Abstract
The development of drug resistance represents a major complication in the effective treatment of breast cancer. Epigenetic therapy, through the use of histone deacetylase inhibitors (HDACi) or demethylation agents, is an emerging area of therapeutic targeting in a number of ontological entities, particularly in the setting of aggressive therapy-resistant disease. Using the well-described HDAC inhibitor trichostatin A (TSA) we demonstrate the suppression of in vitro clonogenicity in the previously described apoptosis-resistant MCF-7TN-R breast carcinoma cell line. Additionally, recent work has demonstrated that these agents can alter the expression profile of microRNA signatures in malignant cells. Using an unbiased microRNA microarray analysis, changes in miRNA expression of MCF-7TN-R cells treated with TSA for 24 h were analyzed. We observed significant up-regulation of 22 miRNAs and down-regulation of 10 miRNAs in response to TSA treatment. Our results demonstrate that the HDACi, TSA, exerts anticancer activity in the apoptosis-resistant MCF-7TN-R breast carcinoma cell line. This activity is correlated with TSA alteration of microRNA expression profiles indicative of a less aggressive phenotype.
Keywords: microRNA, trichostatin A, histone deacetylase, MCF-7, breast cancer, drug resistance
Introduction
Despite significant advancement in the area of endocrine therapies and chemotherapeutics, nearly half of breast cancer patients exhibit de novo resistance, while the majority of remaining patients ultimately progress to drug resistance (1). Drug resistant breast cancer is associated with poor prognoses (2,3), highlighting the critical need to develop novel therapeutics that are effective against these more aggressive forms of the disease. Epigenetic alterations, including aberrant DNA methylation and histone deacetylation, participate in cancer development and progression (4). Epigenetic aberrations lead to breast cancer chemotherapy resistance (5,6); hence, their reversal by inhibitors of DNA methylation and histone deacetylases (DNMTi and HDACi) may overcome it and are at present undergoing clinical testing, either alone or in combination with conventional chemotherapies (7).
Histone deacetylases (HDACs) and histone acetyltransferases (HATs) have important roles in the maintenance and function of chromatin by regulating the acetylation of histones. In addition, these enzymes have recently been shown to regulate the acetylation of many non-histone targets and therefore may represent a key means of post-translational regulation beyond their established roles in transcriptional regulation. The use of HDACi in the clinical setting is currently FDA-approved only for the treatment of progressive or recurrent cutaneous T-cell lymphoma following two other systemic therapies (8). Biologically, HDACi induce growth arrest, differentiation and cell death in breast cancer cells, but the underlying mechanism warrants further investigation.
In addition to direct regulation of mRNA gene expression, HDACis have been shown to alter microRNA (miRNA) expression in several human carcinomas including pancreatic (9), colon (10,11), gastric (12) and breast (13). microRNAs are small non-coding RNAs (18-22 nt) which function as an additional layer of regulation of mRNA stability and translation through 3′-UTR targeting (14). Through their ability to target the 3′-UTR of multiple genes, individual miRNAs can exert vast effects on mRNA-protein expression in cells. In cancer, miRNAs can function as tumor suppressors or oncogenes (15). We examined the effects of the HDACi trichostatin A (TSA) on the survival of the apoptotic-resistant MCF-7TN-R breast carcinoma cell line, as well as its effects on miRNA expression.
Materials and methods
Cell generation and culture
The apoptotic-resistant MCF-7TN-R cells were derived from MCF-7 cells grown in increasing concentrations of tumor necrosis factor α (TNFα) until resistance was established. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (10% DMEM). The detailed methods are described by Weldon et al (16).
Clonogenicity assay
Colony survival assays were performed as previously published (17). Briefly, cells were cultured in 10% FBS-DMEM media. Cells were seeded at a density of 2,000 cells/well in 2 ml of media in 6-well plates. Cells were allowed to adhere overnight at 37°C and treated on the following day with vehicle (DMSO) or TSA (0.1, 1, 10 μM). After 10 days, media was removed and the cells were fixed with gluteraldehyde. Cells were stained with crystal violet (0.1% in 20% methanol) for visualization. Colonies >50 cells were manually counted and treatments normalized to vehicle control. Assays were run in quadruplicate with internal duplicates.
microRNA microarray analysis
MCF-7TN-R cells were plated at a density of 2×106 cells in 25 cm2 flasks in normal culture media (DMEM media supplemented with 5% FBS, 1% penicillin/streptomycin, 1% essential amino acids, 1% non-essential amino acids and 1% sodium pyruvate) and allowed to adhere overnight at 37°C, 5% CO2 and air. The following day the media was changed to phenol red-free media (supplemented as above) and 5% charcoal-stripped serum was substituted for the 5% FBS. Cells were treated with TSA (10 μM) or DMSO for 24 h. Cells were harvested in PBS, collected by centrifugation, and total RNA extracted using the miRNeasy kit (Qiagen) according to manufacturer’s protocol. Enrichment for miRNA was not performed. Quantity and quality of RNA was determined by absorbance (260, 280 nm), and 5 μg total RNA was used for microarray analysis. Microarray analysis was performed as we have previously described (18). Briefly, a custom microarray (19) was used to determine miRNA expression, using three biological replicates for each condition (± TSA). Low intensity probes (signal <100 in more than half samples) were excluded from the analysis. Raw data was log-transformed and normalized by IQR. Clustering of miRNA expression data was performed using CLUSTER (20), with filtering to remove inconsistencies between replicates. For clustering, we first log-transformed the data and median-centered the array and genes, followed by average linkage clustering. Clustering results were visualized by TreeView (http://rana.lbl.gov/EisenSoftware.htm). Full array data is shown in Table I.
Table I.
microRNA microarray results for MCF-7TN-R cells treated with TSA (10 μM) for 24 h.
| ID | Serial no. | Average (TSA) | Average (DMSO) | p-value | logFC |
|---|---|---|---|---|---|
| hsa-miR-215 | 990 | 498.33 | 323.67 | 0.001 | 1.46 |
| hsa-miR-657 | 725 | 206.33 | 573.67 | 0.002 | −1.13 |
| hsa-miR-139 | 524 | 886.33 | 783.67 | 0.003 | 0.88 |
| hsa-miR-155 | 544 | 192.67 | 176.33 | 0.005 | 0.73 |
| hsa-miR-146b | 1501 | 182.67 | 178.00 | 0.005 | 0.66 |
| hsa-miR-645 | 1681 | 181.33 | 519.67 | 0.006 | −1.27 |
| hsa-miR-544 | 127 | 374.67 | 305.67 | 0.006 | 1.01 |
| hsa-miR-194 | 1541 | 362.67 | 319.67 | 0.007 | 0.95 |
| hsa-miR-628 | 696 | 190.67 | 371.67 | 0.007 | −0.60 |
| hsa-miR-144 | 1498 | 148.00 | 115.33 | 0.008 | 0.94 |
| hsa-miR-144 | 530 | 154.67 | 110.67 | 0.008 | 1.10 |
| hsa-miR-559 | 1112 | 128.67 | 111.67 | 0.008 | 0.74 |
| hsa-miR-128b | 511 | 868.00 | 905.00 | 0.009 | 0.62 |
| hsa-miR-143 | 529 | 204.67 | 143.67 | 0.009 | 1.13 |
| hsa-miR-568 | 1121 | 145.67 | 130.33 | 0.009 | 0.74 |
| hsa-miR-769-5p | 744 | 230.33 | 502.33 | 0.010 | −0.69 |
| hsa-miR-1 | 1461 | 226.00 | 130.33 | 0.010 | 1.48 |
| hsa-miR-497 | 645 | 296.67 | 639.67 | 0.011 | −0.66 |
| hsa-miR-632 | 1668 | 217.33 | 603.00 | 0.012 | −1.12 |
| hsa-miR-767-5p | 1708 | 178.00 | 392.67 | 0.012 | −0.73 |
| hsa-miR-512-3p | 660 | 155.33 | 362.67 | 0.013 | −0.82 |
| hsa-miR-191* | 1537 | 206.67 | 138.33 | 0.014 | 1.25 |
| hsa-miR-191* | 569 | 285.33 | 155.33 | 0.015 | 1.65 |
| hsa-miR-155 | 1512 | 163.67 | 150.67 | 0.015 | 0.70 |
| hsa-miR-613 | 1649 | 246.00 | 624.33 | 0.016 | −0.93 |
| hsa-miR-202* | 977 | 479.33 | 378.00 | 0.016 | 1.05 |
| hsa-miR-636 | 704 | 226.67 | 705.33 | 0.018 | −1.20 |
| hsa-miR-486 | 1600 | 192.00 | 129.67 | 0.019 | 1.21 |
| hsa-miR-622 | 1658 | 181.33 | 401.67 | 0.019 | −0.79 |
| hsa-miR-215 | 22 | 469.00 | 289.00 | 0.020 | 1.61 |
| hsa-miR-22 | 27 | 1007.33 | 568.33 | 0.021 | 1.82 |
| hsa-miR-875-5p | 903 | 139.67 | 103.33 | 0.024 | 0.99 |
| hsa-miR-620 | 1656 | 210.00 | 127.33 | 0.024 | 1.36 |
| hsa-miR-370 | 581 | 1265.67 | 1152.67 | 0.026 | 0.88 |
| hsa-miR-200c | 7 | 272.67 | 278.00 | 0.026 | 0.60 |
| hsa-miR-638 | 706 | 1096.67 | 1021.00 | 0.026 | 0.85 |
| hsa-miR-630 | 1666 | 140.33 | 129.00 | 0.028 | 0.71 |
| hsa-miR-208 | 14 | 191.33 | 168.67 | 0.028 | 0.74 |
| hsa-miR-526c | 121 | 321.33 | 215.33 | 0.028 | 1.27 |
| hsa-miR-643 | 711 | 138.33 | 268.33 | 0.029 | −0.61 |
| hsa-miR-651 | 719 | 142.33 | 332.67 | 0.029 | −0.84 |
| hsa-miR-149 | 537 | 733.67 | 711.00 | 0.031 | 0.71 |
| hsa-miR-589 | 1880 | 150.67 | 137.00 | 0.032 | 0.77 |
| hsa-miR-768-3p | 1709 | 994.33 | 1006.00 | 0.033 | 0.79 |
| hsa-miR-22 | 995 | 935.67 | 635.00 | 0.035 | 1.65 |
| hsa-miR-627 | 1663 | 133.67 | 102.00 | 0.035 | 0.96 |
| hsa-miR-450 | 1588 | 270.67 | 571.33 | 0.036 | −0.63 |
| hsa-miR-620 | 688 | 198.33 | 136.67 | 0.036 | 1.17 |
| hsa-miR-211 | 987 | 618.67 | 578.33 | 0.037 | 0.83 |
| hsa-miR-346 | 84 | 625.33 | 604.67 | 0.041 | 0.77 |
| hsa-miR-1 | 493 | 256.00 | 184.67 | 0.041 | 1.11 |
| hsa-miR-607 | 191 | 171.33 | 155.33 | 0.041 | 0.70 |
| hsa-miR-153 | 1509 | 145.67 | 124.00 | 0.041 | 0.85 |
| hsa-miR-621 | 1657 | 277.00 | 196.00 | 0.042 | 1.10 |
| hsa-miR-668 | 1700 | 233.33 | 538.67 | 0.043 | −0.74 |
| hsa-miR-607 | 1159 | 105.33 | 254.00 | 0.043 | −1.04 |
| hsa-miR-129 | 1480 | 533.33 | 495.00 | 0.045 | 0.82 |
| hsa-miR-335 | 76 | 175.67 | 134.00 | 0.045 | 0.96 |
| hsa-miR-640 | 1676 | 188.67 | 447.00 | 0.047 | −0.80 |
| hsa-miR-524* | 116 | 256.00 | 243.00 | 0.048 | 0.61 |
| hsa-miR-663 | 731 | 681.33 | 757.67 | 0.049 | 0.60 |
Statistical analyses
Colony assays were analyzed by one-way ANOVA with the Tukey’s post-test (Graph Pad Prism V.4); p<0.05 was considered to indicate statistically significant differences. The Student’s t-test was performed to evaluate the statistical significance of the cluster selection. For microRNA microarray data, the Welch’s t-test was performed for each probe using their normalized signals, with p-values <0.05 considered significant as previously described (21).
Results
TSA inhibits drug-resistant breast cancer cell clonogenic survival
We have previously described the generation of the MCF-7TN-R cells (16) which have acquired resistance to TNFα- and TRAIL-induced cell death. These cells have been characterized as highly aggressive and metastatic, and have been developed as a model system of chemoresistant breast carcinoma (22,23). To determine the effects of HDACi on apoptotic and clonogenic survival, MCF-7TN-R cells were treated with varying concentrations of TSA (0.1, 1 and 10 μM) or vehicle control (DMSO) for 10 days. MCF-7TN-R cells treated with TSA at 10 μM for 10 days demonstrated a decrease (p<0.01) in colony formation compared to vehicle-treated cells (62.37±6.45%, Fig. 1).
Figure 1.
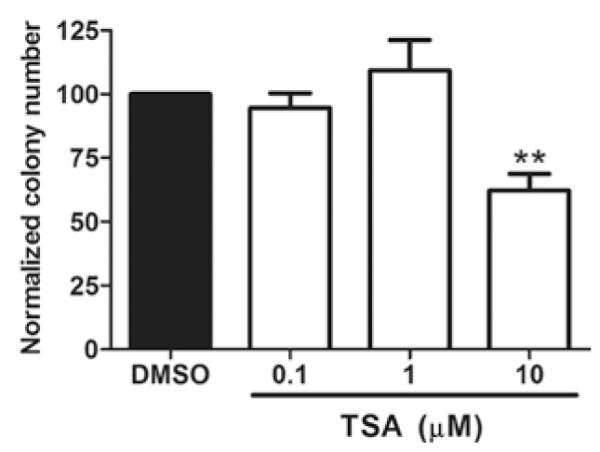
TSA suppression of MCF-7TN-R cell clonogenic survival. MCF-7TN-R cells were plated (2,000 cells/well) in 10% DMEM in 6-well plates and allowed to adhere overnight. Twenty-four hours later the cells were treated with vehicle (DMSO) or TSA (0.1, 1, 10 μM) for 10 days. Colonies of ≥50 cells were counted as positive. Bars represent mean percentage clonogenic survival normalized to DMSO control cells ± SEM. (**p<0.01).
TSA induces microRNA expression in MCF-7TN-R cells indicative of tumor suppressive and anti-metastatic effects
In human breast cancer, we and others have shown that specific miRNAs are significantly altered, as compared with normal breast tissue (21,24-26). Altered expression of specific miRNAs has been associated with poor prognosis (27), as well as breast cancer initiation, invasion and metastasis (28-31). The importance of miRNAs in these advanced breast cancer phenotypes raises the question of their further involvement in apoptotic resistance. Furthermore, based on the above result demonstrating MCF-7TN-R growth-inhibition by TSA, we performed microRNA microarray analysis of MCF-7TN-R cells after treatment with TSA (10 μM for 24 h). As shown in Fig. 2, a number of microRNA expression changes were observed. In addition, the three biological replicates from vehicle-treated MCF-7TN-R clustered together and separately from TSA-treated cells, demonstrating high reproducibility between biological repeats as well as differential microRNA expression induced by TSA. Of the microRNAs significantly altered by HDACi treatment, 22 were up-regulated (Table II) and 10 were down-regulated (Table III). Their predicted (TargetScan and miRanda) or confirmed gene targets are provided.
Figure 2.
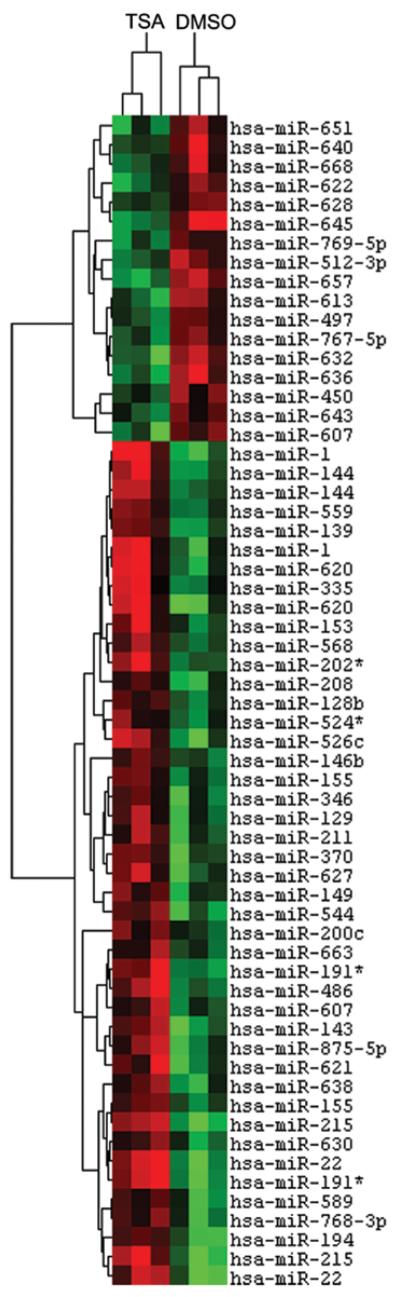
TSA regulation of microRNA expression in MCF-7TN-R. Heatmap of microRNA changes induced by treatment with TSA (10 μM) after 24 h in MCF-7TN-R cells. microRNAs demonstrating statistically significant changes in expression are shown (p<0.01). Green indicates down-regulated expression and red indicates up-regulated expression of microRNAs. Individual samples are represented in columns while specific microRNAs are represented by rows as labeled.
Table II.
Up-regulated miRNA following TSA treatment.
| microRNA | Mean fold change | p-value | Description | Gene targets (ref.) |
|---|---|---|---|---|
| hsa-miR-1 | 2.44 | <0.05 | Tumor suppressor | cMET (39), TAGLN2 (40,41) |
| hsa-miR-22 | 2.69 | <0.05 | Anti-migration, cell cycle arrest | MYCBP, MAX |
| hsa-miR-139 | 1.61 | <0.05 | Anti-metastatic, tumor suppressor | ROCK2 (29), CDK6*, HOXB2* |
| hsa-miR-143 | 1.94 | <0.05 | Tumor suppressor | KRAS (42), BCL2 (43) |
| hsa-miR-144 | 1.97 | <0.05 | Tumor suppressor | Notch1 (44) |
| hsa-miR-153 | 1.70 | <0.05 | Tumor suppressor | BCL2 (45) |
| hsa-miR-155 | 1.53 | <0.01 | OncomiR | FOXO3A (46), SOCS1 (47), FADD and IKKE (48) |
| hsa-miR-191* | 2.51 | <0.05 | Tumor suppressor | SPEN* |
| hsa-miR-194 | 1.70 | <0.01 | Tumor suppressor, anti-metastatic | CDH2, HBEGF, RAC1 and IGF1R (49) |
| hsa-miR-215 | 2.26 | <0.01 | Anti-EMT, cell cycle arrest | ZEB1/2 (50) |
| hsa-miR-202* | 1.82 | <0.05 | Tumor suppressor | TGFBR2*, ROCK1* |
| hsa-miR-335 | 1.83 | <0.05 | Tumor suppressor, cell cycle arrest, metastasis suppressor |
RB1/p105 (51), SOX4 and TNC (28) |
| hsa-miR-486 | 1.97 | <0.05 | Tumor suppressor | CD40 (52) |
| hsa-miR-526c | 2.14 | <0.05 | ||
| hsa-miR-544 | 1.74 | <0.01 | ||
| hsa-miR-559 | 1.61 | <0.05 | Tumor suppressor | ERBB2 (53) |
| hsa-miR-568 | 1.59 | <0.05 | ||
| hsa-miR-620 | 2.31 | <0.05 | ||
| hsa-miR-627 | 1.93 | <0.05 | ||
| hsa-miR-638 | 1.55 | <0.01 | ||
| hsa-miR-641 | 2.02 | <0.05 | ||
| hsa-miR-888 | 1.76 | <0.05 |
Putative targets as indicated by TargetScan and miRanda.
Table III.
Down-regulated miRNA following TSA treatment.
| microRNA | Mean fold change | p-value | Description (ref.) | Gene targets (ref.) |
|---|---|---|---|---|
| hsa-miR-500 | −1.90 | <0.05 | OncomiR (54) | |
| hsa-miR-512-3p | −1.59 | <0.05 | cFLIP (55) | |
| hsa-miR-607 | −1.68 | <0.05 | ||
| hsa-miR-613 | −1.69 | <0.05 | LXR (56) | |
| hsa-miR-622 | −1.57 | <0.05 | ||
| hsa-miR-632 | −1.86 | <0.01 | ||
| hsa-miR-636 | −2.00 | <0.05 | ||
| hsa-miR-645 | −2.03 | <0.01 | OncomiR (57) | |
| hsa-miR-651 | −1.60 | <0.05 | ||
| hsa-miR-657 | −1.91 | <0.01 | IGF2R (58) |
Discussion
Drug resistance, acquired or de novo, remains a major obstacle in the treatment of cancer (1). Progression to resistance represents one of the hallmarks of aggressive carcinomas with limited treatment options and poor prognoses (2,3). Epigenetic therapies, including HDACi, provide a novel class of treatment for therapeutically-resistant cancer patients (32), including breast cancer (33). Here we demonstrate the ability of the HDACi TSA to suppress in vitro clonogenic survival of the apoptotically resistant MCF-7TN-R cells. This cell culture data indicates that in progressive drug resistance or recurrent breast carcinoma, the use of HDACis may exhibit greater inhibitory effects on tumor cell survival. Numerous studies have analyzed gene expression changes in response to HDACi, with the goal of defining specific mechanisms of their anticancer activity (34). Recent studies have also demonstrated the regulation of microRNA expression changes in breast and other cancer cells treated with HDACi alone (9,10,13,35), or in combination with DNMTi (11,36-38). Overall, these studies revealed microRNA expression profiles suggestive of a less aggressive and more tumor-suppressive phenotype after treatment with epigenetic therapies. Consistent with those studies, our microarray results revealed that many of the microRNAs that were significantly up-regulated (Table II) following TSA treatment (compared to control) have been characterized as having tumor suppressive (miR-1, miR-143, miR-144, miR-191*, miR-202*, miR-486, miR-559), anti-migration/anti-metastasis (miR-22, miR-139, miR-194, miR-335), or anti-EMT (miR-215) roles in cancer. Only one known oncomiR, miR-155, was found to be increased following TSA treatment, but with a mean fold change of 1.53 compared to control, this was the lowest change observed, although statistically significant. Of the microRNAs identified as significantly decreased (Table III) following TSA treatment (compared to control), two have been identified as oncomiRs (miR-500, miR-645) and three others have confirmed targets involved in tumorigenesis (miR-512-3p, miR-613, miR-657). Taken together, these data indicate that HDACi treatment may promote an anti-tumor microRNA expression profile in the apoptotically resistant cell line MCF-7TN-R, providing novel therapeutic targets for the treatment of drug resistant breast cancer.
Acknowledgements
This research was supported by The Department of Defense Breast Cancer Research Program BC085426 (B.M. Collins-Burow); The National Institutes of Health/National Center for Research Resources P20RR020152 (B.M. Collins-Burow); The Integrated Cancer Biology Program: Centers for Cancer Systems Biology NIH/NCI U54CA113001 (K. Nephew); and The Walther Cancer Foundation, Indianapolis, IN (S.Y. Nam). The funders did not have any involvement in study design; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
References
- 1.Clarke R, Liu MC, Bouker KB, et al. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22:7316–7339. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- 2.Garcia M, Derocq D, Freiss G, Rochefort H. Activation of estrogen receptor transfected into a receptor-negative breast cancer cell line decreases the metastatic and invasive potential of the cells. Proc Natl Acad Sci USA. 1992;89:11538–11542. doi: 10.1073/pnas.89.23.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Agthoven T, Sieuwerts AM, Meijer-van Gelder ME, et al. Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J Clin Oncol. 2009;27:542–549. doi: 10.1200/JCO.2008.17.1462. [DOI] [PubMed] [Google Scholar]
- 4.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome--components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 5.Lo PK, Sukumar S. Epigenomics and breast cancer. Pharmacogenomics. 2008;9:1879–1902. doi: 10.2217/14622416.9.12.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabnis GJ, Goloubeva O, Chumsri S, Nguyen N, Sukumar S, Brodie AM. Functional activation of the estrogen receptor-alpha and aromatase by the HDAC inhibitor entinostat sensitizes ER-negative tumors to letrozole. Cancer Res. 2011;71:1893–1903. doi: 10.1158/0008-5472.CAN-10-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boumber Y, Issa JP. Epigenetics in cancer: what’s the future? Oncology (Williston Park) 2011;25:220–226. 228. [PubMed] [Google Scholar]
- 8.Wilkes G. Histone deacetylase inhibitors. Oncology (Williston Park) 2007;21:39–40. [PubMed] [Google Scholar]
- 9.Zhang S, Cai X, Huang F, Zhong W, Yu Z. Effect of trichostatin a on viability and microRNA expression in human pancreatic cancer cell line BxPC-3. Exp Oncol. 2008;30:265–268. [PubMed] [Google Scholar]
- 10.Shin S, Lee EM, Cha HJ, et al. MicroRNAs that respond to histone deacetylase inhibitor SAHA and p53 in HCT116 human colon carcinoma cells. Int J Oncol. 2009;35:1343–1352. doi: 10.3892/ijo_00000452. [DOI] [PubMed] [Google Scholar]
- 11.Bandres E, Agirre X, Bitarte N, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 12.Saito Y, Suzuki H, Tsugawa H, et al. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene. 2009;28:2738–2744. doi: 10.1038/onc.2009.140. [DOI] [PubMed] [Google Scholar]
- 13.Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 14.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 15.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 16.Weldon CB, Parker AP, Patten D, et al. Sensitization of apoptotically-resistant breast carcinoma cells to TNF and TRAIL by inhibition of p38 mitogen-activated protein kinase signaling. Int J Oncol. 2004;24:1473–1480. [PubMed] [Google Scholar]
- 17.Zhou C, Nitschke AM, Xiong W, et al. Proteomic analysis of tumor necrosis factor-alpha resistant human breast cancer cells reveals a MEK5/Erk5-mediated epithelial-mesenchymal transition phenotype. Breast Cancer Res. 2008;10:R105. doi: 10.1186/bcr2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xin F, Li M, Balch C, et al. Computational analysis of microRNA profiles and their target genes suggests significant involvement in breast cancer antiestrogen resistance. Bioinformatics. 2009;25:430–434. doi: 10.1093/bioinformatics/btn646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Leva G, Gasparini P, Piovan C, et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102:706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antoon JW, Liu J, Gestaut MM, Burow ME, Beckman BS, Foroozesh M. Design, synthesis, and biological activity of a family of novel ceramide analogues in chemoresistant breast cancer cells. J Med Chem. 2009;52:5748–5752. doi: 10.1021/jm9009668. [DOI] [PubMed] [Google Scholar]
- 23.Meacham WD, Antoon JW, Burow ME, Struckhoff AP, Beckman BS. Sphingolipids as determinants of apoptosis and chemoresistance in the MCF-7 cell model system. Exp Biol Med (Maywood) 2009;234:1253–1263. doi: 10.3181/0902-MR-77. [DOI] [PubMed] [Google Scholar]
- 24.Foekens JA, Sieuwerts AM, Smid M, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 26.Rao X, Di Leva G, Li M, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2010;30:1082–1097. doi: 10.1038/onc.2010.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nana-Sinkam SP, Croce CM. MicroRNAs as therapeutic targets in cancer. Transl Res. 2011;157:216–225. doi: 10.1016/j.trsl.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong CC, Wong CM, Tung EK, et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140:322–331. doi: 10.1053/j.gastro.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 31.Ma L, Weinberg RA. MicroRNAs in malignant progression. Cell Cycle. 2008;7:570–572. doi: 10.4161/cc.7.5.5547. [DOI] [PubMed] [Google Scholar]
- 32.Atadja PW. HDAC inhibitors and cancer therapy. Prog Drug Res. 2011;67:175–195. doi: 10.1007/978-3-7643-8989-5_9. [DOI] [PubMed] [Google Scholar]
- 33.Munster PN, Thurn KT, Thomas S, et al. A phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy-resistant breast cancer. Br J Cancer. 2011;104:1828–1835. doi: 10.1038/bjc.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shankar S, Srivastava RK. Histone deacetylase inhibitors: mechanisms and clinical significance in cancer: HDAC inhibitor-induced apoptosis. Adv Exp Med Biol. 2008;615:261–298. doi: 10.1007/978-1-4020-6554-5_13. [DOI] [PubMed] [Google Scholar]
- 35.Lee EM, Shin S, Cha HJ, et al. Suberoylanilide hydroxamic acid (SAHA) changes microRNA expression profiles in A549 human non-small cell lung cancer cells. Int J Mol Med. 2009;24:45–50. doi: 10.3892/ijmm_00000204. [DOI] [PubMed] [Google Scholar]
- 36.Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220–2222. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- 37.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 38.Lujambio A, Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle. 2007;6:1455–1459. [PubMed] [Google Scholar]
- 39.Yan D, Dong Xda E, Chen X, et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nohata N, Sone Y, Hanazawa T, et al. miR-1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget. 2011;2:29–42. doi: 10.18632/oncotarget.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshino H, Chiyomaru T, Enokida H, et al. The tumoursuppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104:808–818. doi: 10.1038/bjc.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu B, Niu X, Zhang X, et al. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem. 2011;350:207–213. doi: 10.1007/s11010-010-0700-6. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, Cai X, Wang Y, Tang H, Tong D, Ji F. microRNA-143, down-regulated in osteosarcoma, promotes apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol Rep. 2010;24:1363–1369. doi: 10.3892/or_00000994. [DOI] [PubMed] [Google Scholar]
- 44.Sureban SM, May R, Lightfoot S, et al. DCAMKL-1 regulates epithelial-mesenchymal transition in human pancreatic cells through a miR-200a-dependent mechanism. Cancer Res. 2011;71:2328–2338. doi: 10.1158/0008-5472.CAN-10-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J, Liao X, Wong C. Downregulation of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer. 2010;126:1029–1035. doi: 10.1002/ijc.24823. [DOI] [PubMed] [Google Scholar]
- 46.Kong W, He L, Coppola M, et al. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869–17879. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Jiang S, Zhang HW, Lu MH, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 48.Tili E, Michaille JJ, Cimino A, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 49.Meng Z, Fu X, Chen X, et al. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52:2148–2157. doi: 10.1002/hep.23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kato M, Zhang J, Wang M, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scarola M, Schoeftner S, Schneider C, Benetti R. miR-335 directly targets Rb1 (pRb/p105) in a proximal connection to p53-dependent stress response. Cancer Res. 2010;70:6925–6933. doi: 10.1158/0008-5472.CAN-10-0141. [DOI] [PubMed] [Google Scholar]
- 52.Mees ST, Mardin WA, Sielker S, et al. Involvement of CD40 targeting miR-224 and miR-486 on the progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol. 2009;16:2339–2350. doi: 10.1245/s10434-009-0531-4. [DOI] [PubMed] [Google Scholar]
- 53.Chen H, Sun JG, Cao XW, et al. Preliminary validation of ERBB2 expression regulated by miR-548d-3p and miR-559. Biochem Biophys Res Commun. 2009;385:596–600. doi: 10.1016/j.bbrc.2009.05.113. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto Y, Kosaka N, Tanaka M, et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers. 2009;14:529–538. doi: 10.3109/13547500903150771. [DOI] [PubMed] [Google Scholar]
- 55.Chen F, Zhu HH, Zhou LF, Wu SS, Wang J, Chen Z. Inhibition of c-FLIP expression by miR-512-3p contributes to taxol-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2010;23:1457–1462. doi: 10.3892/or_00000784. [DOI] [PubMed] [Google Scholar]
- 56.Ou Z, Wada T, Gramignoli R, et al. MicroRNA hsa-miR-613 targets the human LXR(alpha) gene and mediates a feedback loop of LXR(alpha) autoregulation. Mol Endocrinol. 2011;25:584–596. doi: 10.1210/me.2010-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shih KK, Qin LX, Tanner EJ, et al. A microRNA survival signature (MiSS) for advanced ovarian cancer. Gynecol Oncol. 2011;121:444–450. doi: 10.1016/j.ygyno.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 58.Lv K, Guo Y, Zhang Y, Wang K, Jia Y, Sun S. Allele-specific targeting of hsa-miR-657 to human IGF2R creates a potential mechanism underlying the association of ACAA-insertion/ deletion polymorphism with type 2 diabetes. Biochem Biophys Res Commun. 2008;374:101–105. doi: 10.1016/j.bbrc.2008.06.102. [DOI] [PubMed] [Google Scholar]


