogba N, Chaplin LJ, Doughman YQ, Fujinaga K, Montano MM. Cancer Res. 2008;68:7015. doi: 10.1158/0008-5472.CAN-08-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montano MM, Doughman YQ, Deng H, Chaplin L, Yang J, Wang N, Zhou Q, Ward NL, Watanabe M. Circ Res. 2008;102:415. doi: 10.1161/CIRCRESAHA.107.157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q, Yik JHN, Chen R. US10/928,009 2006
- 12.Espinoza-Derout J, Wagner M, Salciccioli L, Lazar JM, Bhaduri S, Mascareno E, Chaqour B, Siddiqui MA. Circ Res. 2009;104:1347. doi: 10.1161/CIRCRESAHA.108.191726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mascareno E, Galatioto J, Rozenberg I, Salciccioli L, Kamran H, Lazar JM, Liu F, Pedrazzini T, Siddiqui MA. J Biol Chem. 2012;287:13084. doi: 10.1074/jbc.M111.288944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ketchart W, Smith KM, Krupka T, Wittmann BM, Hu Y, Rayman PA, Doughman YQ, Albert JM, Bai X, Finke JH, Xu Y, Exner AA, Montano MM. Oncogene. 2012 doi: 10.1038/onc.2012.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruefli AA, Smyth MJ, Johnstone RW. Blood. 2000;95:2378. [PubMed] [Google Scholar]
- 16.Mascareno EJ, Belashov I, Siddiqui MA, Liu F, Dhar-Mascareno M. Prostate. 2012;72:1035. doi: 10.1002/pros.21510. [DOI] [PubMed] [Google Scholar]
- 17.Richon VM, Webb Y, Merger R, Sheppard T, Jursic B, Ngo L, Civoli F, Breslow R, Rifkind RA, Marks PA. Proc Natl Acad Sci U S A. 1996;93:5705. doi: 10.1073/pnas.93.12.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breslow R, Jursic B, Yan ZF, Friedman E, Leng L, Ngo L, Rifkind RA, Marks PA. Proc Natl Acad Sci U S A. 1991;88:5542. doi: 10.1073/pnas.88.13.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]